Abstract
Molecular cloning of the “old” but still unclassified Euonymus europaeus agglutinin (EEA) demonstrated that the lectin is a homodimeric protein composed of 152 residue subunits. Analysis of the deduced sequence indicated that EEA is synthesized without a signal peptide and undergoes no posttranslational processing apart from the removal of a six-residue N-terminal peptide. Glycan array screening confirmed the previously reported high reactivity of EEA toward blood group B oligosaccharides but also revealed binding to high mannose N-glycans, providing firm evidence for the occurrence of a plant carbohydrate-binding domain that can interact with structurally different glycans. Basic Local Alignment Search Tool searches indicated that EEA shares no detectable sequence similarity with any other lectin but is closely related evolutionarily to a domain that was first identified in some abscisic acid- and salt stress-responsive rice (Oryza sativa) proteins, and, according to the available sequence data, might be ubiquitous in Spermatophyta. Hence, EEA can be considered the prototype of a novel family of presumably cytoplasmic/nuclear proteins that are apparently ubiquitous in plants. Taking into account that some of these proteins are definitely stress related, the present identification of the EEA lectin domain might be a first step in the recognition of the involvement and importance of protein-glycoconjugate interactions in some essential cellular processes in Embryophyta.
Plant lectins have been studied for more than a century. Nevertheless, the inventory of all carbohydrate-binding domains occurring in plant cells is still incomplete. Until a few years ago, virtually all known plant lectins could be classified into seven families of structurally and evolutionarily related proteins (Van Damme et al., 1998). However, the identification of three novel sugar-binding domains/proteins during the last 2 years (Kaku et al., 2006; Peumans et al., 2007; Van Damme et al., 2007) leaves little doubt that more carbohydrate-binding domains remain to be discovered in plants. Two major problems hamper the discovery of the remaining sugar-binding motifs in plants. First, unless homologous lectins have been identified in other organisms, no relevant information is provided by genome/proteome analyses. Second, evidence is accumulating that the expression level of lectins with a specific endogenous role is so low that they escape detection by the currently available activity assays (Van Damme et al., 2004a, 2004b).
Though at present virtually all abundant plant lectins can be classified into well-defined protein families, there are still a few exceptions for which sufficient sequence information is not available. One of these “orphan” lectins is the Euonymus europaeus agglutinin (EEA). As early as 1954, Schmidt (1954) reported that the fleshy arils surrounding the seeds of E. europaeus contain a lectin with a clear preference for B-type erythrocytes within the human ABO system. The lectin was isolated for the first time in 1975 by conventional protein purification techniques (Pacak and Kocourek, 1975) and later by affinity chromatography on immobilized polyleucyl hog A+H blood group substance (Petryniak et al., 1977). Though the data shown in both articles indicated that the lectin consisted predominantly of partly disulfide-linked subunits of approximately 17 kD, the molecular structure of the native agglutinin remained unclear. According to Pacak and Kocourek (1975), the lectin is a mixture of isoforms that have a similar molecular mass (varying between 119 and 127 kD) but differ in carbohydrate content (1.9%–4.7%). Petryniak et al. (1977) also distinguished multiple molecular forms but reported a higher molecular mass (166 kD) and higher carbohydrate content (approximately 10%) for the native lectin. Later studies of both the E. europaeus lectin (Petryniak and Goldstein, 1987) and a lectin from the closely related species Euonymus sieboldiana (Yamamoto and Sakai, 1981) yielded no additional information about the molecular structure of the native agglutinins. In contrast to the molecular structure, fairly detailed information was reported about the sugar-binding specificity of the E. europaeus lectin, which was found to be directed against the blood group B substance Galα1-3(Fucα1-2)Galβ1-4GlcNAc (Petryniak et al., 1977; Petryniak and Goldstein, 1987).
This report describes a detailed reinvestigation of the E. europaeus agglutinin (EEA) using a combination of biochemical, molecular, and cellular-biological approaches. EEA represents a novel lectin family that shares no significant sequence similarity with any other known lectin family. Glycan array screening experiments confirmed that EEA recognizes the blood group B antigen but also demonstrated that the lectin interacts with high Man N-glycans. Interestingly, EEA shares high sequence identity with some previously identified salt stress/ABA responsive proteins from rice (Oryza sativa; Moons et al., 1995, 1997), which are apparently expressed in all terrestrial plants but in no other organisms.
RESULTS AND DISCUSSION
Purification and Biochemical Characterization of EEA
Because EEA purification using a classical protocol for plant lectin isolation was hampered by the formation of insoluble complexes with endogenous glycoconjugates, the crude extract was first fractionated by ion-exchange chromatography and gel filtration under conditions whereby the carbohydrate-binding activity of EEA was reversibly inhibited. The resulting protein fraction was fully soluble in an aqueous buffer at neutral pH and could readily be chromatographed on a column of immobilized ovomucoid to yield a pure water-soluble lectin preparation.
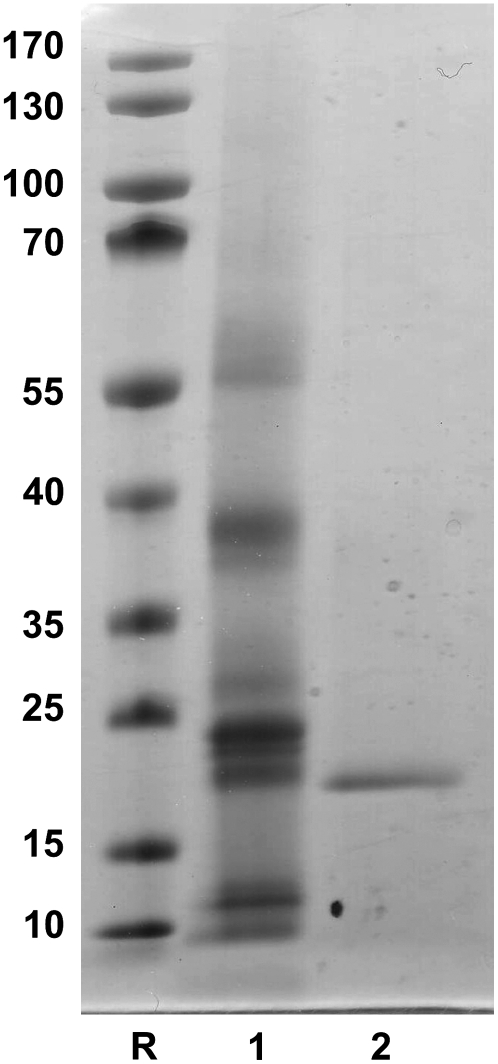
SDS-PAGE of the purified lectin in the presence of β-mercaptoethanol yielded a single polypeptide band of 17 kD (Fig. 1). The lectin did not contain any covalently bound sugar. Mass spectrometry of the lectin yielded a single peak with a molecular mass of 16,907 ± 2 D. Edman degradation of the electroblotted 17-kD polypeptide yielded a single sequence (ATGPTYRVYXRAAPNYNMTV; Supplemental Fig. S1).
