Abstract
A systematic comparison of six sugar indicators for their sensitivity, specificity, cross reactivity and suitability in the context of crude lysates revealed para-hydroxybenzoic acid hydrazide (pHBH) to be best suited for application in a plate-based phosphatase-assisted universal sugar-1-phosphate nucleotidyltransferase assay. The addition of a general phosphatase to nucleotidyltransferase reaction aliquots enabled the conversion of remaining sugar-1-phosphate to free sugar, the concentration of which could be rapidly assessed via the pHBH assay. The assay was validated using the model glucose-1-phosphate thymidylyltransferase from Salmonella enterica (RmlA) and compared favorably to a previously reported HPLC assay. This coupled discontinuous assay is quantitative, high-throughput and robust; relies only on commercially available enzymes and reagents; does not require chromatography, specialized detectors (e.g. mass or evaporative light scattering detectors) or radioisotopes; and is capable of detecting less than 5 nmol of sugar-1-phosphate. It is anticipated this high throughput assay system will greatly facilitate nucleotidyltransferase mechanistic and directed evolution/engineering studies.
Keywords: carbohydrate, sugar, nucleotide, glycosyltransferase, enzyme
Introduction
Sugar nucleotides are critical to the survival of all living organisms. As substrates for Leloir-dependent processes in primary metabolism, sugar nucleotides play an integral role in a variety of essential processes including glycogen biosynthesis, carbohydrate metabolism, protein glycosylation and cell wall biosynthesis [1–5]. Equally important in secondary metabolism, sugar nucleotides are essential for the biosynthesis of a vast array of therapeutically important glycosylated natural products [6–9]. In nature, sugar nucleotidyltransferases (E.C. 2.7.7.-; also sometimes referred to NDP-sugar pyrophosphorylases) catalyze the formation of these vital reagents from nucleotide triphosphates (NTPs) and sugar-1-phosphates. Based upon the TrEMBL databank [10], more than 8,000 non-polymerase nucleotidyltransferases have been reported. Given the predominance of nucleotidyltransferase and NDP-sugars in nature, the study of nucleotidyltransferases remains an active area of research and continues to offer many exciting opportunities ranging from potential targets for the design of anti-infectives [11–13] to new tools for the synthesis of glycoconjugates [6, 14–19].
Despite the overwhelming importance of nucleotidyltransferases, their study has been limited to a degree by the lack of flexible, high-throughput assays for enzyme activity. Most of the commonly used nucleotidyltransferase assays to date are discontinuous and rely upon radioactive isotopes and/or chromatographic approaches [20–24]. A variety of coupled assays exist [25], but most are restricted to reactions which employ specific sugars (usually Glc) and/or NTPs. Among the ‘universal’ nucleotidyltransferase assays reported over the past two decades, the nucleotidyltransferase substrate screening assay (NUSSA) developed by Elling and coworkers is a coupled assay which ultimately correlates pyrophosphate production (a product of all nucleotidyltransferase reactions) to the spectrophotometric determination of NADH levels [26]. This assay offers a major advance over the prior pyrophosphate detection techniques which are prone to high backgrounds from phosphate contamination due to their dependence upon pyrophosphate hydrolysis and subsequent orthophosphate detection [27–30]. Yet, the NUSSA requires an uncommon enzyme (pyrophosphate-dependant phosphofructokinase) and may still suffer some interference from crude lysate components. Recently, Pohl and coworkers developed an alternative electrospray ionization mass spectrometry (ESI-MS)-based nucleotidyltransferase assay [31]. A notable advantage of the ESI-MS assay over pyrophosphate detection methods is the ability to monitor depletion of essentially any substrate and formation of effectively any product simultaneously. While this assay should also accommodate moderate to high throughput, the method requires an appropriate mass spectrometer, access to a standard for the substance being monitored, and has yet to be evaluated in the context crude extracts or a wide range of biological buffers.
We previously described the development and application of a robust, high-throughput colorimetric ‘DNS assay’ for the kinetic characterization, directed-evolution and structure-based engineering promiscuous anomeric kinases [32–36]. This assay strategy, based upon an oxidation-reduction reaction between dinitrosalicylic acid (DNS, the oxidant) and a reducing sugar, was readily applicable to a wide range of free sugars in the context of crude lysates and presented a general assay platform potentially amenable to a variety of sugar-processing enzymes. For example, the addition of a general phosphatase at various timepoints (to selectively convert unreacted sugar-1-phosphate to free sugar), followed by a colorimetric sugar determination should present an opportunity for a universal nucleotidyltransferase activity assay. To explore this potential application, herein we report a comparison of available free sugar assays for their sensitivity, specificity, cross reactivity and suitability in the context of crude lysates. The subsequent development of a high-throughput assay for nucleotidyltransferases and other sugar-1-phosphate utilizing enzymes, based on the detection of remaining sugar-1-phosphate, is presented. Specifically, the addition of sufficient alkaline phosphatase at any given time point rapidly terminates the nucleotidyltransferase reaction by converting residual sugar-1-phosphate to the reducing sugar, which can be detected by a free sugar assay capable of detecting less than 5 nmol of sugar-1-phosphate. The assay was validated using the model glucose-1-phosphate thymidylyltransferase from Salmonella enterica (RmlA) and compared favorably to a previously reported HPLC assay. The coupled discontinuous assay is quantitative, relies only on readily available enzymes and reagents, and does not require chromatography, specialized detectors (e.g. mass or evaporative light scattering detectors) or radioisotopes.
Materials and methods
General materials and methods
Unless otherwise specified, all chemicals and enzymes were reagent grade or better obtained from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) and used without further purification. Recombinant Salmonella enterica N-His6-RmlA, and corresponding mutants, were purified as previously described [37]. HPLC was performed using a Varian (Palo Alto, CA) ProStar HPLC system. Absorbance readings were performed on a Beckman Coulter (Fullerton, CA) DU 800 spectrophotometer (cuvette scale) or a BMG Labtech (Durham, NC) FLOUstar Optima plate reader (microtiter plate scale).
Reducing sugar assays
Previously reported assays were modified from to fit a standard final volume of 1 mL (in 1.5 mL microcentrifuge tubes) and heating was accomplished in a Barnstead/Thermolyne (Dubuque, IA) dry well heat block. Assay solutions were subsequently transferred to 1.5 mL semi-micro polystyrene cuvettes for absorbance measurements. For all assays, water-only assay reactions served as the spectrophotometer blank.
Dinitrosalicylic acid (DNS) assay [32, 38]
To 0.25 mL of sample was added 0.75 mL of a solution of 1% (w/v) 3,5-dinitrosalycilic acid, 0.2% (w/v) phenol, and 0.05% (w/v) sodium bisulfite in 1% (w/v) sodium hydroxide. The mixture was heated for 5 min at 100 °C and the absorbance of the cooled solution was measured at 575 nm.
Tetrazolium blue assay [39]
To 0.01 mL of sample was added 0.5 mL of 2% (w/v) tetrazolium blue in 0.1 M sodium hydroxide and 0.5 mL of 1 M sodium potassium tartrate. The mixture was heated for 3 min at 100 °C and the absorbance of the cooled solution was measured at 660 nm.
