Summary
Pluripotent human embryonic stem (hES) cells can differentiate into various cell types derived from the three embryonic germ layers and extra-embryonic tissues such as trophoblasts. The mechanisms governing lineage choices of hES cells are largely unknown. Here, we report that we established two independent hES cell clones lacking a group of cell surface molecules, glycosyl-phosphatidyl-inositol anchored proteins (GPI-APs). The GPI-AP deficiency in these two hES clones is due to the deficiency in the gene expression of PIG-A (phosphatidyl-inositol-glycan class A), which is required for the first step of GPI synthesis. GPI-AP deficient hES cells were capable of forming embryoid bodies and initiating cell differentiation into the three embryonic germ layers. However, GPI-AP deficient hES cells failed to form trophoblasts after differentiation induction by embryoid body formation or by adding exogenous BMP4. The defect in trophoblast formation was due to the lack of GPI-anchored BMP co-receptors, resulting in the impairment of full BMP4 signaling activation in the GPI-AP deficient hES cells. These data reveal for the first time that GPI-AP enhanced full activation of BMP signaling is required for human trophoblast formation.
Keywords: human embryonic stem cells, ES cells, GPI, PIG-A, trophoblast, BMP signaling, PNH
Introduction
PIG-A is required for the first step in GPI anchor biosynthesis (Kinoshita et al., 1997). The human (X chromosome-linked) PIG-A gene is found mutated in hematopoietic stem cells (HSCs) of patients with Paroxysmal Nocturnal Hemoglobinuria (PNH), a clonal disorder of the blood system that causes intravascular hemolysis, venous thrombosis and bone marrow failure (Takeda et al., 1993; Luzzatto et al., 1997; Kinoshita et al., 1997; Dunn et al., 1999). Inactivation of PIG-A in HSCs results in the lack of all GPI-APs including two complement inhibitors CD55 and CD59; the lack of these two cell surface proteins explains the complement-mediated intravascular hemolysis associated with PNH. However, other clinical features of PNH, such as clonal expansion and the associated bone marrow failure, remain poorly understood (Kinoshita et al., 1996; Luzzatto et al., 1997; Dunn et al., 1999). Members of dozens of GPI-APs function as co-receptors, co-ligands, ecto-enzymes and cell adhesion molecules (Kinoshita et al., 1997; Minchiotti et al., 2000; Chesebro et al., 2005). The importance of the GPI anchor moiety in linking the protein to the cell membrane has been demonstrated for several GPI-APs (Minchiotti et al., 2000; Chesebro et al., 2005).
To establish a prospective experimental system for PNH, a somatic disease, mouse models have been established by disrupting the Pig-a gene in mouse ES (mES) cells (Dunn et al., 1996; Rosti et al., 1997; Keller et al., 2001). Although the Pig-a gene (also X-linked) is dispensable for the growth of undifferentiated mES cells in culture, the inactivation of the mouse Pig-a gene is embryonic lethal (Rosti et al., 1997; Keller et al., 2001). Conditional Pig-a null mice lacking GPI-APs in all the lineages of blood and immune cells were later achieved (Keller et al., 2001). However, these mice have a normal life span and do not recapitulate the PNH symptoms seen in human patients. Because of the current limited ability to expand human HSCs in culture that are required for selecting and expanding rare clones after stable genetic modification, it has been impossible to make a PIG-A null mutation by knocking out or down the PIG-A gene in normal human HSCs.
Our initial goal of this project was to make PIG-A deficient hES cells that can be subsequently induced to differentiate into hematopoietic cells (Kaufman et al. 2001; Zhan et al., 2004; Lensch et al., 2006), which may serve as a novel genetic model for PNH. After trials with several methods, we established two independent clones of hES cells lacking the expression of the PIG-A gene and GPI-APs on hES cell surface. Although full characterizations of these GPI-AP deficient hES cells such as differentiation to hematopoietic and other somatic lineages are still in progress, our data reveal an unexpected but critical role of GPI-APs in potentiating cellular signaling by bone morphogenetic protein 4 (BMP4) and trophoblast development of hES cells.
Results
Establishment of clonal hES cells lacking GPI-APs
Consistent with previous studies, we found that several GPI-APs such as alkaline phosphatase (APase), CD90/Thy1 and Cripto are preferentially expressed on cell surface of undifferentiated hES cells (Fig 1). The mRNA expression profile of known GPI-AP genes in undifferentiated and differentiated hES cells is provided in Table S1. We have attempted several approaches to knock out or down the X chromosome-linked PIG-A gene in XY hES cell line such as H1. The most successful approach to date was to use pro-aerolysin for counter selection of cells lacking GPI-APs. Pro-aerolysin is a bacterial toxin that uses GPI-APs as a cellular receptor. It is converted by cell surface proteases to aerolysin that potently kills mammalian cells normally expressing various GPI-APs (Brodsky et al., 1999; Hu et al., 2005). Cells lacking GPI-APs such as PIG-A null mutants escape aerolysin-mediated cell killing. We used the H1 hES cell population that had been transduced by a GFP-expressing lentiviral vector (Dravid et al., 2005; Zhou et al., 2007). After serial aerolysin selections of these GFP-expressing hES cells (called G-GFP thereafter), aerolysin resistant (AR) hES cells lacking GPI-APs were obtained from two independent batch selections. By limiting dilution, 6 independent AR hES cell clones were established (Table S2). Clones that lack GPI-APs but can be restored by PIG-A transgene expression were further expanded. One clone from each batch selection, AR1-clone 1 (AR1-c1) or AR2-c1, retained undifferentiated morphology after serial expansion and was characterized (Fig 1).
Fig 1. Analyses of two hES cell clones lacking GPI-anchored proteins (GPI-APs).