Figure 1.
SDS-PAGE of crude extract and purified E. europaeus agglutinin. Samples were loaded as follows: lane 1, crude extract of mature arilli; lane 2, reduced EEA. Molecular mass reference proteins (lane R): PageRuler prestained protein ladder (Fermentas), mixture of 10 recombinant proteins with apparent molecular masses of approximately 10, 15, 25, 35, 40, 55, 70, 100, 130, and 170 kD.
Because gel filtration experiments yielded no conclusive results, the molecular mass of native EEA was estimated by dynamic light scattering (DLS). DLS of the sample revealed that the lectin was largely monodisperse. The scattering peak corresponded to particles having an average hydrodynamic diameter of 5.6 nm, consistent with globular protein assemblies of 37 kD. Given a molecular mass of 16.9 kD for the monomer, the DLS data indicate that native EEA occurs as a dimer.
Though our data confirm the size of the EEA subunits reported before, our lectin preparation did not contain any covalently bound sugars. Moreover, as is demonstrated below, EEA is synthesized on free ribosomes and hence cannot be N-glycosylated. Therefore, the relatively high carbohydrate content (2%–10%) of the EEA preparations described in previous articles can hardly be ascribed to the lectin itself. Taking into account that EEA tends to form aggregates with endogenous glycoconjugates present in crude extracts, it is likely that the previously purified preparations consisted at least partly of lectin-glycoprotein complexes. The presence of such complexes not only accounts for the carbohydrate found in the lectin preparations described by Pacak and Kocourek (1975) and Petryniak et al. (1977) but also explains why these preparations sedimented with an apparent molecular mass of 119 to 127 kD and 166 kD, respectively, upon analytical centrifugation.
Molecular Cloning of EEA
Screening of a cDNA library prepared from mRNA isolated from developing arilli allowed isolating a cDNA clone with a deduced sequence that perfectly matched the N-terminal sequence of the EEA polypeptide. The Cys, which is degraded during Edman degradation if it is not alkylated prior to the analysis, corresponded to the blank in the experimentally determined sequence. The cDNA clone comprised an open reading frame of 474 nucleotides corresponding to an EEA precursor sequence (LECEEA) of 158 amino acid residues that contains six extra residues preceding the N terminus of the mature polypeptide (Supplemental Fig. S1). Calculation of the molecular mass of the polypeptide spanning residues A7 to G158 yielded a value of 16,903.8 D, which is in good agreement with the value obtained by mass spectrometry of the lectin (16,907 D). This nearly perfect match in molecular mass and the occurrence of a 20-amino acid sequence identical to the N terminus of the mature lectin polypeptide at the N terminus of the deduced amino acid sequence of the cDNA shows that the isolated cDNA clone encodes EEA.
No putative signal peptide could be identified in the deduced sequence, indicating that the protein is synthesized on free ribosomes. After synthesis, the first six residues are apparently removed from the primary translation product. In silico analyses predict a cytoplasmic location of the Euonymus lectin.
To check for the presence of intron(s), a genomic sequence corresponding to the EEA gene was amplified and sequenced. Alignment of the genomic and cDNA sequence demonstrated that the lectin gene contains three introns (Supplemental Fig. S1).
EEA Recognizes Two Classes of Structurally Different Glycans
A reinvestigation of the carbohydrate-binding specificity of EEA using glycan array screening experiments confirmed its interaction with blood group B substance as previously described (Petryniak et al., 1977; Petryniak and Goldstein, 1987; Teneberg et al., 2003), but at the same time also revealed a previously unobserved interaction with N-linked, high Man-type glycans. The binding of fluorescent-labeled EEA to glycans on the microarray is shown in Supplemental Figure S2, in which average relative fluorescence units (RFU) bound by each glycan are plotted versus glycan numbers that correspond to structures identified in Supplemental Table S1. The unusual carbohydrate-binding specificity is summarized in Table I, in which the glycan array data at five different concentrations of the labeled lectin are selected for the highest binding to blood group B-related structures and N-linked, high Man-type structures. The glycans are ranked in approximate order of apparent affinity. The data indicate that EEA specifically binds to the blood group B oligosaccharides with highest affinity for B-type II structures, with lower affinity for blood group H and structures with terminal Galα1,3. EEA interacts with glycan 79, the only blood group A-type I structure on the array. No binding was observed to A-type II structures (glycans 80–83). EEA also has specificity for N-linked, high Man-type glycans (192–193, 197–198) as shown in Table I at concentrations of 10 to 200 μg/mL. Because other linear oligomannosides on the array showed no binding, the binding of EEA toward N-linked glycans apparently requires the core pentasaccharide [Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAc]. To assess the relative affinity of EEA for blood group B oligosaccharides and high Man-type N-glycans, the binding assays were carried out at decreasing lectin concentrations to reveal the higher affinity structures. At 50 μg/mL and 10 μg/mL EEA, the blood group B structures demonstrated highest affinity for EEA, while the fluorescence values for the high Man N-glycans were roughly 10-fold lower (Table I). These data indicate that EEA has a much higher affinity for blood group B oligosaccharides than for high Man N-glycans.
Table I.