Ferricyanide (Prussian blue) assay [40]
To 0.111 mL of sample was added 0.22 mL of a solution of 1.5 mM potassium ferricyanide, 50 mM sodium carbonate and 10 mM potassium cyanide. The mixture was heated for 15 min at 100 °C. To the cooled solution was added 0.555 mL of 1.36 mM ferric ammonium sulfate in 25 mM sulfuric acid. This was heated for 15 min at 50 °C. To the cooled solution was added 0.111 mL of 125 mM oxalic acid and the absorbance was measured at 690 nm.
Honda 3-methyl-2-benothiazolinonehydrazone (MBTH) assay [41]
To 0.25 mL of sample was added 0.125 mL of 2% (w/v) 3-methyl-2-benothiazolinonehydrazone, 0.125 mL of 0.3 M sodium hydroxide, and 0.5 mL of 2-methoxyethanol. The mixture was heated for 10 min at 100 °, and the absorbance of the cooled solution was measured at 390 nm.
Anthon MBTH assay [42]
To 0.1 mL of sample was added 0.1 mL of 0.5 M sodium hydroxide and 0.1 mL of a freshly prepared 1:1 mixture of 0.3% (w/v) 3-methyl-2-benothiazolinonehydrazone and 1 mg/mL dithiothreitol. The mixture was heated for 4 min at 100 °C and 0.2 mL of 0.5% (w/v) ferric ammonium sulfate, 0.5% (w/v) sulfamic acid, and 0.25 M hydrochloric acid added to the warm mixture. The solution was cooled, 0.5 mL of water was added, and the absorbance was measured at 620 nm.
para-Hydroxybenzoic acid hydrazide (pHBH) assay [43]
For the 1.5 mL microcentrifuge tube format, to 0.01 mL of sample was added 1 mL of a freshly prepared 9:1 mixture of 0.5 M sodium hydroxide and 5% (w/v) para-hydroxybenzoic acid hydrazide in 0.5 M hydrochloric acid. The mixture was heated for 5 min at 100 °C, and the absorbance of the cooled solution was measured at 410 nm.
For the 96-well plate-based format, to 40 μL of sample in a 96 well PCR plate was added 120 μL of a freshly prepared 1:1 mixture of 2 M sodium hydroxide and 2% (w/v) para-hydroxybenzoic acid hydrazide in 0.5 M hydrochloric acid. The mixture was heated for 5 min at 100 °C, the reaction mixture subsequently cooled thoroughly on ice and the solutions transferred to a clear bottomed 96 well polystyrene plate (Nunc, Rochester, NY). The absorbance in each well was measured on a BMG FLOUstar Optima plate reader using a 410 nm filter (10 nm bandwith).
Selectivity and cross-reactivity of reducing sugar assays
To determine the sensitivity, specificity, and cross-reactivity of the above assays, the appropriate volume of solution containing glucose (0.05–20 mM), glucose-1-phosphate (0.5–10 mM), UDP-glucose (0.5–10 mM), methyl-α-D-glucopyranoside (0.5–10 mM), 2-deoxyglucose (0.5–10 mM), glucosamine (0.5–10 mM), or bovine serum albumen (BSA) (0.03–100 mg/mL) in water were assayed in the appropriate assay reaction conditions. To validate the plate scale pHBH reactions, 40 μL of glucose, galactose, glucosamine (each 0.1–2 mM), buffers (100 mM), glycerol (5%), MgCl2 (25 mM), ZnSO4 (2.5 mM), or BSA (1–10 mg/mL) were assayed in the appropriate assay reaction conditions.
Nucleotidyltransferase assays
High throughput pHBH-based nucleotidyltransferase assays
Master mixes consisting of 100 mM MOPS pH 7.5, ([UTP] + 5 mM) MgCl2, 2 mM α-D-glucose-1-phosphate, 0.5–16 mM UTP, and 10 U/mL inorganic pyrophosphatase (final concentrations) were pre-warmed at 37 °C in 0.2 mL PCR tubes. Reactions were initiated by the addition of S. enterica N-His6-RmlA (0.1–0.6 μM). Aliquots (20 μL each) were removed every 30 s and added to an equal volume of phosphatase solution [100 mM sodium carbonate pH 10.0, 2 mM ZnSO4, and 8 U bovine intestinal phosphatase (Sigma-Aldrich cat. P0114; ≥7,500 DEA units/mg protein in 50% glycerol) in 20 μL]. The reactions were subsequently incubated at 37 °C for ≥ minutes and reducing sugar detected via the plate-scale pHBH assay. Absorbance measurements were converted to [sugar-1-phosphate] via direct correlation to a sugar-1-phosphate standard curve (0.4–2 mM) generated in parallel.
Nucleotidyltransferase kinetic analysis
For single-substrate kinetic analysis, nucleotidyltransferase activity assays with purified S. enterica N-His6-RmlA (0.3 μM) were performed as described above for varying UTP concentration (0.5–16 mM) at a constant [Glc-1-P] of 2 mM (the KM for Glc-1-P reacting with dTTP was previously measured at 0.3 mM [44]). Unreacted sugar-1-phosphate substrate was quantified with the plate-form pHBH assay. Initial reaction velocities were obtained as the slope of best fit to the initial linear portion of the reaction time course, and fit to the Michaelis-Menten equation using SigmaPlot (Systat, San Jose, CA).
Crude Lysate analysis
For use of this assay format with crude extracts, overnight cultures of Escherichia coli BL21(DE3) cells overproducing S. enterica N-His6-RmlA wild type, Q83A, Q83S, Q83D, Q83N and Q83E variants [37] were diluted 1:100 in 1 mL of LB media in a 96-well deep-well plate. After 4 hours of growth (37 °C, 350 rpm), IPTG was added to 1 mM and incubation continued for another 4 hours. The pelleted cells were resuspended in 100 μL of 100 mM MOPS pH 7.5 with 20 mg/mL lysozyme. The resuspended cells were frozen 1 h at −80 °C, thawed for 1 h at 30 °C, and clarified by centrifugation. Clarified lysate (10 μL) was added to 30 μL of master mix (final concentration: 100 mM MOPS pH 7.5, 7 mM MgCl2, 2 mM glucose-1-phosphate, 2 mM UTP, and 10 U/mL inorganic pyrophosphatase) and the assay incubated at 37 °C. At 6, 15 or 60 min, two identical 20 μL aliquots were removed from each reaction. One was used to assess product formation via the high-throughput pHBH nucleotidyltransferase assay (described above) while the second was employed as a direct HPLC comparator. Plate-form pHBH absorbance measurements were converted to percent conversion using the equation 1-(Absex-Abspos)/(Absneg-Abspos), where Absex is the experimental absorbance, Abspos is the absorbance of a no-sugar-phosphate control, and Absneg is the absorbance for an empty vector control reaction.