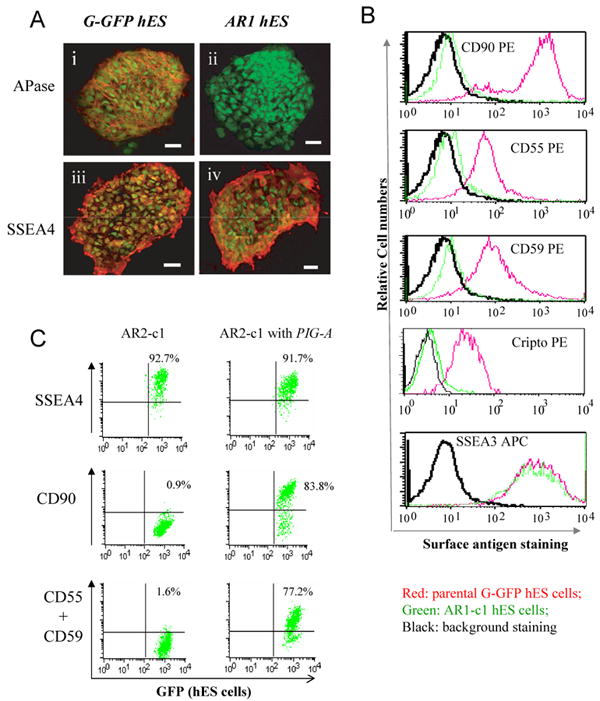
After aerolysin selection, G-GFP derived hES cells that lacked GPI-APs were sorted and expanded. 6 clones were obtained from two large-scale experiments. Two (AR1-c1 and AR2-c1) were further characterized since they can form undifferentiated colonies (Supplement Table S2). (A). Cell surface staining for alkaline phosphatase (APase). The AR1-c1 or its parental G-GFP hES cells (both constitutively expressing GFP) were cultured under a feeder-free condition and fixed before cell surface staining. Mouse monoclonal IgG recognizing either the APase (i and ii) or SSEA-4 (iii and iv) was used as primary antibodies. Then Alexa 555-conjugated antibodies recognizing anti-mouse IgG were used to light up detected antigens. The micrographic images of stained were superimposed with that of cellular GFP signals. While both APase and SSEA4 were detected on cell surface of G-GFP cells (i and iii), only SSEA-4 (iv) but not the APase (ii) was detected on AR1-c1 cells. (B). Flow cytometric analysis of other GPI-APs such as CD90, CD55, CD59 and Cripto using PE-conjugated specific IgGs. The red lines represent staining profiles of the G-GFP control whereas the green lines represent the AR1-c1 hES cells. The black lines represent the background staining (using an irrelevant antibodies). The both hES cell types expressed a high level of surface marker SSEA3. (C). Similar analysis of the AR2-c1 hES cell clone before or after transduction by a lentiviral vector expressing a PIG-A transgene. AR2-c1 cells express SSEA4, but lack CD90, CD55 or CD59. After one round of transduction, ∼80% AR2-c1 cells stably expressed GPI-APs.
We found that both AR clones completely lack GPI-APs such as APase, CD90, CD55, CD59 and Cripto on cell surface (Fig 1), even though the protein portion was detected inside hES cells (Supplement Fig S1). Both AR hES clones can maintain undifferentiated morphology and express undifferentiated cell-surface markers such as SSEA3 and SSEA4 that are not GPI-APs (Fig 1). Both AR clones have a similar proliferation rate as compared to the parental G-GFP hES cells when cultured on feeder cells or under feeder-free conditions (data not shown). GPI-APs such as CD90, CD55 or CD59 can be restored by transduction of a lentiviral vector expressing the PIG-A coding sequence both in AR2-c1 (Fig 1C) and AR1-c1 cells (Zhou et al., 2007), indicating that the GPI-AP deficiency is due to the absence of PIG-A.
Once sufficient hES cells were obtained by expansion, both clones were confirmed to have a normal karyotype (Supplement Fig S2). The AR1-c1 clone is capable of maintaining a normal karyotype over two years although karyotypically abnormal cells occasionally arise. The AR2-c1 clone gained an extra Chromosome 12 in a small fraction (2/20) initially soon after clonal derivation. As happens often with wildtype hES cells after prolonged culture (Draper et al., 2004), mutated AR2-c1 cells that gain selective growth advantage soon took over the whole population and acquired additional mutations. Early-passage AR2-c1 cells with or without an extra chromosome 12 gave essentially the same results (see below). In this report, we present data obtained from karyotypically normal hES cells, mainly AR1-c1 hES cells, and G-GFP (H1) and H9 hES cells as controls.
The GPI-AP deficient hES cell clones lack PIG-A mRNA
We analyzed directly the PIG-A expression in normal and GPI-AP deficient undifferentiated hES cells cultured under a feeder-free condition with or without subsequent differentiation induction. As compared to somatic cell lines and differentiated (teratoma-derived) cells, undifferentiated normal (G-GFP) hES cells express a low level of PIG-A mRNA, as assayed by both conventional and quantitative RT-PCR analyses (Fig 2). Notably, PIG-A mRNA in AR1-c1 or AR2-c1 hES clone showed much a lower level than that in G-GFP hES cells (Fig 2A-B), consistent with the fact that the AR cells lack GPI-APs on cell surface. PIG-A mRNA levels in differentiated progeny after BMP4 induction or embryoid body (EB) formation was elevated significantly in the parental G-GFP cells, but remained low in AR1-c1 derivatives (Fig 2). Quantitative RT-PCR analysis using a different primer set revealed that the PIG-A mRNA level in the differentiated AR1-c1 cells is 450- and 52-fold lower than that in the parental G-GFP hES cells, after BMP4-induced differentiation or EB formation (Fig 2C). As expected, the AR1-c1 hES cells transduced with a PIG-A expressing vector expressed PIG-A mRNA at a very high level, before or after differentiation (Fig 2A-C). Northern blot further confirmed the RT-PCR results: PIG-A mRNA of any size was not detected in AR1-c1 cells before the transgene restoration, whereas G-GFP cells expressed the wildtype (∼3.6 kb) PIG-A transcript (Fig 2D). This is consistent with the fact that we did not detect any genetic mutation in all 6 exons and intron/exon junctions of the PIG-A gene in either AR hES clone after extensive sequencing (data not shown). The exact mechanisms of the observed PIG-A deficiency in the AR hES clones remain to be fully determined; nonetheless, we have obtained two PIG-A null hES clones lacking GPI-APs, permitting us to examine the roles of GPI-APs using hES cell-initiated developmental models.