Glycan array binding assay of different concentrations of EAA
Avg, Average RFU. The histogram of the binding of EEA at a concentration of 200 μg/mL without addition of an inhibitor is shown in Supplemental Figure S2. The list of structures (version 3.0 of the Glycan Array) ranked from highest RFU to lowest can be found in Supplemental Table S1.
| Glycan No. | Glycan Structure | Avg 200 μg/mL | Avg l00 μg/mL | Avg 50 μg/mL | Avg 10 μg/mL | Avg 5 μg/mL |
|---|---|---|---|---|---|---|
| Blood group B-related structures | ||||||
| 99 | Galα1-3(Fucα1-2)Galβ-Sp8 | 49,370 | 37,555 | 22,185 | 32,195 | 177 |
| 290 | Galα1-3(Fucα1-2)Galβ-Sp18 | 45,917 | 43,578 | 46,448 | 46,764 | 244 |
| 97 | Galα1-3(Fucα1-2)Galβ1-4GlcNAc-Sp0 | 35,329 | 38,351 | 39,913 | 41,304 | 2,234 |
| 95 | Galα1-3(Fucα1-2)Galβ1-3GlcNAcβ-Sp0 | 35,176 | 33,212 | 41,881 | 42,788 | 2,036 |
| 98 | Galα1-3(Fucα1-2)Galβ1-4Glcβ-Sp0 | 47,659 | 45,717 | 54,776 | 48,208 | 1,208 |
| 105 | Galα1-3Galβ1-4GlcNAcβ-Sp8 | 6,872 | 9,570 | 4,375 | 1,685 | 153 |
| 112 | Galα1-4GlcNAcβ-Sp8 | 3,204 | 1,518 | 961 | 409 | 99 |
| 103 | Galα1-3Galβ1-4(Fucα1-3)GlcNAcβ-Sp8 | 4,514 | 2,228 | 1,278 | 387 | 49 |
| 70 | Fucα1-2Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1- 4GlcNAcβ-Sp0 | 5,641 | 5,369 | 2,950 | 1,746 | 106 |
| 61 | Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glcβ-Sp10 | 24,615 | 22,118 | 13,985 | 8,012 | 61 |
| 63 | Fucα1-2Galβ1-3GlcNAcβ-Sp0 | 21,920 | 23,138 | 15,808 | 9,243 | 178 |
| 69 | Fucα1-2Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc-Sp0 | 19,799 | 10,747 | 6,702 | 3,689 | 85 |
| 64 | Fucα1-2Galβ1-3GlcNAcβ-Sp8 | 7,994 | 18,933 | 11,425 | 7,305 | 53 |
| 62 | Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glcβ-Sp8 | 9,804 | 32,376 | 19,942 | 9,081 | 77 |
| 72 | Fucα1-2Galβ1-4GlcNAcβ-Sp8 | 4,881 | 3,851 | 1,483 | 626 | 145 |
| 270 | Fucα1-2Galβ1-4[6OSO3]GlcNAc-Sp8 | 2,543 | 2,694 | 972 | 961 | 53 |
| 241 | Galβ1-4GlcNAcβ1-2Manα1-3(Fucα1-3(Galβ1-4)GlcNAcβ1- 2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp20 | 23,203 | 12,628 | 3,012 | 899 | 131 |
| 172 | (GlcNAcβ1-4)5β-Sp8 | 12,800 | 3,781 | 1,755 | 798 | 59 |
| 201 | Fucα1-3(Galβ1-4)GlcNAcβ1-2Manα1-3(Fucα1-3(Galβ1-4) GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp20 | 11,885 | 2,456 | 1,270 | 626 | 47 |
| 173 | GlcNAcβ1-4GlcNAcβ1-4GlcNAcβ-Sp8 | 7,530 | 6,594 | 1,287 | 999 | 49 |
| 79 | GalNAcα1-3(Fucα1-2)Galβ1-3GlcNAcβ-Sp0 | 7,512 | 8,659 | 6,660 | 2,681 | 138 |
| 87 | GalNAcα1-4(Fucα1-2)Galβ1-4GlcNAcβ-Sp8 | 6,044 | 2,163 | 1,209 | 596 | 111 |
| 171 | (GlcNAcβ1-4)6β-Sp8 | 6,402 | 4,909 | 946 | 740 | 46 |
| 52 | Galβ1-4GlcNAcβ1-2Manα1-3(Galβ1-4GlcNAcβ1- 2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp13 | 9,560 | 2,213 | 1,134 | 805 | 20 |
| N-linked high Man-type structures | ||||||
| 193 | Manα1-2Manα1-6(Manα1-3)Manα1-6(Manα2Manα2Manα1- 3)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 24,511 | 3,242 | 1,785 | 751 | 33 |
| 192 | Manα1-6(Manα1-2Manα1-3)Manα1-6(Manα2Manα1-3)Manβ1- 4GlcNAcβ1-4GlcNAcβ-Sp12 | 36,914 | 11,766 | 5,950 | 3,345 | 145 |
| 197 | Manα1-6(Manα1-3)Manα1-6(Manα2Manα1-3)Manβ1-4GlcNAcβ1- 4GlcNAcβ-Sp12 | 40,788 | 24,583 | 5,037 | 1,995 | 88 |
| 310 | Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 37,819 | 32,564 | 6,034 | 4,719 | 110 |
| 198 | Manα1-6(Manα1-3)Manα1-6(Manα1-3)Manβ1-4GlcNAcβ1- 4GlcNAcβ-Sp12 | 50,258 | 27,893 | 11,061 | 3,030 | 154 |
| 50 | Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp13 | 30,930 | 46,287 | 32,315 | 10,055 | 129 |
Although the results of the glycan array screening experiments are only semiquantitative, they indicate that EEA binds two structurally unrelated glycans. To determine if the lectin possibly possesses two independent binding sites with different specificities, the glycan array screening experiment was repeated in the presence of inhibitory oligosaccharides. The inhibition data are graphically presented in Supplemental Figure S3 and summarized in Table II. For this analysis, a high concentration of lectin (200 μg/mL) was used so that strong binding to both the blood group B-related structures and the N-linked high Man-type structures were tested simultaneously. Addition of B-active oligosaccharide (at a concentration of 3 mg/mL) completely abolished the interaction of EEA with all B-active glycans on the array (95,97,98,99,290) but had little effect on the binding to the Man containing oligosaccharides (Table II; Supplemental Fig. S3). However, when the assay was done in the presence of 3 mg/mL RNase B glycopeptides (a mixture of glycopeptides containing the N-linked high Man-type structures), binding of EEA to all glycans on the array was inhibited, indicating that the RNase B glycopeptides can also displace the B-active glycans from the carbohydrate-binding site of EEA. Based on these data, no final conclusion can be drawn with respect to the possible presence of two distinct binding sites. The results of the inhibition experiments with the high affinity ligand (B-active oligosaccharide), however, strongly argue for the occurrence of two different binding sites, whereas those obtained with the lower affinity ligand (the Man containing oligosaccharides) cannot be reconciled with the same concept. It is also difficult to explain why the lower affinity ligand displaces the high affinity ligand (and not the other way around). It can be expected that only structural data can give a definitive answer to the question of the possible presence of two distinct sites in the EEA domain.