For HPLC analysis, aliquots from the nucleotidyltransferase reaction were quenched by the addition of 80 μL of prewarmed HPLC buffer, heat inactivated (100 °C, 5 min), deproteinated via centrifugation, and analyzed by analytical HPLC as previously described (Supelcosil LC18-T, Supelco, Bellefonte, PA; 5 μm, 250 × 3 mm, 40 mM phosphoric acid, adjusted to pH 6.5 with triethylamine, with a gradient of 0–7.5% MeOH over 15 min, 0.8 ml/min, A254) [37]. HPLC integration was converted to percent conversion with the formula (UDP-Glcex/Totalex)/(UTPctrl/Totalctrl), where UDP-Glcex is the peak area of UDP-Glc in the experimental samples, Totalex is the total area of uridine containing peaks for the experimental sample, UTPctrl is the amount of UTP in an empty vector control reaction, and Totalctrl is the total area of uridine containing peaks for the empty vector control reaction.
Results and discussion
The prior successful applications of the DNS reducing sugar assay for kinetic analysis and substrate specificity determinations in the context of sugar kinase engineering and directed evolution inspired the investigation of a similar strategy for nucleotidyltransferase activity assays [32–36]. Specifically, we hypothesized that the addition of sufficient levels of phosphatase to the nucleotidyltransferase reaction mixture at any given time point would rapidly hydrolyze remaining sugar-1-phosphate to provide free reducing sugar, a substance directly detectable with available free sugar assays (Fig. 1A). As described herein, a variety of sugar indicators were first directly compared for sensitivity, range of detectable sugars and cross reactivity with various biological contaminants (e.g. proteins, buffers, cellular lysates). Using this crucial indicator comparison as a foundation, the subsequent development and validation of the corresponding nucleotidyltransferase assay was pursued.
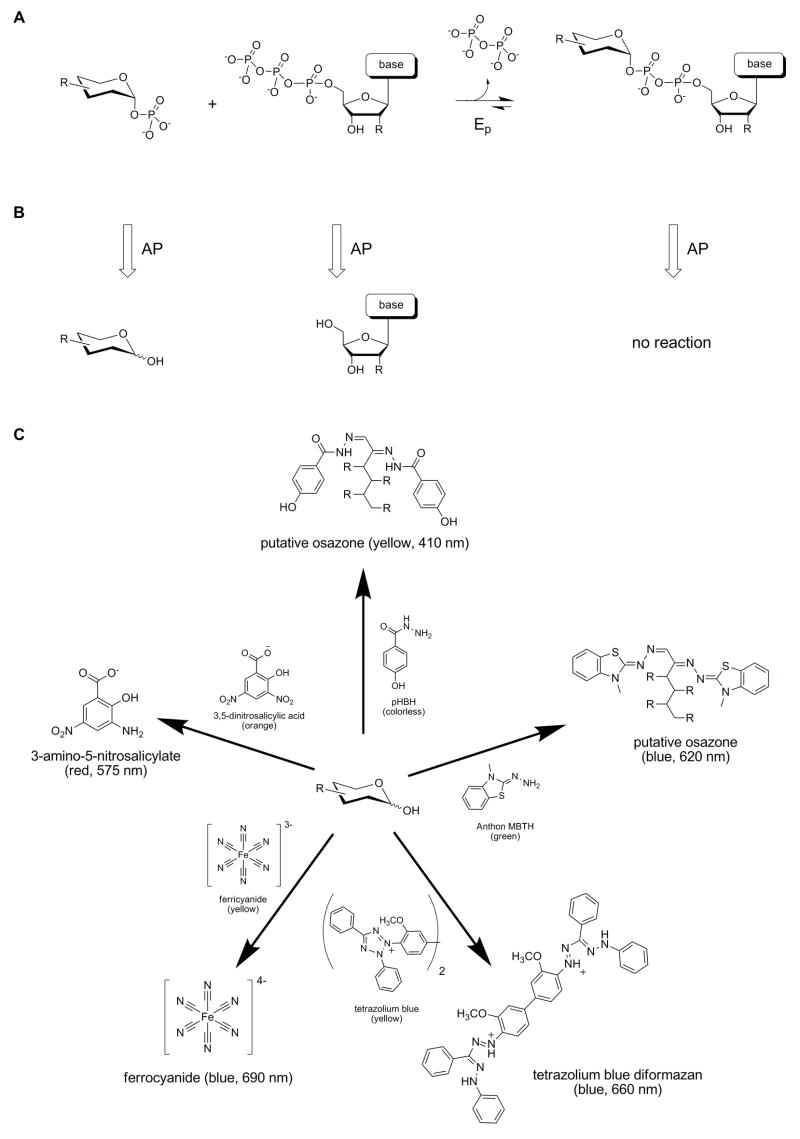
Fig. 1.
Overview of reactions. (A) The reaction catalyzed by a nucleotidyltransferase (Ep). (B) Reaction products after treatment of a nucleotidyltransferase reaction with alkaline phosphatase (AP). (C) General reactions of free sugar with the selected sugar indicators. For each selected indicator, reaction progress can be monitored visually (qualitative) or spectrophotometrically at the indicated wavelength (quantitative).
Selection of a reducing sugar indicator
From the many reported reducing sugar indicators, we restricted our in-depth evaluation to six assays on the basis of i) wavelength of indicator absorbance, ii) availability of reagents, and, in some cases, iii) known cross-reactivity with glycosides or proteins. Of the six selected sugar indicators, reduction of DNS [32, 38], tetrazolium blue [39], or ferricyanide [40] via reaction with sugar leads to products with distinct colorimetric properties. In comparison, reaction of pHBH [43] or MBTH [41, 42] with free sugars leads to covalent osazone adducts [45, 46]. To select the best free sugar indicator for nucleotidyltransferase assay development, the sensitivity, specificity and potential for cross reactivity of the selected six potential reducing sugar indicators were directly compared.
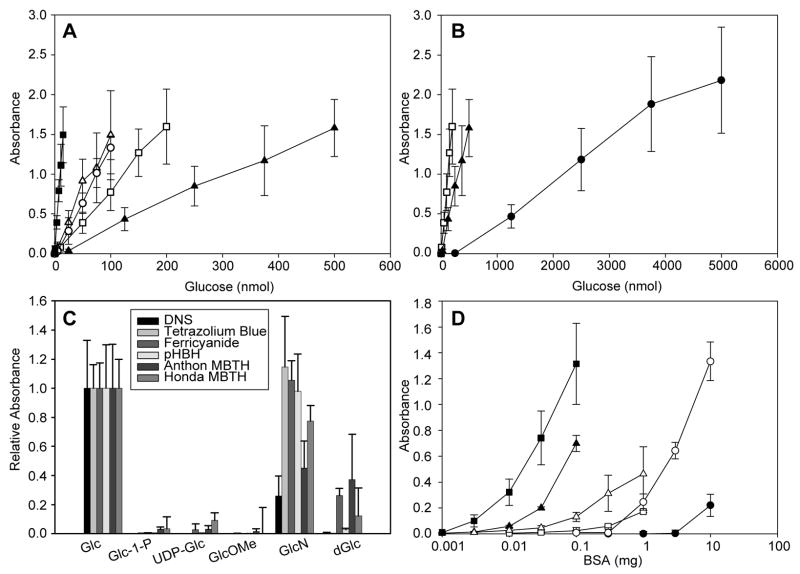
Indicator sensitivities were tested against a range of glucose concentrations in a standardized 1 mL format (Fig. 2A). From this direct comparison, the lower limits of sensitivity ranged from 1 nmol (ferricyanide) – 500 nmol (DNS) with a ranked sensitivity order of ferricyanide > Anthon MBTH Honda MBTH > pHBH > tetrazolium blue > DNS. In comparison, the estimated detection limits for the previously reported NUSSA, HPLC or MS assays range from 10–100 pmol [26, 31, 37]. The usable range of detection for sugar indicator assays (0.8–15 nmol for ferricyanide, 2–300 nmol for pHBH, 5–200 nmol for MBTH, 25–500 nmol for tetrazolium blue, or 500–5000 nmol for DNS) is comparable to that of NUSSA, MS, and HPLC assays (0.2–8 nmol, 50–1250 pmol and 20–10,000 pmol, respectively [26, 31, 37]). While a comparison of both MBTH assays revealed similar detection limits, the longer wavelength detection offered by the Anthon variant (620 nm for Anthon MBTH versus 390 nm for Honda MBTH) may be advantageous [42]. It is interesting to note that, for the redox-based indicators, a comparison of redox potentials (ferricyanide, E0 ~ 0.4–0.5 V; tetrazolium, E0 ~ −0.1 V; standard reducing sugar, E0 ~ −0.4–0.7 V [47–51]) were not good predictors for sensitivity.