Fig 2. Analysis of PIG-A gene expression in human ES cells lacking GPI-APs.

(A). Conventional RT-PCR of the PIG-A gene expression in undifferentiated ES cells (undiff. ES) and differentiated hES cells after BMP4 induction or embryoid body formation (EB) after 10 days. G-GFP hES cells (samples 1) and AR1-c1 (samples 2) clonal hES cells were analyzed side by side. In addition, we analyzed reconstituted AR1-c1 hES cells after transduction of PIG-A transgene that restored GPI-AP expression (samples 3). (B). Similar analysis of AR2-c1 hES cells (sample 4) and the AR2-c1 cells restored by the PIG-A transgene expression (sample 5). (c) Quantitative RT-PCR analysis of the PIG-A gene in AR1-c1 hES cells before and after differentiation. The relative level of PIG-A mRNA is first normalized by that of beta-actin, and then by the level in teratoma (defined as 100), which was used as a common positive control in quantitative RT-PCR analyses. The mean and SD (n=4) were plotted in a log scale. The PIG-A mRNA in undifferentiated G-GFP cells is found at a low level (1-2% of teratoma), but elevated significantly after differentiation by BMP4 induction or EB formation (A and C-D). Notably, the PIG-A mRNA level in undifferentiated AR1-c1 hES cells (sample 2) and AR2-c1 (sample 4) was much lower (∼ 10 fold) than the control G-GFP cells by both assays in (A-C). The deficiency of PIG-A mRNA in AR1-c1 cells is more obvious after differentiation (A, C). (D). Northern blot of PIG-A mRNA in AR1-c1 cells confirmed RT-PCR data that the PIG-A deficiency is due to the lack of PIG-A mRNA.
Two GPI-AP deficient hES clones formed morphologically normal EBs and hematopoietic cells in vitro
We next examined differentiation potentials of both AR1-c1 and AR2-c1 clones, in comparison with the parental G-GFP hES cells. Similarly to other normal hES cells, G-GFP hES cell aggregates cultured in suspension (with serum factors) developed into more structured (cystic) EBs. By day 8, enlarged EBs showed visible cavity. These cystic EBs are more evident by day 10-15 (Fig 3A). AR1-c1 and AR2-c1 both formed morphologically normal EBs at a rate similar to G-GFP hES cells (Fig 3A). Cells derived from EBs up to day 15 were further analyzed. The ubiquitously expressed GPI-APs, CD55 and CD59, remained absent in EB-derived cells from AR1-c1 and AR2-c1 clones (Fig 3B). This contrasts to previous studies that mES cells lacking PIG-A formed EBs poorly (Dunn et al., 1996).
Fig 3. Embryoid body (EB) formation and differentiation from the parental (G-GFP) and two independent PIG-A/GPI-AP deficient hES cell clones.
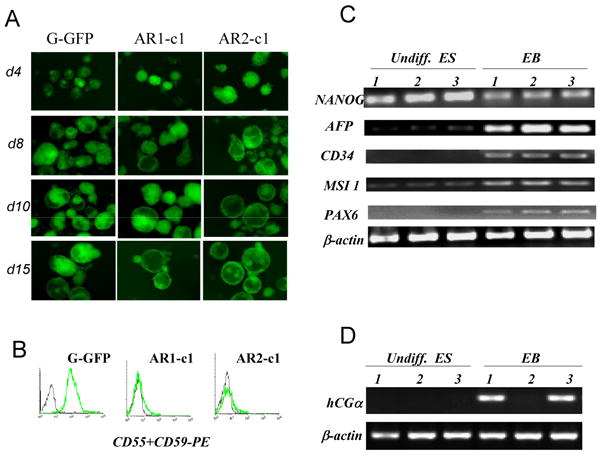
(A). Three ES cell types were cultured in suspension as aggregates to form EBs. The morphology and numbers of EBs were monitored daily for 15 days. The EB morphology is better illustrated by constitutively-expressed GFP signal. The rate as well as morphology of EBs by 3 hES cell types was similar. (B). Confirmation of the GPI-AP deficiency in EB-derived cells. Antibodies recognizing CD55 and CD59, were mixed and used to stain single cell suspension. (C). RT-PCR analysis of marker gene expression before and after EB formation at day 10. As expected, Nanog expression in G-GFP control cells (samples 1) is high in undifferentiated ES cells (undiff. ES) and visibly reduced after spontaneous differentiation by EB formation (EB). The gene expression of differentiated markers such as AFP (endoderm), CD34 (mesoderm), MSI 1 and PAX6 (ectoderm) were significantly elevated as compared to undifferentiated ES cells. AR1-c1 (samples 2) showed similar pattern to the G-GFP control, so did the AR1-c1 cells expressing PIG-A transgene (samples 3). (D). RT-PCR analysis of a marker gene expression for trophectoderm. hCGα expression from AR1-c1 hES cells (samples 2) failed to elevate after EB formation, but restored by PIG-A transgene expression. See more data by real-time quantitative RT-PCR in Supplement Fig S3.
To examine the differentiation commitment to the 3 embryonic germ layers, we analyzed the acquired expression of lineage specific markers in EB-derived cells. The expression of AFP (endoderm), PAX6 and Musashi (MSI) 1 (ectoderm), and CD34 (mesoderm) marker genes within the EBs from AR1-c1 hES cells were comparable or moderately higher than within the G-GFP EBs (Fig 3c). Similar results were obtained using quantitative RT-PCR with different primer sets and detection probes (Fig S3). Although interesting differences were observed between two groups (such as levels of CD34 and NANOG expression), it appears AR1-c1 cells are overall competent in differentiation commitment to the 3 embryonic germ layers at this early stage. We further analyzed hematopoietic differentiation of AR1-c1 and AR2-c1 hES cells after EB formation (Fig S4). The level of produced CD34+ cells (hematopoietic and/or endothelial) or colony-forming hematopoietic progenitor cells from AR1-c1 clone appears comparable to the G-GFP control or the isogenic control where the PIG-A expression is restored by a transgene (Fig S4). Our results indicate that GPI-APs in hES cells are dispensable for entry to the 3 embryonic germ layers or the hematopoietic lineage commitment. The effects of GPI-APs on the progression along ectoderm and endoderm lineages are under investigation.