Table II.
Glycan array binding assay of the EAA in the presence and absence of the blood group B tetrasaccharide or a mixture of glycopeptides derived from RNase B
Avg RFU, Average RFU for control (EEA without addition of inhibitor); Avg + B, average RFU for EEA in the presence of blood group B tetrasaccharide; Avg + MAN, average RFU for EEA in the presence of a mixture of glycopeptides derived from RNase B; %CV, coefficient of variation expressed as percentage. A summary of the results for EEA inhibition by blood group B tetrasaccharide or a mixture of glycopeptides derived from RNase B can be found in Supplemental Figure S3.
| Glycan No. | Glycan Name | Avg RFU | %CV | Avg + B | %CV | Avg + MAN | %CV |
|---|---|---|---|---|---|---|---|
| Blood group B-related structures | |||||||
| 99 | Galα1-3(Fucα1-2)Galβ-Sp8 | 62,332 | 1 | 155 | 69 | 1,243 | 43 |
| 290 | Galα1-3(Fucα1-2)Galβ-Sp18 | 54,525 | 12 | 357 | 55 | 845 | 41 |
| 97 | Galα1-3(Fucα1-2)Galβ1-4GlcNAc-Sp0 | 52,093 | 14 | 1,822 | 54 | 2,615 | 55 |
| 95 | Galα1-3(Fucα1-2)Galβ1-3GlcNAcβ-Sp0 | 37,688 | 8 | 582 | 124 | 839 | 49 |
| 98 | Galα1-3(Fucα1-2)Galβ1-4Glcβ-Sp0 | 37,318 | 26 | 2,252 | 37 | 1,468 | 66 |
| 105 | Galα1-3Galβ1-4GlcNAcβ-Sp8 | 15,512 | 27 | 649 | 44 | 246 | 37 |
| 112 | Galα1-4GlcNAcβ-Sp8 | 11,351 | 6 | 341 | 96 | 1,053 | 120 |
| 103 | Galα1-3Galβ1-4(Fucα1-3)GlcNAcβ-Sp8 | 7,077 | 16 | 518 | 47 | 689 | 65 |
| 70 | Fucα1-2Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1- 3Galβ1-4GlcNAcβ-Sp0 | 30,494 | 11 | 1,120 | 58 | 500 | 20 |
| 61 | Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glcβ-Sp10 | 27,090 | 6 | 1,090 | 61 | 1,168 | 49 |
| 63 | Fucα1-2Galβ1-3GlcNAcβ-Sp0 | 21,403 | 21 | 357 | 82 | 1,410 | 64 |
| 69 | Fucα1-2Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc-Sp0 | 12,647 | 2 | 1,117 | 58 | 783 | 76 |
| 64 | Fucα1-2Galβ1-3GlcNAcβ-Sp8 | 10,659 | 8 | 131 | 118 | 1,280 | 65 |
| 62 | Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glcβ-Sp8 | 9,753 | 29 | 786 | 21 | 989 | 63 |
| 72 | Fucα1-2Galβ1-4GlcNAcβ-Sp8 | 5,787 | 27 | 1,446 | 56 | 712 | 17 |
| 270 | Fucα1-2Galβ1-4[6OSO3]GlcNAc-Sp8 | 5,200 | 57 | 858 | 88 | 5,007 | 19 |
| 241 | Galβ1-4GlcNAcβ1-2Manα1-3(Fucα1-3(Galβ1-4) GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1- 4GlcNAcβ-Sp20 | 23,607 | 5 | 21,032 | 2 | 206 | 111 |
| 172 | (GlcNAcβ1-4)5β-Sp8 | 13,453 | 2 | 8,212 | 8 | 680 | 56 |
| 201 | Fucα1-3(Galβ1-4)GlcNAcβ1-2Manα1-3(Fucα1- 3(Galβ1-4)GlcNAcβ1-2Manα1-6)Man β1-4GlcNAcβ1-4GlcNAcβ-Sp20 | 11,274 | 21 | 12,266 | 11 | 451 | 84 |
| 173 | GlcNAcβ1-4GlcNAcβ1-4GlcNAcβ-Sp8 | 10,622 | 7 | 8,940 | 6 | 4,003 | 11 |
| 79 | GalNAcα1-3(Fucα1-2)Galβ1-3GlcNAcβ-Sp0 | 9,274 | 54 | 742 | 72 | 799 | 58 |
| 87 | GalNAcα1-4(Fucα1-2)Galβ1-4GlcNAcβ-Sp8 | 7,365 | 13 | 866 | 85 | 1,464 | 16 |
| 171 | (GlcNAcβ1-4)6β-Sp8 | 6,891 | 11 | 5,635 | 4 | 1,096 | 40 |
| 52 | Galβ1-4GlcNAcβ1-2Manα1-3(Galβ1-4GlcNAcβ1- 2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp13 | 6,132 | 12 | 5,310 | 7 | 1,265 | 43 |
| N-linked high Man-type structures | |||||||
| 193 | Manα1-2Manα1-6(Manα1-3)Manα1-6 (Manα2Manα2Manα1-3)Manβ1-4GlcNAc β1-4GlcNAcβ-Sp12 | 11,861 | 15 | 15,956 | 10 | 1,888 | 46 |
| 192 | Manα1-6(Manα1-2Manα1-3)Manα1-6 (Manα2Manα1-3)Manβ1-4GlcNAcβ1- 4GlcNAcβ-Sp12 | 25,386 | 4 | 20,390 | 20 | 1,335 | 24 |
| 197 | Manα1-6(Manα1-3)Manα1-6(Manα2Man α1-3)Manβ1-4GlcNAcβ1-4GlcNAcβ-Sp12 | 29,927 | 14 | 22,023 | 13 | 1,893 | 10 |
| 310 | Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1- 4GlcNAcβ-Sp12 | 31,136 | 31 | 20,644 | 6 | 545 | 15 |
| 198 | Manα1-6(Manα1-3)Manα1-6(Manα1-3)Manβ1- 4GlcNAcβ1-4 GlcNAcβ-Sp12 | 52,567 | 19 | 31,661 | 6 | 456 | 20 |
| 50 | Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1- 4GlcNAcβ-Sp13 | 59,884 | 5 | 58,680 | 6 | 3,559 | 5 |
Nonetheless, the results of the glycan array screening have two important consequences. First, they demonstrate that the resolving power of this novel technique allows distinguishing a previously undetectable weak interaction with high Man N-glycans against a background of strong interactions with blood group B substances. Secondly, the finding that EEA recognizes high Man N-glycans is highly relevant because it implies that the previously observed biological activities of the lectin (depending on the concentration used) are not necessarily due to binding of the lectin to Galα1-3(Fucα1-2)Galβ1-4GlcNAc or related glycans and that the distribution pattern obtained upon staining tissue sections with labeled EEA cannot simply be linked to the presence of the same sugars (Teneberg et al., 2003).