Fig. 2.
Comparison of sugar indicator sensitivity, specificity and cross reactivity. (A) The absorbance responses of six reducing sugar assays (tetrazolium blue, ▲; ferricyanide, ■; Honda MBTH, ○ Anthon MBTH, △; pHBH, □) toward varying amounts of glucose. (B) The absorbance response of the DNS assay (●), in comparison to tetrazolium blue (▲) and pHBH (□), toward varying amounts of glucose. (C) Cross reactivity of assays toward representative sugars (Glc, glucose; GlcN, glucosamine; dGlc, 2-deoxyglucose) and glycosides (UDP-Glc uridine 5′diphospho-α-D-glucose; GlcOMe, methyl-α-D-glucopyranoside, Glc-1-P, α-D-glucose-1-phosphate). Substrate amounts for each indicator were adjusted to be equivalent to the amount of glucose required for a 1 AU response (DNS, 2500 nmol; tetrazolium blue, 250 nmol; terricyanide, 7.5 nmol; Honda MBTH, 50 nmol; Anthon MBTH, 50 nmol; pHBH, 100 nmol). (D) Cross reactivity with BSA (tetrazolium blue, ▲; ferricyanide, ■; Honda MBTH, ○; Anthon MBTH, △; pHBH, □; DNS, ●). The x-axis (BSA) is plotted on a logarithmic scale. Turbidity was observed with the tetrazolium blue and Anthon MBTH assays at high levels of protein.
Specificity studies relied upon two distinct model sugar sets – representative non-reducing sugars [the anomerically-’blocked’ sugars α-D-glucose-1-phosphate (Glc-1-P), UDP-Glc, and methyl-α-D-glucopyranoside (GlcOMe)] and representative free sugars [glucosamine (GlcN) and 2-deoxyglucose (dGlc)] (Fig. 2C). As expected, all non-reducing sugars were unreactive. Particularly important among this set were Glc-1-P and UDP-Glc as models for nucleotidyltransferase substrate and product, respectively. Of the representative free sugars, the reactivity of GlcN exceeded that of dGlc for all indicators with the exception of ferricyanide (where they were roughly equivalent). From this analysis, GlcN performed best with tetrazolium blue ≥ ferricyanide ≥ pHBH, while dGlc performed best with Anthon MBTH ≥ ferricyanide > Honda MBTH. It should be noted that some non-reducing sugars (e.g. ketoses) are detectable via the osazone-based assays.
Proteins can also be detected through redox methods (e.g. the Lowry and bicinchoninic acid assays), and methods similar to both the Lowry and bicinchoninic acid assays have been used to detect free sugars [52–55]. However, for the development of a robust enzyme assay platform, particularly one amenable to crude cell lysates, a selected indicator must be minimally reactive toward proteins. To assess the impact of protein upon the panel of candidate sugar indicators, different concentrations of bovine serum albumen (BSA) in water were examined via each assay format. As illustrated in Fig 2D, the most universally sensitive indicator (ferricyanide) was also the most highly reactive toward protein. Both the tetrazolium blue and Anthon MBTH displayed intermediate levels of response with turbidity observed at higher protein amounts, presumably due to protein precipitation. Honda MBTH and pHBH rendered lower responses toward BSA and displayed acceptable background absorbance (<0.2 AU) in the presence of up to 1 mg protein. Finally, the least sensitive assay (DNS) was also the least influenced by protein, with acceptable background absorbances (<0.05) with up to 5 mg protein in the assay.
On the basis of the sensitivity, the interference with protein, the cost of reagents, and the ease of preparation, pHBH was selected as the indicator of choice and the pHBH assay subsequently translated to a microtiter plate-based format. Examination of the published variants of the pHBH assay [43, 56–62] served as a basis for the final 1:3 sample:indicator ratio in the assay, wherein the freshly-prepared indicator was conveniently comprised of a 1:1 mixture of stock solutions (2 M NaOH and 2% pHBH in 0.5 M HCl). The plate-based assay format was validated with a set of representative reducing sugars (glucose, glucosamine, mannose, galactose, fructose, arabinose and 2-deoxyglucose) in a 96-well plate format. (Fig. 3A) Importantly, the average standard error of the microtiter plate-based pHBH assay (Fig. 3A) was significantly lower than the standard 1 mL format (Fig. 2C), possibly indicative of incomplete sample heating with the larger assay volumes. In contrast to the 1 mL scale trials, it was noticed that 2-deoxyglucose (dGlc) gave a low but detectable absorbance reading. Increasing the 100 °C incubation to 20 minutes improved the sensitivity toward dGlc ~5 fold. The response of this assay to various biological buffers (Fig. 3B) revealed compatibility with a variety of common buffers and additives with the notable exceptions of piperazine-based (HEPES, PIPES), tris(hydroxymethyl)aminomethane-based (Tris, Bis-Tris, TES, Bis-Tris Propane, Tricine), or imidazole. It is curious, that, despite their chemical similarity, the piperazine-based buffers (HEPES, PIPES) showed the highest absorbances, whereas the morpholine based buffers (MOPS, MES) gave some of the lowest responses. Whether assay interference in these cases derive from the buffer components, or potentially trace contamination from buffer manufacturing [63], remains undetermined.
Figure 3.