Defect in trophoblast differentiation from GPI-AP deficient hES cells
Unlike mES cells, hES cells readily gave rise to trophoblasts after spontaneous or induced differentiation (Xu et al., 2002; Pera et al., 2004; Gerami-Naini et al., 2004). So we also examined the expression of trophoblast or trophectoderm (TE) markers from the same EBs (Fig 3). To our surprise, the expression of human chorionic gonadotropin alpha (hCGα), a TE marker, failed to elevate after EB formation from AR1-c1 hES cells (Fig 3D). Similar results were obtained with the expression of CDX2 (Fig S3), a marker of TE lineage in early embryos (Strumpf et al., 2005; Niwa et al. 2000; Niwa et al., 2005). The defect appears specifically due to the PIG-A and GPI-AP deficiency, because the failure to turn on hCGα and CDX2 expression in AR1-c1 cells can be fully rescued by the PIG-A transgene (Fig 3d; Fig S3).
To confirm the link between defects of GPI-APs and trophoblast formation, we also used a second method of trophoblast formation from hES cells. The addition of exogenous BMP4 or a related family member to monolayer culture of hES cells that are cultured otherwise as undifferentiated cells (with feeder-derived factors and bFGF) rapidly induced morphological changes (Xu et al., 2002). Consistent with the previous report, we found that G-GFP hES cells differentiated into cells resembling trophoblasts when exogenous BMP4 (10-50 ng/ml) were added for 5 days. Nearly all the cells flattened after 7-10 days and hCG hormone was readily detected as previously reported (Xu et al., 2002). Ten days after BMP4 induction of G-GFP hES cells, NANOG mRNA decreased and hCGα mRNA increased over 100 fold (Fig S3 and Fig 4A). However, hCGα gene expression in AR1-c1 hES cells failed to elevate after BMP4 treatment. Similar patterns were observed with other trophoblast markers such as hCGβ and CDX2 (Fig 4A). The deficiency of hCGα gene up-regulation was rescued by the PIG-A transgene (Fig 4B). We further confirmed the deficiency of trophoblast formation at the protein level: the β subunit of secreted hCG in the AR1-c1 group was absent (Fig 4C). The lack of trophoblast formation in AR1-c1 cells after BMP treatment was further confirmed by the lack of induced placenta–cadherin (CDH3) and TROMA-I that are normally accompanied with trophoblast differentiation (Niwa et al., 2006; Brulet et al., 1982), as seen with G-GFP parental hES cells (Fig 4D).
Fig 4. BMP4 induced trophoblast differentiation of normal G-GFP hES cells, but not AR1-c1 hES cells lacking GPI-APs.
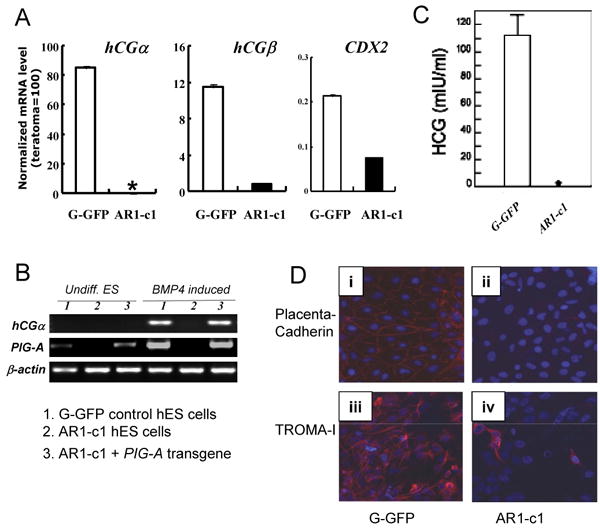
The parental (G-GFP) and AR1-c1 hES cells were cultured under a feeder-free condition and treated with BMP4 (50 ng/ml) for 10 days. Then cells were harvested for RT-PCR analyses (A and B) after the conditioned medium was collected and used to detect hCG hormone by ELISA (C). (A). Quantitative RT-PCR for trophoblast markers such as hCGα, hCGβ and CDX2. * undetectable as in undifferentiated hES cells after 40 cycle PCR. Profiles of the gene expression of other markers (such as NANOG, AFP, CD34 and PAX6) after BMP4 induction are shown in Supplement Fig S3. (C). Conventional RT-PCR using a different primer set for hCGα (as in Fig 3D) and a specific primer set for PIG-A as in Fig 2A. RNA from undifferentiated ES cells (undiff. ES) or after BMP4 induction (BMP4 induced) from either G-GFP (samples 1) or AR1-c1 (samples 2) hES cells was used. In addition, we also included AR1-c1 derivatives in which a PIG-A transgene is expressed (samples 3) in this experiment. (D). immuno-fluorescent staining for trophectoderm markers placenta-cadherin (upper row, i-ii) and TROMA-I (lower row, iii-iv) from the differentiated G-GFP (left) and AR1-c1 cells (right), 11 days after BMP4 induction. The presence of antigen is visualized by anti-mouse IgG conjugated with Alexa 595 (red) and nuclei were stained by DAPI (blue). While the trophectoderm marker induction is obvious in the differentiated G-GFP cells, it is much lower in the GPI-AP deficient AR1-c1 cell population.