EEA Shares High Sequence Similarity with a Domain Found in Some Abscisic Acid- and Salt Stress-Responsive Rice Proteins
Even though EEA cannot be classified into any of the currently known lectin families, it definitely shares a high sequence similarity with several other (hypothetical) plant proteins. BLASTp searches with the deduced complete sequence of LECEEA revealed that the rice protein OSR40g3 scored best (Expect value = 1e-28) sharing 46% and 62% sequence identity and similarity, respectively, with EEA within a 151-residue overlap (Fig. 2A). OSR40g3 was identified as an abscisic acid- and salt stress-responsive protein (Moons et al., 1997). The protein is encoded by the rice gene Os07g0684000 (National Center for Biotechnology Information [NCBI] annotation)/Os07g48500 (The Institute for Genomic Research annotation). Four additional genes were identified in the rice genome, three of which encode proteins comprising two in-tandem arrayed domains equivalent to OSR40g3 (Fig. 2B). Interestingly, these rice proteins are annotated as a “Ricin B-related lectin domain containing protein.” This annotation is based on the presence in their sequence of two QXW repeats, which are considered typical motifs of the ricin-B domain. However, it is questionable whether OSR40g3 can be classified in the ricin-B family because, according to BLASTp searches, it shares no significant overall sequence similarity with any protein comprising a ricin-B domain. Moreover, alignment of the amino acid sequences of OSR40g3 and, for example, the B-chain of the Ricinus communis agglutinin (AAA33869.1) yields a very low sequence identity/similarity (Supplemental Fig. S4).
Figure 2.
A, Alignment of the amino acid sequences of LECEEA and OSR40g3 (CAA70175.1). Asterisk, colon, and period denote identical, conserved, or semiconserved amino acid residues, respectively. The amino acid sequence corresponding to the N-terminal sequence of EEA is underlined. B, Schematic representation of the domain architecture of the rice proteins with domain(s) homologous to EEA.
Besides the five members of the rice OSR40 family, 20 other plant proteins were retrieved by the BLASTp searches (E-value <0.1; Supplemental Table S2). One of these proteins is a wheat (Triticum aestivum) ortholog of OSR40g3. Another is a putative OSR40g3 homolog from Arabidopsis (Arabidopsis thaliana). This Arabidopsis protein (At2g39050) is annotated as a “Hydroxyproline-rich glycoprotein family protein, contains QXW lectin repeat domain, Pfam:PF00652 (=Ricin-type beta-trefoil lectin domain).” In this case, the annotation is also primarily based on the presence of two QXW repeats. Orthologs of At2g39050 were also identified in Populus trichocarpa, Vitis vinifera, the gymnosperm Picea sitchensis, and the moss Physcomitrella patens. Proteins with two domains (similar to OSR40cl and OSR40g2 from rice) were found in P. sitchensis and P. patens.
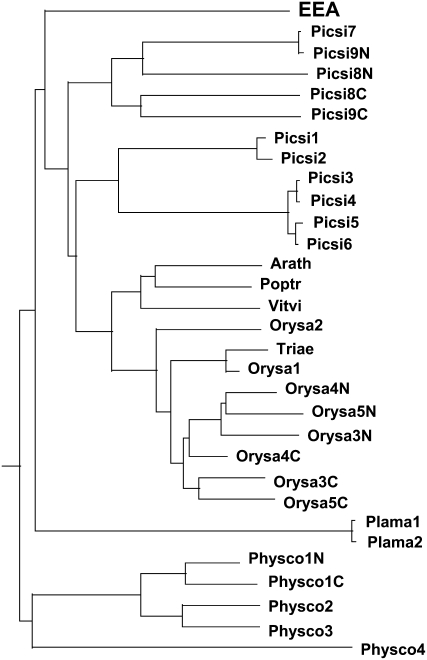
Though all these proteins share a high sequence similarity with EEA, there is an important difference concerning the overall structure of the proteins. Apart from the two proteins expressed in the vascular tissues of Plantago major (CAH59433.1 and CAH59435.1, which are annotated as lectin-like protein 1 and lectin-like protein 2, respectively), all other proteins contain an N-terminal extension varying in length from approximately 10 to >150 residues. The presence of highly variable extra N-terminal sequences combined with the occurrence of proteins with two tandemly arrayed lectin domains makes it difficult to make a phylogenetic analysis using the complete sequences of the proteins. To circumvent this problem, the alignment was confined to the sequences spanning the (putative) lectin domains only (Supplemental Fig. S5). In addition, proteins with a double lectin domain were split up in an N-terminal and a C-terminal domain. A phylogenetic analysis indicates that EEA (and also the presumed lectins from P. major) do not cluster with the sequences from the angiosperms but are placed in two separate branches outside the Spermatophyta group (Fig. 3). Due to the limited number of sequences, no definitive conclusions can be drawn with respect to the aberrant position of EEA in the dendrogram. However, it seems likely that the Euonymus agglutinin, which for the time being is the only identified lectin of this protein family, does not belong to the main evolutionary line but represents a small side group. A similar conclusion was drawn, for example, for the Gal-specific jacalin-related lectins, which are confined to a small taxonomic group within the family Moraceae and are believed to represent a small side group of a ubiquitous family of Man-specific plant lectins (Van Damme et al., 2004a).
Figure 3.
Phylogenetic analysis of EEA domain sequences retrieved from the NCBI database.
Proteins Comprising Domain(s) Homologous to EEA Are Expressed in All Embryophyta for Which Adequate Genomic Coverage Is Available
None of the hits of the BLASTp search with the EEA sequence corresponds to a previously isolated and characterized (plant) lectin. In addition, the lists comprise exclusively plant sequences. PSI-BLAST searches yielded no additional positive hits. Accordingly, one can reasonably conclude that EEA cannot be classified into any of the previously identified plant lectin families and hence represents a novel lectin family that, based on the currently available information, seems to be confined to plants.