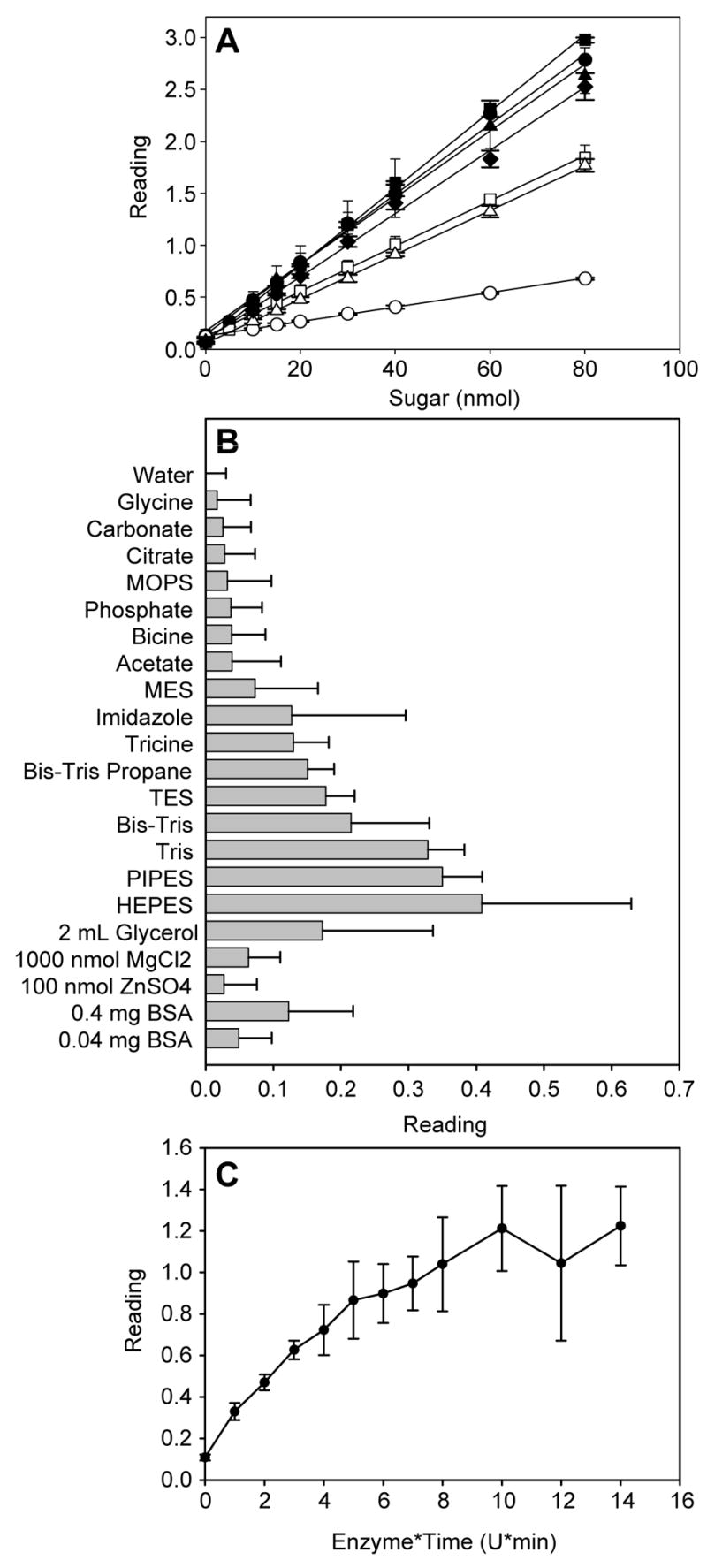
Optimization of the pHBH plate-based format. (A) Sensitivity of the plate-based pHBH assay to various sugars (glucose, ●; glucosamine, ▲; mannose ◆; fructose, ■; galactose, □; arabinose, △; 2-deoxyglucose, ○;). To maximize sensitivity, 2-deoxyglucose was incubated with the pHBH reagent at 100 °C for 20 min (in comparison to a 5 min incubation for all other sugars examined). (B) Cross reactivity of the plate-based pHBH assay with typical buffers and additives. (C) Optimization of the phosphatase-assisted detection of sugar-1-phosphate. Up to 7 U alkaline phosphatase was incubated with 40 nmol of glucose-1-phosphate for 1–3 min and the amount of sugar formed was subsequently determined by the pHBH plate-based assay.
Development and validation of the pHBH-based nucleotidyltransferase assay
Fig. 2C clearly revealed pHBH to be unreactive to representative nucleotidyltransferase substrates (Glc-1-P) or products (UDP-Glc). The addition of sufficient levels of phosphatase to any mixture containing sugar-1-phosphates and/or sugar nucleotides (e.g. a nucleotidyltransferase reaction mixture, Fig. 1B) should therefore lead to the selective hydrolysis of sugar-1-phosphate to provide free reducing sugar - a reagent directly detectable with available free sugar assays as illustrated in Fig. 1C. Optimization of the required amount of alkaline phosphatase to achieve rapid, selective hydrolysis of Glc-1-P (Fig. 3C) revealed complete hydrolysis of 40 nmol Glc-1-P in 1 min with 8–10 U of alkaline phosphatase. Substantiation of the corresponding pHBH microtiter plate-based nucleotidyltransferase assay thus relied upon combining three simple, validated steps – the nucleotidyltransferase reaction, reaction termination via phosphatase hydrolysis, and pHBH-based [sugar] determination –wherein the rate of substrate (sugar-1-phosphate) disappearance over time directly correlates to nucleotidyltransferase activity.
Using the pHBH microtiter plate-based nucleotidyltransferase assay, the turnover rates with purified N-His6-RmlA were linear to 50% conversion of substrate (data not shown) and the initial reaction rates increased with the amount of enzyme (Fig. 4A). Single substrate kinetics for UTP at a constant [Glc-1-P] (2 mM) (Fig. 4B) revealed an apparent kcat of 1440 min−1 and KM of 1.2 mM which, given the differences between buffer conditions and [Glc-1-P], match well with previously reported values (2460 min−1, and 4.6 mM, respectively) [37]. With the newly validated assay in hand, the applicability of the assay to detect varying levels of activity in crude lysate was investigated. A series of S. enterica RmlA mutants representing a range of catalytic efficiencies (wild type, Q83A, Q83S, Q83D, Q83N and Q83E) were overproduced in E. coli BL21(DE3) and the activities of the corresponding crude lysates assessed by both the pHBH microtiter plate-based nucleotidyltransferase assay and HPLC. A comparison of the percent conversions determined by the two assay methods (Fig. 4C) revealed a strong correlation (slope = 1.02, R2= 0.9884) and thereby validates the pHBH microtiter assay in the context of crude extracts representing a wide range of enzymatic activity.
Figure 4.