BMP signaling is impaired in AR1-c1 hES cells lacking GPI-APs
We next investigated underlying mechanisms of defective trophoblast formation from GPI-AP deficient hES cells by employing the BMP4-mediated differentiation system. BMP4 functions through BMP receptor-mediated intracellular signaling by activating one or more members of SMAD family (SMAD1, 5 and 8) and other targets (Zhang and Li, 2005). Two BMP receptors, type I and II, have been identified. Type II BMP receptors are primarily involved in ligand binding and activating a BMP type I receptor that has intrinsic serine/threonine kinase activity (Zhang and Li, 2005). One of three type I BMP receptors, ALK2, ALK3 (BMPR-Ia) or ALK6 (BMPR-Ib), subsequently activates (phosphorylates) SMAD 1, 5 or 8 and other targets, and turns on downstream signaling cascades. In conjunction with other transcriptional factors, the BMP-activated SMAD complex modulates the expression of an array of genes including four members of Inhibitor of differentiation or Id genes (Hollnagel et al, 1999; Lopez-Rovira et al., 2002; Ying et al., 2003). We found that in hES cells the expression of human Id or ID genes (ID1 to ID4), especially that of ID1 and ID2, is indeed activated immediately by BMP4 but not by TGFβ (Fig S5). Our data extended the previous report with the ID1 gene (Xu et al., 2005). The direct activation of ID1 and ID2 promoters by BMP4 signaling was further confirmed in both H1 and H9 hES cell lines by a Luciferase (Luc) reporter assay (Fig S5).
We next examined directly BMP4 signaling activation in AR1-c1 hES cells using two assays. In the first assay, the Id1-Luc reporter was transfected into either AR1-c1 or G-GFP control hES cells followed by overnight BMP4 stimulation (Fig 5A). While the exogenous BMP4 stimulates the reporter activity up to ∼12 fold in G-GFP cells in a dose-dependent manner, the stimulation on GPI-AP defective AR1-c1 cells could reach only ∼3 fold (Fig 5A). Notably, the PIG-A transgene expression in AR1-c1 cells largely restored the BMP4-mediated activation in this assay as in BMP4-induced trophoblast differentiation (Fig 5A). In the second assay, we measured earlier events of BMP signaling activation by western blot. We used a specific antibody recognizing the activated (phosphorylated) form of 3 highly related SMAD proteins (SMAD 1/5/8) that are direct targets of activated BMP receptors type I (BMPR-I) after BMP4 stimulation. Consistent with previous reports, phosphorylated forms of SMAD 1/5/8 were low in undifferentiated hES cells before BMP4 induction (Fig 5B). BMP4 induction drastically increased the level of phosphorylated forms of SMAD 1/5/8 in G-GFP cells, but not in AR1-c1 cells. Based on the results obtained from the two independent assays, we conclude that BMP signaling is defective in AR1-c1 cells, likely at the level of BMPR-I activation.
Fig 5. BMP signaling activation is significantly reduced in AR1-c1 hES cells due to the PIG-A and GPI-AP deficiency.
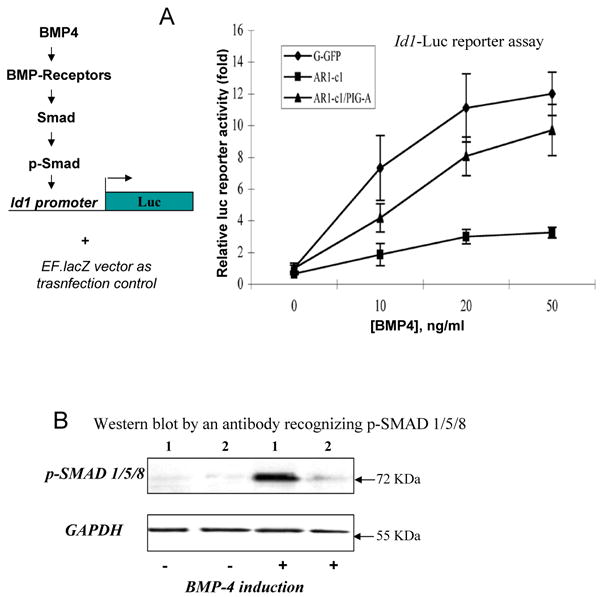
(A). BMP4 signaling activation in G-GFP or AR1-c1 hES cells was measured by a Luciferase reporter (Id1-Luc) controlled by the Id1 gene promoter. Six hours after transfection of the reporter plasmid, cells were stimulated with various concentration of BMP4 for 20 hours and harvested. The Id1-Luc activity was measured, normalized, and calculated relative to that of G-GFP without BMP4 stimulation (defined as 1). The normalized mean and SD (n≥5) were plotted. In one experiment, the Id1-Luc activity in AR1-c1 hES cells that have been transduced with PIG-A transgene was also measured before and after BMP induction (n=5). Combined from two experiments (Σn=7), the mean of the reconstituted AR1-c1/PIG-A cells is statistically greater than the AR1-c1 cells (p<0.05) and insignificant from G-GFP hES cells (p>0.05). (B). Western blot to detect the phosphorylated (activated) form of 3 highly related SMAD proteins (1/5/8) without or with BMP4 induction. G-GFP (1) or AR1-c1 (2) hES cells were treated with BMP4 for 6 hours, and cell lysates were analyzed by Western blotting, using purified specific antibodies recognizing the phosphorylated SMAD1/5/8. After stripping, the blot was re-probed by a specific antibody recognizing the housekeeping protein GAPDH.
We also examined the requirement of BMP signaling for trophoblast differentiation from a different hES cell line H9, using a known BMP antagonist NOGGIN. As previously reported (Xu et al., 2002), NOGGIN blocked BMP4-induced trophoblast differentiation from H9 as well as H1 (G-GFP) hES cells in monolayer cultures. We next tested if NOGGIN could also prevent preferentially trophoblast differentiation following EB formation (in the absence of exogenous BMP). Ten days after EB formation from H9 hES cells, undifferentiated markers such as NANOG reduced whereas lineage (differentiation) markers such as CDX2, hCGα and PAX6 increased (Fig S6). Increasing concentrations of NOGGIN resulted in a sharp reduction of CDX2 and hCGα up-regulation, but affected little on NANOG down-regulation. In fact, the upregulation of PAX6 (an ectoderm marker) was further enhanced with increased concentrations of NOGGIN. Mesoderm marker up-regulation following EB formation is less sensitive to NOGGIN blocking (Fig S6). Taken together, we show that in two distinct hES cell lines the dependency on BMP signaling for trophoblast formation is greater than some other lineages.