To check whether the EEA protein family is more widespread than can be inferred from the BLASTp and PSI-BLAST searches, the publicly accessible transcriptome databases were screened for cDNA/expressed sequence tags encoding proteins with EEA domain(s). tBLASTn searches yielded a very high number (several thousands) of hits with an Expect value <1e-20 (Supplemental Table S3). A quick overview of the 20 best matches already illustrates that homologs of EEA are expressed in a variety of species covering both monocots (rice, Panicum virgatum, Saccharum officinarum, Sorghum bicolor) and dicots (Glycine max, Gossypium hirsutum, Ipomoea nil, Poncirus trifoliata, V. vinifera). More detailed analyses revealed that proteins with one or two domains equivalent to EEA are expressed in all Embryophyta for which a reasonable number of sequences have been deposited (including liverworts, ferns, lycopods, cycads, gymnosperms, and angiosperms). All these proteins are, like EEA, synthesized without a signal peptide and accordingly destined to reside in the cytoplasmic/nuclear compartment of the plant cell. Preliminary experiments aiming at the localization of a fusion protein of EEA coupled to GFP have confirmed the nucleocytoplasmic location (E. Fouquaert and E.J.M. Van Damme, unpublished data).
Similar BLAST searches in nonplant protein, genome, and transcriptome databases did not yield a single positive hit, indicating that the EEA domain is absent from other Eukaryota (e.g. animals and fungi) as well as from Prokaryota. Accordingly, one can reasonably conclude that the EEA domain is confined to plants.
CONCLUSION
A reinvestigation of EEA indicated that the previously reported molecular structure has to be revised. In addition, glycan array screening revealed that EEA interacts with two structurally unrelated glycans, namely the blood group B oligosaccharide and high Man N-glycans. Molecular cloning demonstrated that EEA cannot be classified into any of the currently known (plant) lectin families but shares a high sequence similarity with a domain found in some previously identified abscisic acid- and salt stress-responsive rice proteins. Although no similar lectins have been isolated yet, searches in the databases leave no doubt that all Spermatophyta express one or more proteins comprising either a single or two in-tandem domains equivalent to the EEA subunit. We therefore propose that EEA represents a novel family of proteins that is apparently ubiquitous in Spermatophyta. Moreover, because no homologous genes/proteins are present in other eukaryotes or in prokaryotes, the EEA lectin family can be considered plant specific. At present, the physiological role of the EEA family remains unclear. It has been proposed that the rice OSR40 protein family plays a role in the adaptive response of roots to a hyperosmotic environment and most probably has structural functions (Moons et al., 1995, 1997). The latter assumption was based primarily on the presence at the N terminus of some OSR40 proteins of a His-rich sequence that was believed to mediate protein-protein interactions. Evidently, the finding that the OSR40 proteins contain one or two EEA domains sheds new light on their function in the plant cells because they might be bifunctional proteins possessing both a protein-binding and a carbohydrate-binding domain. This is of paramount importance because OSR40 proteins are at least in principle capable of cross-linking proteins and glycoconjugates. Taking into consideration that OSR40 proteins are presumably located in the cytoplasmic/nuclear compartment and, in addition, are apparently involved in responses to stress, the identification of EEA eventually leads to the conclusion that lectin-mediated protein-glycoconjugate interactions are essential for some important cellular processes in Embryophyta. Moreover, the fact that the EEA domain is apparently confined to plants strongly suggests that there are fundamental differences between plants and other eukaryotes at the level of intracellular glycobiology.
MATERIALS AND METHODS
Purification of EEA
EEA was purified from arillus tissue using a combination of conventional protein purification techniques and affinity chromatography. Seeds were collected from local Euonymus europaeus trees at the end of October and were air-dried. Arilli were removed by gently crushing the seeds, extracted for 24 h in ethanol, and air-dried. Twenty-five grams of dried arillus tissue was powdered with mortar and pestle and extracted with 500 mL of 20 mm unbuffered 1,3-diaminopropane. The homogenate was centrifuged (9,000g for 15 min) and the supernatant filtered through filter paper (Whatman 3MM). The clarified filtrate was diluted with an equal volume of distilled water and loaded on a column of Q Fast Flow (GE Healthcare; 2.6 cm × 5 cm; approximately 25-mL bed volume) equilibrated with 20 mm unbuffered 1,3-diaminopropane. The column was washed with 20 mm unbuffered 1,3-diaminopropane until the A280 fell below 0.01 and the bound proteins eluted with a gradient (500 mL) of increasing NaCl (0–1 m) in the same buffer. Estimation of the protein and lectin content of the fractions (5 mL each) by measuring the A280 and agglutination activity, respectively, indicated that EEA eluted in the main peak. The peak fractions were pooled, diluted with 10 volumes of 20 mm unbuffered 1,3-diaminopropane, and applied onto a small column (1.6 cm × 5 cm; approximately 10-mL bed volume) of Q Fast Flow. Desorption with 1 m NaCl yielded 5 mL of a concentrated solution of partially purified EEA that was directly applied onto a column (2.6 cm × 70 cm; approximately 350-mL bed volume) of Sephacryl 100 equilibrated with 0.2 m NaCl in 20 mm unbuffered 1,3-diaminopropane for subsequent gel filtration. Fractions were collected and assayed for protein and lectin content. Most of the protein eluted in a single symmetrical peak that contained virtually all agglutinating activity and according to SDS-PAGE consisted almost exclusively of a single 17-kD polypeptide. Peak fractions (40 mL in total) were pooled and dialyzed against phosphate-buffered saline. Unlike in previously described purification protocols (Pacak and Kocourek, 1975; Petryniak et al., 1977), no precipitation occurred during dialysis, indicating that the ion-exchange and gel filtration chromatography steps effectively removed some interfering compounds from the crude extract. Final purification of the lectin was achieved by affinity chromatography. The lectin fraction was mixed with an equal volume of 2 m ammonium sulfate and applied on a column of ovomucoid-Sepharose 4B (2.6 cm × 10 cm; 50-mL bed volume) equilibrated with 1 m ammonium sulfate. After loading, the column was washed with 1 m ammonium sulfate until the A280 fell below 0.01 and the bound lectin desorbed with 20 mm Tris-HCl, pH 10. The resulting affinity-purified lectin was dialyzed against an appropriate buffer and used immediately or stored at −20°C until use. Following this procedure, approximately 50 mg of pure EEA was obtained from 100 g of dry arillus material, with an overall recovery of roughly 75%.