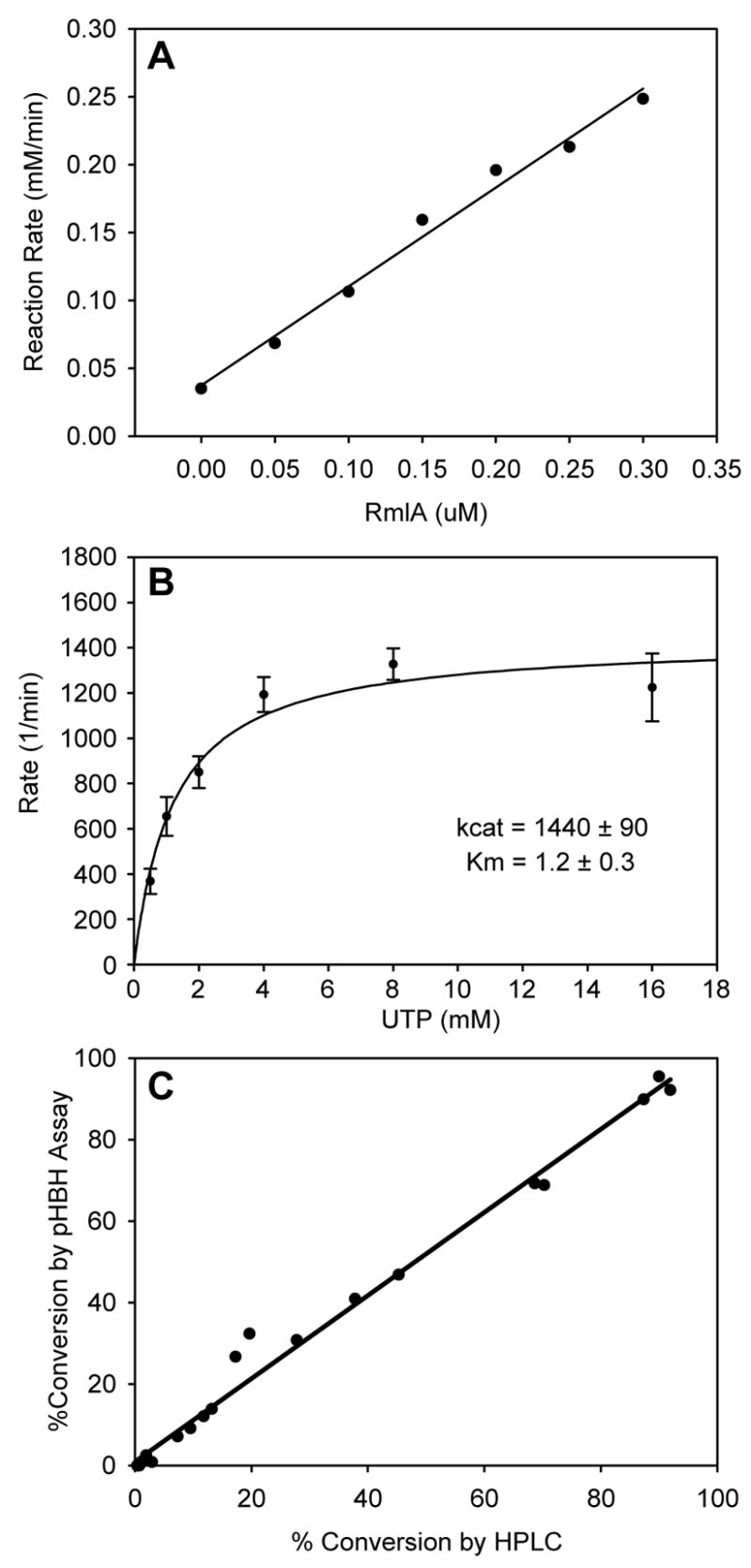
Validation of the phosphatase-assisted plate-based pHBH nucleotidyltransferase assay. (A) The observed rate of glucose-1-phosphate disappearance, under constant substrate concentrations (2 mM glucose and 2.5 mM UTP), plotted against enzyme concentration. (B) Determination of steady-state kinetics via the pHBH assay. Purified S. enterica RmlA (0.3 μM) was incubated with 2 mM glucose-1-phosphate and varying amounts of UTP. Aliquots were taken every 30s, and assayed by the plate-form pHBH assay. The rates of glucose-1-phosphate disappearance (corrected for the constant 0.3 μM enzyme concentration) versus UTP were fit to the Michaelis-Menten equation. (C) Comparison of activities determined by plate-form pHBH assay and HPLC. The activities of crude lysates from E. coli BL21(DE3) cells expressing RmlA variants representing a range of catalytic efficiencies (wild type, Q83A, Q83S, Q83D, Q83N and Q83E) were assessed by both by plate-form pHBH and standard HPLC methods at three different time points (2 mM glucose-1-phosphate, 2 mM UTP; reaction times - 10, 30 and 60 min). The results of the two sets of assays (pHBH versus HPLC) were plotted against one another (slope of line = 1.02, R2 = 0.9884).
Conclusions
A comparison of the performance of variety of sugar indicators revealed pHBH to be most advantageous in the context of composition simplicity, sensitivity, cross reactivity with biological contaminants and range of applicability. Using pHBH as the selected indicator, the subsequent development and validation of the corresponding nucleotidyltransferase assay relied upon the addition of sufficient levels of phosphatase to the nucleotidyltransferase reaction mixture at selected time points to convert remaining sugar-1-phosphate to free sugar – a substance directly detectable via reaction with pHBH. This phosphatase-assisted pHBH assay was found to be quantitative, amenable to a high throughput plate-based format, suitable for both purified enzymes or crude lysates with a sensitivity/accuracy comparable to that of chromatographic (HPLC) or MS-based techniques. It is anticipated this readily accessible, high throughput assay system will facilitate a wide range of nucleotidyltransferase mechanistic studies and enable the directed evolution/engineering of new catalysts [64].
Acknowledgments
This work was supported, in part, by National Institutes of Health Grants GM70637 and U19 CA113297 (JST). RM was a National Institutes of Health Molecular Bioscience Training Grant trainee (GM07215). JST is an H. I. Romnes fellow.