Over-expression of an activated BMPR-I gene in AR1- c1 hES cells restored BMP signaling and trophoblast differentiation
Using the unique AR1-c1 (H1) hES cells, we examined if the BMP signaling as well as trophoblast differentiation could be restored by the over-expression of an activated form of BMPR-I in the deficient hES cells. We used an activated form of ALK3 (also known as BMPR-Ia) gene which contains a mutation (Q233D), rendering enhanced activation without or with less BMP ligands (Chen et al., 1998). We transfected AR1-c1 and G-GFP cells with such ALK3 vector together with the Id1-Luc reporter, and treated cells with or without BMP4 after transfection (Fig 6A). Twenty hours after BMP4 induction following co-transfection with the control plasmid, the BMP4 signaling measured as the Id1-Luc activation was as expected: BMP4 (50 ng/ml) stimulated the reporter activity by ∼14 fold in G-GFP cells, but only ∼4 fold in AR1-c1 cells as in Fig 5a. Transfection of the activated ALK3 gene to AR1-c1 cells moderately stimulated the reporter in the absence of exogenous BMP4. When BMP4 was added, however, the level of BMP4 activation in AR1-c1 cells with ALK3 transfection is comparable to that of G-GFP cells without transfection (Fig 5a) or transfected by a control plasmid (Fig 6A), even with a transfection efficiency of 40-50%.
Fig 6. Transient transfection of an activated BMP-RI (ALK3) gene restored the BMP signaling and trophoblast formation.
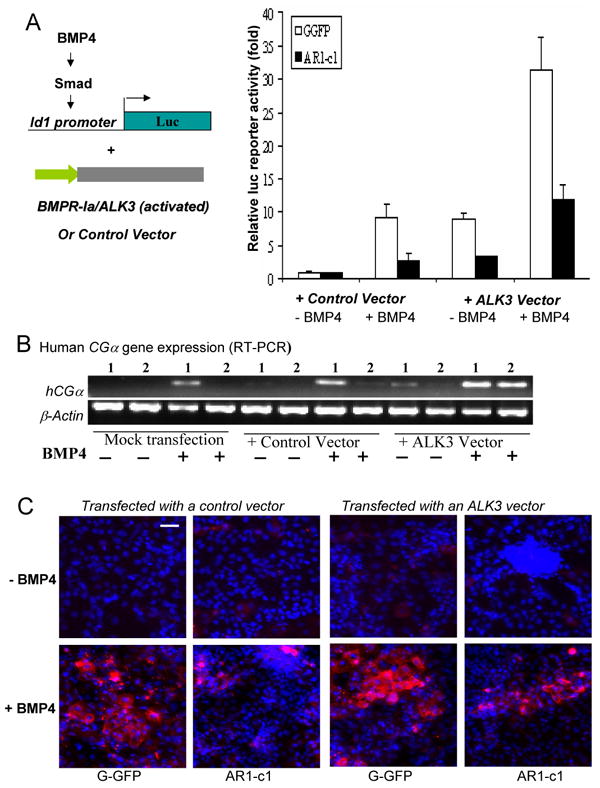
(A). G-GFP or AR1-c1 hES cells were transfected with an activated ALK3 gene or control vector, together with the Id1-luciferase reporter plasmid and the EF.lacZ as in Fig 5a. Six hours after transfection, cells were stimulated in the absence (-) or presence (+) of BMP4 (50 ng/ml) for 20 hours and harvested. The Id1-luciferase activity was measured, normalized by the lacZ activity, and calculated relative to the level without BMP4 stimulation (defined as 1) for either G-GFP or AR1-c1 hES cells. The normalized mean and SD values from 3 independent experiments were combined and plotted (Σn=8). The difference of the ALK3-transfected, BMP4 activated AR1-c1 cells vs. any of other 3 AR1-c1 groups is significant (p<0.05). (B). Human CGα gene expression in AR1-c1 cells after ALK3 transfection and BMP4 stimulation. G-GFP (samples 1) or AR1-c1 (samples 2) hES cells in pair were transfected with the activated ALK3 gene or control vector at day 0 for six hours, and treated in the absence or presence of BMP4 (50 ng/ml) for 2 days. Two days after, the same treatment was repeated. After a total of 5 days, various types of transfected cells were either harvested for RT-PCR (b) or fixed for immuno-staining (c). Human hCGα gene expression was measured by RT-PCR as in Fig 3d and Fig 4b. (C). TROMA-I staining of the double-transfected hES cells at day 5 as in (b). Scale bar: 50 μm.
To test whether the over-expression of the activated ALK3 transgene in AR1-c1 cells can also restore trophoblast differentiation, we monitored up-regulation of the hCGα gene expression (Fig 6B). After two rounds of transfection (at day 0 and 2) with the ALK3 or control vector, G-GFP or AR1-c1 cells treated with or without BMP4 were harvested at day 5. The levels of hCGα gene expression in the control vector-transfected cells were similar to the mock-transfected cells as expected: the hCGα gene expression was only turned on after BMP4 induction, and it was much weaker in AR1-c1 cells even after BMP4 induction. When AR1-c1 hES cells were transfected with the activated ALK3 gene, however, the BMP4-mediated hCGα gene up-regulation is restored (Fig 6B). The presence of acquired TROMA-I protein, a trophoblast marker, further supported that trophoblast formation in AR1-c1 hES cells can be restored by the activated ALK3 gene transfection (Fig 6C). Similar results were observed with an activated form (Q203D) of ALK6 also known as BMPR-Ib (Chen et al., 1998; data not shown). Based on BMPR-I reconstitution assays, we conclude that one or more GPI-APs are critical for full activation of BMP4 receptors and human trophoblast differentiation.