Analytical Methods
The purified lectin was analyzed by SDS-PAGE in a 4% to 12% (w/v) BisTris acrylamide gel (Invitrogen) and visualized by staining with Coomassie Brilliant Blue. Glycoproteins were distinguished after SDS-PAGE and electroblotting using periodic acid Schiff's staining following the instructions of Sigma-Aldrich. Alternatively, total neutral sugar was determined by the phenol/H2SO4 method with d-Glc as standard (Dubois et al., 1956).
For N-terminal amino acid sequencing, the EEA polypeptides were separated by SDS-PAGE and electroblotted on a polyvinylidene difluoride membrane. Polypeptides were excised from the blots and sequenced on a model Procise 491cLC protein sequencer without alkylation of Cys (Applied Biosystems).
DLS measurements were carried out using a Zetasizer Nano S (Malvern Instruments) equipped with a 633-nm He-Ne laser and a temperature-controlled measuring chamber. Purified EEA at 0.45 mg/mL in distilled water was clarified by centrifugation for 2 h at 16,000g, and the supernatant was then subjected to DLS measurements at 20°C.
Glycan Array Screening
The microarrays were printed as described before (Blixt et al., 2004), and version 3.0 (see https://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8.shtml) was used for the analyses reported here. Lyophilized lectin preparations were dissolved in phosphate-buffered saline at 1 mg/mL and labeled with tetrafluorophenyl-Alexa Fluor 488 using the Invitrogen protein labeling kit following the manufacturer's instructions. Assuming an extinction coefficient of 1.85 for a 1.0 mg/mL solution, the molar ratios of Alexa488 to protein were 0.3 or 0.7 in two separate labelings.
The labeled lectin was diluted to 0.2 mg/mL in Tris-buffered saline (20 mm Tris, 150 mm NaCl, 2 mm CaCl2, 2 mm MgCl2, pH 7.4) containing 1% bovine serum albumin and 0.05% Tween 20. An aliquot (70 μL) of the labeled lectin solution with or without oligosaccharide or glycopeptide inhibitors was applied to separate microarray slides and incubated under a coverslip for 60 min in a dark, humidified chamber at room temperature. After the incubation, the coverslips were gently removed in a solution of Tris-buffered saline containing 0.05% Tween 20 and washed by gently dipping the slides four times in successive washes of Tris-buffered saline containing 0.05% Tween 20, Tris-buffered saline, and deionized water. After the last wash, the slides were spun in a slide centrifuge for approximately 15 s to dry and immediately scanned in a PerkinElmer ProScanArray MicroArray scanner using an excitation wavelength of 488 nm and ImaGene softweare (BioDiscovery) to quantify fluorescence. The data were reported as average RFU of four to six replicates (after removal of the highest and lowest values) for each glycan represented on the array.
Inhibition of EEA binding to the array at 200 μg/mL was carried out using a 3 mg/mL human blood group B-active tetrasaccharide (Galα1-3[Fucα1-2]Galβ1-4GlcNAcβ) obtained from the Glycan Array Synthesis Core D of the Consortium for Functional Glycomics or 3 mg/mL of a mixture of glycopeptides from bovine ribonuclease B (Sigma) containing N-linked, high Man oligosaccharides (Man5-8GlcNAc2) obtained by pronase (Calbiochem) digestion of RNase B and affinity purification of the glycopeptides on a column of concanavalin A as previously described (Lang et al., 1984).
RNA Isolation and Construction of a cDNA Library
Total RNA was prepared from the arils of E. europaeus as described by Van Damme and Peumans (1993). The plant material was ground to a fine powder in liquid nitrogen using a prechilled mortar and pestle and extracted in 20 mL/g fresh weight cold homogenization buffer (100 mm Tris-HCl, pH 9.0, 5 mm EDTA, 100 mm NaCl, 1% β-mercaptoethanol). After centrifugation, SDS was added to a final concentration of 0.5%. RNA was extracted with phenol (preheated to 60°C):chloroform:isoamylalcohol (25:24:1). The phenol/chloroform extraction was repeated twice. Nucleic acids in the supernatant were precipitated overnight at −20°C by adding 0.3 m sodium acetate and 2.5 volumes cold ethanol. RNA was collected by centrifugation and washed with ethanol. After centrifugation, the pellet was dried and dissolved in a small volume of 10 mm Tris-HCl containing 1 mm EDTA, pH 8.0. The redissolved RNA was precipitated overnight at −20°C at a final concentration of 2 m lithium chloride, collected by centrifugation, washed with ethanol, and dissolved in water. Poly(A+)-enriched RNA was obtained using the Oligotex mRNA Maxi kit (Qiagen). The quality of the RNA was checked by spectrophotometric analysis. A cDNA library was constructed from 2 μg poly(A)-rich RNA by using the cDNA Synthesis System from Roche (Roche Diagnostics) following the instruction manual. cDNA fragments were inserted into the pJET1/blunt cloning vector with the use of the GeneJET PCR Cloning kit (MBI Fermentas), and the ligation mixture was transformed into Escherichia coli Top 10 F′ competent cells using the heat shock protocol.
Screening of cDNA Library
Clones were screened by colony hybridization using a 32P-end labeled synthetic oligonucleotide probe derived from the N-terminal amino acid sequence of the mature EEA polypeptide. In subsequent screenings, a cDNA clone encoding the EEA was used as a probe, as described previously (Van Damme et al., 1996). The radioactive signal was visualized using the FujiFilm Fluorescent Image Analyzer FLA-5100 (Fuji).
Colonies that yielded positive signals were selected and rescreened at low density under the same conditions. Plasmids were isolated from purified single colonies on a miniprep scale using the QIAprep Spin MiniPrep kit (Qiagen) and sequenced at the VIB Genetic Service Facility (Antwerp, Belgium).
PCR Amplification of Genomic DNA Fragment
Genomic DNA was isolated from 300 mg of E. europeaus seeds using the Fast DNA Spin kit in a homogenizer (FastPrep Instrument, MP Biomedicals and Qbiogene) following the manufacturer's recommendations. The genomic sequence encoding the E. europeaus agglutinin was amplified by PCR. The forward primer was complementary to the 5′ end (5′-ATGGCTTCAACAATCATCGCAA-3′) of the coding sequence of the cDNA clone encoding EEA and the reverse primer complementary to the 3′ untranslated sequence (5′-TTC CAA AGC TAT AAG GAA AGG -3′). PCR was performed in a 50-μL reaction volume containing 200 ng of genomic DNA, 10× DNA polymerase buffer, 1.5 mm MgCl2, 0.4 mm dNTPs, 0.2 μm of each primer, and 1.25 units of Taq polymerase (Invitrogen). The PCR program consisted of 25 repetitive cycles with a denaturation step at 94°C for 15 s, an annealing step at 50°C for 30 s, and an elongation step at 72°C for 1 min. The PCR cycles were preceded by an extra denaturation step of 2 min at 94°C and ended with an extra elongation step of 5 min at 72°C.