Abbreviations
- GlcN
Glucosamine
- dGlc
2-deoxyglucose
- GlcOMe
methyl-α-D-glucopyranoside
- NTP
nucleotide triphosphate
- NDP-sugar
nucleoside diphosphosugar
- NUSSA
nucleotidyltransferase substrate screening assay
- DNS
3,5-dinitrosalycilic acid
- MBTH
3-methyl-2-benothiazolinonehydrazone
- pHBH
para-hydroxybenzoic acid hydrazide
- BSA
bovine serum albumen
- Glc-1-P
α-D-glucose-1-phosphate
- UDP-Glc
uridine 5′diphospho-α-D-glucose
- ESI-MS
electrospray ionization mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- 3.Trefzer A, Salas JA, Bechthold A. Genes and enzymes involved in deoxysugar biosynthesis in bacteria. Nat Prod Rep. 1999;16:283–299. doi: 10.1039/a804431g. [DOI] [PubMed] [Google Scholar]
- 4.Bulter T, Elling L. Enzymatic synthesis of nucleotide sugars. Glycoconj J. 1999;16:147–159. doi: 10.1023/a:1026444726698. [DOI] [PubMed] [Google Scholar]
- 5.Freeze HH, Aebi M. Altered glycan structures: the molecular basis of congenital disorders of glycosylation. Curr Opin Struct Biol. 2005;15:490–498. doi: 10.1016/j.sbi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Blanchard S, Thorson JS. Enzymatic tools for engineering natural product glycosylation. Curr Opin Chem Biol. 2006;10:263–271. doi: 10.1016/j.cbpa.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Thorson JS, Hosted TJ, Jiang JQ, Biggins JB, Ahlert J. Nature’s carbohydrate chemists: The enzymatic glycosylation of bioactive bacterial metabolites. Current Organic Chemistry. 2001;5:139–167. [Google Scholar]
- 8.Thibodeaux CJ, Melancon CE, Liu HW. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 9.Kren V, Martinkova L. Glycosides in medicine: “The role of glycosidic residue in biological activity”. Curr Med Chem. 2001;8:1303–1328. doi: 10.2174/0929867013372193. [DOI] [PubMed] [Google Scholar]
- 10.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blankenfeldt W, Asuncion M, Lam JS, Naismith JH. The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA) Embo J. 2000;19:6652–6663. doi: 10.1093/emboj/19.24.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soltero-Higgin M, Carlson EE, Phillips JH, Kiessling LL. Identification of inhibitors for UDP-galactopyranose mutase. J Am Chem Soc. 2004;126:10532–10533. doi: 10.1021/ja048017v. [DOI] [PubMed] [Google Scholar]
- 13.Desvergnes S, Desvergnes V, Martin OR, Itoh K, Liu HW, Py S. Stereoselective synthesis of beta-1-C-substituted 1,4-dideoxy-1,4-imino-D-galactitols and evaluation as UDP-galactopyranose mutase inhibitors. Bioorg Med Chem. 2007;15:6443–6449. doi: 10.1016/j.bmc.2007.06.059. [DOI] [PubMed] [Google Scholar]
- 14.Griffith BR, Langenhan JM, Thorson JS. ‘Sweetening’ natural products via glycorandomization. Curr Opin Biotechnol. 2005;16:622–630. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Elhalabi JM, Rice KG. Synthesis and applications for unnatural sugar nucleotides. Curr Med Chem. 1999;6:93–116. [PubMed] [Google Scholar]
- 16.Rupprath C, Schumacher T, Elling L. Nucleotide deoxysugars: essential tools for the glycosylation engineering of novel bioactive compounds. Curr Med Chem. 2005;12:1637–1675. doi: 10.2174/0929867054367167. [DOI] [PubMed] [Google Scholar]
- 17.Song J, Zhang HC, Li L, Bi ZS, Chen M, Wang W, Yao QJ, Guo HJ, Tian M, Li HF, Yi W, Wang PG. Enzymatic biosynthesis of oligosaccharides and glycoconjugates. Curr Org Synth. 2006;3:159–168. [Google Scholar]
- 18.Weigel PH, Deangelis PL. Hyaluronan Synthases: A Decade-plus of Novel Glycosyltransferases. J Biol Chem. 2007;282:36777–36781. doi: 10.1074/jbc.R700036200. [DOI] [PubMed] [Google Scholar]
- 19.Ichikawa Y, Look GC, Wong CH. Enzyme-catalyzed oligosaccharide synthesis. Anal Biochem. 1992;202:215–238. doi: 10.1016/0003-2697(92)90099-s. [DOI] [PubMed] [Google Scholar]
- 20.Goulard F, Diouris M, Deslandes E, Floc’h JY. An HPLC method for the assay of UDP-glucose pyrophosphorylase and UDP-glucose-4-epimerase in Solieria chordalis (Rhodophyceae) Phytochem Anal. 2001;12:363–365. doi: 10.1002/pca.604. [DOI] [PubMed] [Google Scholar]
- 21.Yep A, Bejar CM, Ballicora MA, Dubay JR, Iglesias AA, Preiss J. An assay for adenosine 5′-diphosphate (ADP)-glucose pyrophosphorylase that measures the synthesis of radioactive ADP-glucose with glycogen synthase. Anal Biochem. 2004;324:52–59. doi: 10.1016/j.ab.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 22.Salvucci ME, Crafts-Brandner SJ. A high-performance liquid chromatography-based radiometric assay for sucrose-phosphate synthase and other UDP-glucose requiring enzymes. Anal Biochem. 1991;194:365–368. doi: 10.1016/0003-2697(91)90242-l. [DOI] [PubMed] [Google Scholar]
- 23.Evers CA, Palatnik CM. A simplified radioactive assay for the enzyme UDP-glucose pyrophosphorylase. Anal Biochem. 1981;118:108–112. doi: 10.1016/0003-2697(81)90164-0. [DOI] [PubMed] [Google Scholar]
- 24.Hames BD. An improved radiochemical assay for uridine diphosphoglucose pyrophosphorylase. Anal Biochem. 1976;73:215–219. doi: 10.1016/0003-2697(76)90157-3. [DOI] [PubMed] [Google Scholar]
- 25.Duggleby RG, Peng HL, Chang HY. An improved assay for UDPglucose pyrophosphorylase and other enzymes that have nucleotide products. Experientia. 1996;52:568–572. doi: 10.1007/BF01969730. [DOI] [PubMed] [Google Scholar]
- 26.Ritter JE, Berlin C, Elling L. A continuous microtiter plate assay for screening nucleotide sugar-synthesizing nucleotidyltransferases. Anal Biochem. 1996;234:74–82. doi: 10.1006/abio.1996.0052. [DOI] [PubMed] [Google Scholar]
- 27.Upson RH, Haugland RP, Malekzadeh MN. A spectrophotometric method to measure enzymatic activity in reactions that generate inorganic pyrophosphate. Anal Biochem. 1996;243:41–45. doi: 10.1006/abio.1996.0479. [DOI] [PubMed] [Google Scholar]
- 28.Cogan EB, Birrell GB, Griffith OH. A robotics-based automated assay for inorganic and organic phosphates. Anal Biochem. 1999;271:29–35. doi: 10.1006/abio.1999.4100. [DOI] [PubMed] [Google Scholar]