Over-expression of membrane-bound DRAGON in AR1- c1 hES cells also restored BMP signaling and trophoblast differentiation
We then focused on a group of GPI-APs that are recently identified as accessory co-receptors for BMP2 and BMP4 (Samad et al., 2005; Babbitt et al., 2005; Babbitt et al., 2006). This GPI-AP subfamily include 3 structurally related proteins called Repulsive Guidance Molecule a (RGMa), DRAGON (also called RGMb) and HFE2 (RGMc). It has been demonstrated that GPI-anchored (but not soluble) RGM proteins facilitate the binding of a BMP2 or BMP4 ligand to one or more forms of type II BMP receptors and potentiate the downstream BMP signaling (Samad et al., 2005; Babbitt et al., 2005; Babbitt et al., 2006; Xia et al, 2007).
By RT-PCR, we confirmed the gene microarray data that DRAGON and to a less extent RGMa gene is expressed in all the hES cells used (Table S1; Fig S7a). Similar to previous reports with other cell types, soluble forms of DRAGON or RGMa proteins purchased did not activate BMP4 signaling in hES cells (data not shown). Among several approaches we tested, we observed that transgene expression of a modified DRAGON cDNA in AR1-c1 hES cells made functional constitution as did by the PIG-A transgene (Fig 7). Because the PIG-A deficiency results in a post-translational defect for GPI-APs, transgene expression of the wildtype DRAGON cDNA (like the endogenous gene) could not produce a GPI anchored product on cell surface, even the precursor protein is made in cytoplasm. Therefore, we explored the reconstitution approach by making a cell-surface form of DRAGON using the coding sequence of the mature DRAGON domain fused to a transmembrane (TM) domain provided by an expression vector called pDisplay (Fig S7b). When overexpressed in 293T cells, the pDisplay vector encoding the fusion TM protein (DRAGON™) indeed directed the cell surface expression as measured by flow cytometry (Fig S7) or immuno-staining (not shown). When the same vector was used to transfect AR1-c1 hES cells as we did previously with the ALK3 vector (Fig 6a), the DRAGON™ vector reconstituted the BMP4-mediated Id1-Luc activation (Fig 7A). Similarly, serial (transient) transfection of DRAGON™ vector in AR1-c1 cells resulted in the up-regulation of trophoblast marker expression at day 5, such as hCGα by RT-PCR (Fig 7B) and TROMA-I by antibody staining (Fig 7C). Finally, we generated knockdown H9 hES cells that are stably transduced with a lentiviral vector expressing a small hairpin RNA targeting the DRAGON or RGMa gene (Fig S8). The H9 hES cells with reduced DRAGON or RGMa expression were impaired in BMP4-mediated Id1-Luc activation, hCGα up-regulation and acquired TROMA-I expression after BMP stimulation, a phenotype similar to AR1-c1 (H1) hES cells lacking all the GPI-APs.
Fig 7. Transient transfection of a transmembrane form of DRAGON restored the BMP signaling activation and trophoblast formation.
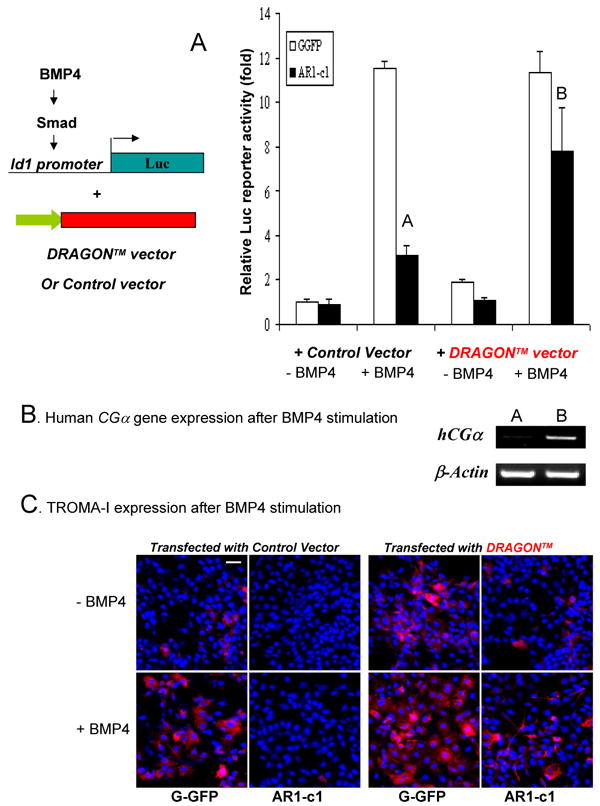
(A). G-GFP or AR1-c1 hES cells were transfected with the engineered DRAGON™ vector or the control pDisplay vector, together with the Id1-luciferase reporter plasmid and the EF.lacZ as in Fig 6a. Six hours after transfection, cells were stimulated in the absence (-) or presence (+) of BMP4 for 20 hours and harvested. The Id1-luc activity was measured, normalized by the lacZ activity, and calculated relative to the level without BMP4 stimulation (defined as 1) for each cell type. The normalized mean and SD values (n=4) were plotted. The difference between Samples A and B is significant (p<0.05). (B). Human CGα gene expression in AR1-c1 cells after DRAGON™ transfection and BMP4 stimulation. G-GFP (samples 1) or AR1-c1 (samples 2) hES cells in pair were transfected with the DRAGON™ or control vector at day 0 for six hours, and treated in the absence or presence of BMP4 (50 ng/ml) for 2 days. Two days after, the same treatment was repeated. After a total of 5 days, various types of transfected cells were either harvested for RT-PCR (B) or fixed for TROMA-I immuno-staining (C). Scale bar: 50 μm.