PCR fragments were cloned into the pCR2.1-TOPO vector using the TOPO TA Cloning kit from Invitrogen and the ligation mixture transformed into E. coli Top 10 F′ heat shock competent cells. Transformed clones were selected on Luria-Bertani agar plates containing ampicillin (100 μg/mL), and PCR screening was used to check for positive clones. Plasmid DNA of a positive colony was purified and its sequence analyzed.
Retrieval of Sequences
Sequences encoding proteins with a domain homologous to EEA were retrieved (in the NCBI database) by BLASTp and PSI-BLAST searches using the complete deduced amino acid sequences of EEA as a query.
Sequence Alignment and Preliminary Phylogenetic Analysis
The amino acid sequences of EEA and homologous domains from other plant proteins were aligned by ClustalW (1.81) Multiple Sequence Alignments (http://align.genome.jp/). A dendrogram (N-J tree with branch length) was generated by the same program for a phylogenetic analysis.
Accession numbers for the sequence data: EF990655 and EF990656.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genomic sequence of EEA.
Supplemental Figure S2. Determination of the specificity of EEA by screening on the printed array version 3 of the Consortium for Functional Glycomics.
Supplemental Figure S3. Summary of EEA inhibition by B-active oligosaccharides and RNase B glycopeptides.
Supplemental Figure S4. Alignment of the amino acid sequences of OSR40g3 (CAA70175.1, Os07g0684000) and the individual ricin-B domains of R. communis agglutinin (AAA33869.1).
Supplemental Figure S5. Sequence alignment of EEA domains retrieved from the NCBI database.
Supplemental Table S1. Glycan array binding of EEA at a concentration of 200 μg/mL; list of glycan structures ranked from highest RFU to lowest.
Supplemental Table S2. List of best hits (E < 0.01) of a BLASTp search using the deduced sequence of LECEEA as a query.
Supplemental Table S3. List of top 50 hits of a tBLASTn search using the deduced sequence of LECEEA as a query (EST database).
Supplemental Signature Alignment Data S1. Sequence alignment of EEA domains.
Supplementary Material
This work was supported by Ghent University and the Fund for Scientific Research-Flanders (FWO grant nos. G.0201.04 and G.0022.08) and by the National Institute of General Medical Sciences GM62116 (to The Consortium for Functional Glycomics for the glycan array analysis).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Els J.M. Van Damme (elsjm.vandamme@ugent.be).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al (2004) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA 101 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugar and related substances. Anal Chem 28 350–356 [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang L, Reitman M, Tang J, Roberta RM, Kornfeld S (1984) Lysosomal enzyme phosphorylation. Recognition of a protein-dependent determinant allows specific phosphorylation of oligosaccharides present on lysosomal enzymes. J Biol Chem 259 14663–14671 [PubMed] [Google Scholar]
- Moons A, Bauw G, Prinsen E, Van Montagu M, Van der Straeten D (1995) Molecular and physiological responses to abscisic acid and salts in roots of salt-sensitive and salt-tolerant Indica rice varieties. Plant Physiol 107 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons A, Gielen J, Vandekerckhove J, Van der Straeten D, Gheysen G, Van Montagu M (1997) An abscisic-acid- and salt-stress-responsive rice cDNA from a novel plant gene family. Planta 202 443–454 [DOI] [PubMed] [Google Scholar]
- Pacak F, Kocourek J (1975) Studies on Phytohemagglutinins. XXV. Isolation and characterization of hemagglutinins of the spindle tree seeds (Evonymus europaea L.). Biochim Biophys Acta 400 374–386 [PubMed] [Google Scholar]
- Petryniak J, Goldstein IJ (1987) Evonymus europaea lectin. Methods Enzymol 138 552–561 [DOI] [PubMed] [Google Scholar]
- Petryniak J, Pereira ME, Kabat EA (1977) The lectin of Euonymus europeus: purification, characterization, and an immunochemical study of its combining site. Arch Biochem Biophys 178 118–134 [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Fouquaert E, Jauneau A, Rougé P, Lannoo N, Hamada H, Alvarez R, Devreese B, Van Damme EJM (2007) The liverwort Marchantia polymorpha expresses orthologs of the fungal Agaricus bisporus agglutinin family. Plant Physiol 144 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt VG (1954) Die hämagglutination, im besonderen menschlicher B-Blutzellen durch Extrakte aus Samen von Euonymus vulgaris (Phaffen-Hütchen). Z Immunitätsforschung 11 432–439 [PubMed] [Google Scholar]
- Teneberg S, Alsen B, Angstrom J, Winter HC, Goldstein IJ (2003) Studies on Galα3-binding proteins: comparison of the glycosphingolipid binding specificities of Marasmius oreades lectin and Euonymus europaeus lectin. Glycobiology 13 479–486 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Barre A, Rougé P, Peumans WJ (2004. a) Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci 9 484–489 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Barre A, Verhaert P, Rougé P, Peumans WJ (1996) Molecular cloning of the mitogenic mannose/maltose-specific rhizome lectin from Calystegia sepium. FEBS Lett 397 352–356 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Culerrier R, Barre A, Alvarez R, Rougé P, Peumans WJ (2007) A novel family of lectins evolutionarily related to class V chitinases. An example of neofunctionalization in legumes. Plant Physiol 144 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Lannoo N, Fouquaert E, Peumans WJ (2004. b) The identification of inducible cytoplasmic/nuclear carbohydrate-binding proteins urges to develop novel concepts about the role of plant lectins. Glycoconj J 20 449–460 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Peumans WJ (1993) Cell-free synthesis of lectins. In H-J Gabius, S Gabius, eds, Lectins and Glycobiology. Springer-Verlag, Berlin, pp 458–468
- Van Damme EJM, Peumans WJ, Barre A, Rougé P (1998) Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit Rev Plant Sci 17 575–692 [Google Scholar]
- Yamamoto S, Sakai I (1981) Composition and immunochemical properties of glycoproteins with anti-B agglutinin activity isolated from Euonymus sieboldiana seeds. J Immunogenet 8 271–279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.