- 29.Fusari C, Demonte AM, Figueroa CM, Aleanzi M, Iglesias AA. A colorimetric method for the assay of ADP-glucose pyrophosphorylase. Anal Biochem. 2006;352:145–147. doi: 10.1016/j.ab.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 30.Mok MT, Edwards MR. Critical sources of error in colorimetric assay for UDP-N-acetylglucosamine pyrophosphorylase. Anal Biochem. 2005;343:341–343. doi: 10.1016/j.ab.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 31.Zea CJ, Pohl NL. General assay for sugar nucleotidyltransferases using electrospray ionization mass spectrometry. Anal Biochem. 2004;328:196–202. doi: 10.1016/j.ab.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, Fu X, Jia Q, Shen J, Biggins JB, Jiang J, Zhao J, Schmidt JJ, Wang PG, Thorson JS. Studies on the substrate specificity of Escherichia coli galactokinase. Org Lett. 2003;5:2223–2226. doi: 10.1021/ol034642d. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmeister D, Yang J, Liu L, Thorson JS. Creation of the first anomeric D/L-sugar kinase by means of directed evolution. Proc Natl Acad Sci U S A. 2003;100:13184–13189. doi: 10.1073/pnas.2235011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Liu L, Thorson JS. Structure-based enhancement of the first anomeric glucokinase. Chembiochem. 2004;5:992–996. doi: 10.1002/cbic.200400041. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmeister D, Thorson JS. Mechanistic implications of Escherichia coli galactokinase structure-based engineering. Chembiochem. 2004;5:989–992. doi: 10.1002/cbic.200400003. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Fu X, Liao J, Liu L, Thorson JS. Structure-based engineering of E. coli galactokinase as a first step toward in vivo glycorandomization. Chem Biol. 2005;12:657–664. doi: 10.1016/j.chembiol.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Moretti R, Thorson JS. Enhancing the latent nucleotide triphosphate flexibility of the glucose-1-phosphate thymidylyltransferase RmlA. J Biol Chem. 2007;282:16942–16947. doi: 10.1074/jbc.M701951200. [DOI] [PubMed] [Google Scholar]
- 38.Miller GL. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 39.Jue CK, Lipke PN. Determination of reducing sugars in the nanomole range with tetrazolium blue. J Biochem Biophys Methods. 1985;11:109–115. doi: 10.1016/0165-022x(85)90046-6. [DOI] [PubMed] [Google Scholar]
- 40.Porro M, Viti S, Antoni G, Neri P. Modifications of the Park-Johnson ferricyanide submicromethod for the assay of reducing groups in carbohydrates. Anal Biochem. 1981;118:301–306. doi: 10.1016/0003-2697(81)90586-8. [DOI] [PubMed] [Google Scholar]
- 41.Honda S, Nishimura Y, Chiba H, Kakehi K. Determination of Carbohydrates by Condensation with 3-Methyl-2-Benzothiazolinonehydrazone. Analytica Chimica Acta. 1981;131:293–296. [Google Scholar]
- 42.Anthon GE, Barrett DM. Determination of reducing sugars with 3-methyl-2-benzothiazolinonehydrazone. Anal Biochem. 2002;305:287–289. doi: 10.1006/abio.2002.5644. [DOI] [PubMed] [Google Scholar]
- 43.Lever M. A new reaction for colorimetric determination of carbohydrates. Anal Biochem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- 44.Barton WA, Lesniak J, Biggins JB, Jeffrey PD, Jiang J, Rajashankar KR, Thorson JS, Nikolov DB. Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization. Nat Struct Biol. 2001;8:545–551. doi: 10.1038/88618. [DOI] [PubMed] [Google Scholar]
- 45.Barry VC, Mitchell PW. Mechanism of osazone formation. Nature. 1955;175:220. doi: 10.1038/175220a0. [DOI] [PubMed] [Google Scholar]
- 46.Capon B. Mechanism in Carbohydrate Chemistry. Chemical Reviews. 1969;69:407. [Google Scholar]
- 47.O’Reilly JE. Oxidation-reduction potential of the ferro-ferricyanide system in buffer solutions. Biochim Biophys Acta. 1973;292:509–515. doi: 10.1016/0005-2728(73)90001-7. [DOI] [PubMed] [Google Scholar]
- 48.Rutenburg AM, Gofstein R, Seligman AM. Preparation of a new tetrazolium salt which yields a blue pigment on reduction and its use in the demonstration of enzymes in normal and neoplastic tissues. Cancer Res. 1950;10:113–121. [PubMed] [Google Scholar]
- 49.Kuhn R, Jerchel D. UeberInvertseifen. VIII Mitteil: Reduktion von Tetrazoliumsalzen durch Bakterien, gaerende Hefe, und keimende Samen. Ber Deut Chem Ges. 1941;74B:949–952. [Google Scholar]
- 50.Blackburn RS, Harvey A. Green chemistry methods in sulfur dyeing: application of various reducing D-sugars and analysis of the importance of optimum redox potential. Environ Sci Technol. 2004;38:4034–4039. doi: 10.1021/es0498484. [DOI] [PubMed] [Google Scholar]
- 51.Yao SJ, Appleby AJ, Geisel A, Cash HR, Wolfson SK., Jr Anodic oxidation of carbohydrates and their derivatives in neutral saline solution. Nature. 1969;224:921–922. doi: 10.1038/224921a0. [DOI] [PubMed] [Google Scholar]
- 52.Fox JD, Robyt JF. Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal Biochem. 1991;195:93–96. doi: 10.1016/0003-2697(91)90300-i. [DOI] [PubMed] [Google Scholar]
- 53.Waffenschmidt S, Jaenicke L. Assay of reducing sugars in the nanomole range with 2,2′-bicinchoninate. Anal Biochem. 1987;165:337–340. doi: 10.1016/0003-2697(87)90278-8. [DOI] [PubMed] [Google Scholar]
- 54.Green F, 3rd, Clausen CA, Highley TL. Adaptation of the Nelson-Somogyi reducing-sugar assay to a microassay using microtiter plates. Anal Biochem. 1989;182:197–199. doi: 10.1016/0003-2697(89)90578-2. [DOI] [PubMed] [Google Scholar]
- 55.Somogyi M. Notes On Sugar Determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- 56.Koziol MJ. An Evaluation of the Alkaline Para-Hydroxybenzoic Acid Hydrazide Procedure for the Determination of Reducing Sugars. Analytica Chimica Acta. 1981;128:195–205. [Google Scholar]
- 57.Morrison IM, Smith EH. Observations on the 4-Hydroxybenzoylhydrazide Methods for the Determination of Carbohydrates. Analyst. 1988;113:841–842. [Google Scholar]
- 58.Lever M, Walmsley TA, Visser RS, Ryde SJ. Optimal conditions for 4-hydroxybenzoyl-and 2-furoylhydrazine as reagents for the determination of carbohydrates, including ketosamines. Anal Biochem. 1984;139:205–211. doi: 10.1016/0003-2697(84)90406-8. [DOI] [PubMed] [Google Scholar]
- 59.Lever M. Colorimetric and fluorometric carbohydrate determination with p-hydroxybenzoic acid hydrazide. Biochem Med. 1973;7:274–281. doi: 10.1016/0006-2944(73)90083-5. [DOI] [PubMed] [Google Scholar]
- 60.Powell JC, Lever M. A new automated procedure for the colorimetric determination of glucose. Biochem Med. 1972;6:543–547. doi: 10.1016/0006-2944(72)90008-7. [DOI] [PubMed] [Google Scholar]
- 61.Lever M, Powell JC, Killip M, Small CW. A comparison of 4-hydroxybenzoic acid hydrazide (PAHBAH) with other reagents for the determination of glucose. J Lab Clin Med. 1973;82:649–655. [PubMed] [Google Scholar]
- 62.Lever M. Carbohydrate determination with 4-hydroxybenzoic acid hydrazide (PAHBAH): effect of bismuth on the reaction. Anal Biochem. 1977;81:21–27. doi: 10.1016/0003-2697(77)90594-2. [DOI] [PubMed] [Google Scholar]
- 63.Smith BD, Soellner MB, Raines RT. Potent inhibition of ribonuclease A by oligo(vinylsulfonic acid) J Biol Chem. 2003;278:20934–20938. doi: 10.1074/jbc.M301852200. [DOI] [PubMed] [Google Scholar]
- 64.Thorson JS, Barton WA, Hoffmeister D, Albermann C, Nikolov DB. Structure-based enzyme engineering and its impact on in vitro glycorandomization. Chembiochem. 2004;5:16–25. doi: 10.1002/cbic.200300620. [DOI] [PubMed] [Google Scholar]