Discussion
Specific gene inactivation in hES cells may provide novel genetic and developmental models for human diseases. Previously two reports described the inactivation of the X-linked HPRT gene in a XY hES cell line as a potential model for Lesch-Nyhan disease (Zwaka and Thomson 2003; Urbach et al., 2004). Although these HPRT knockout hES cells indeed lacked the encoded enzyme, the biological or pathological consequences remain to be determined (Zwaka and Thomson 2003; Urbach et al., 2004). In contrast, the present study reports the establishment of hES cells lacking PIG-A gene expression, resulting in clonal hES cells deficient of a group of cell surface (GPI-anchored) proteins including APase, Cripto and CD90. Our data demonstrate that GPI-APs are not essential for self-renewal of hES cells in culture, although they are preferentially expressed in undifferentiated hES cells.
PIG-A and GPI-AP deficient hES cells also appeared normal in the initial differentiation into derivatives of the three embryonic germ layers. Among mesoderm-derived lineages we examined, the production of CD34+ hematopoietic precursor cells in vitro from GPI-AP deficient hES cells also appeared normal, thus allowing us to next examine the effect of PIG-A and GPI-AP deficiency on downstream steps of hematopoiesis. In addition, further neural cell development may require multiple GPI-APs based on previous mouse studies. The availability of hES cells lacking PIG-A and GPI-APs, and their derivatives expressing an inducible PIG-A transgene, may help us to study roles of various GPI-APs in the development of human cell types other than trophoblasts as demonstrated in this study.
We unexpectedly found that PIG-A/GPI-AP deficient hES cells failed to differentiate into trophoblasts. One of the prominent innovations in early mammalian embryogenesis is the formation of the trophoblast, the specialized tissue that subsequently forms trophic interface between the embryo and the mother. The first overt differentiation event in mammalian embryos is the formation of TE (the outer epithelial layer of the blastocyst), which goes on to develop into placenta. The cells isolated from TE, including trophoblast stem (TS) cell lines, can only commit to the cell fate of trophoblastic tissues. In contrast, the inner cell mass in the mouse blastocyst maintains pluripotency, and later develops into epiblasts and then the three embryonic germ layers and other extra-embryonic tissues. Numerous studies showed that mES cells, derived from the inner cell mass of mouse blastocysts, rarely form trophoblasts naturally. A typical exception is when the level of master transcriptional factors is altered by genetic manipulation, e.g. by enforced reduction of Oct4 expression or Cdx2 over-expression (Niwa et al., 2000; Niwa et al., 2006; Zhang et al., 2006). It was recently reported, however, that some mES cell lines, but not others, can differentiate into trophoblasts (or TS cells) at a low level together with many mesodermal cells when cultured on collagen IV (Schenke-Layland et al., 2007). It remains to be determined whether hES cells can also be induced to trophoblasts or TS cells by culturing on collagen IV or under other conditions, in BMP-dependent or independent manners.
It is now clear that most, if not all, hES cell lines readily form trophoblasts and other extra-embryonic lineages after differentiation induction. With the BMP4 addition into a feeder-free culture condition for ≥ 5 days, undifferentiated hES cells cultured as a monolayer differentiate preferentially into trophoblasts (Xu et al., 2002). This unique feature of hES cells helped us to discover that one or more GPI-APs are required for trophoblast differentiation. Mechanistically, our data reveal for the first time that GPI-AP mediated high-level BMP activation is required for human trophoblast formation. It is of interest to determine whether this mechanism also holds true in other placenta-bearing mammals such as mice. It is possible that species difference may exist between humans and mice in this regard as seen between hES and mES cells (Ginis et al., 2004). For example, BMP signaling mediated by Alk3 (Bmpr-Ia) is required for derivation of mES cells (Qi et al., 2004). BMP4 also cooperate with LIF to maintain the proliferation of undifferentiated mES cells (Ying et al., 2003; Qi et al., 2004). However, adding BMP4 stimulates differentiation of hES cells under culture conditions that otherwise promote self-renewal of hES cells as shown here and in previous studies (Xu et al, 2002; Dravid et al., 2006; Xu et al., 2005). Blocking endogenous BMP (and those made by feeder cells) by soluble BMP antagonists enhanced the self-renewal of hES cells (Pera et al., 2004; Xu et al., 2005). Thus, it appears that in undifferentiated hES cells BMP activation is kept at an inactive or low level. This is in a sharp contrast to mES cells where BMP signaling is required for their self-renewal (Ying et al., 2004; Qi, et al., 2004). However, hES cells can respond to BMP stimulation acutely and differentiate.
It has been known that some cell-surface proteins (also called as type III receptors) are important to the signaling by some members of the TGFβ/Activin/Nodal/ BMP ligand superfamily, positively regulating ligand activities (Zhang and Li, 2005; Mishra et al., 2005). For example, cell-surface protein β-glycan or CD105 (endoglin) functions as a co-receptor for TGFβ. Cripto, another GPI-AP, functions as a co-receptor for Nodal that utilizes activin receptors (Minchiotti et al., 2000). Very recently, DRAGON and related proteins RGM were identified as co-receptors for BMP2 and BMP4 (Samad et al., 2005; Babbitt et al., 2005; Babbitt et al., 2006). Notably, DRAGON and other RGM members such as RGMa and RGMc (HFE2) are GPI-APs. One possibility is that a membrane-anchored RGM protein, although not absolutely essential for BMP signaling, may help to sensitize all or some forms of type II BMP receptors (BMPRII, actRIIA or ActRIIB) to the binding of a BMP ligand: thus enhancing or extending downstream BMP (type I receptor) signaling (Xia et al, 2007). Among these 3 RGMs, DRAGON/RGMb is expressed in mouse E2.5 pre-implantation embryos and in later stages of embryos from E7 to E17 (Samad et al., 2005). DRAGON is also found expressed in human reproductive axis (Xia et al., 2005). We found that DRAGON and to a less extent RGMa genes are expressed in hES cells, and postulated that the defect of GPI-AP deficient (AR1-c1) hES cells in BMP signaling is primarily due to the lack of a DRAGON/RGM (GPI-AP) subfamily member that serves as a co-receptor for BMP4. To express a functional form of this subfamily on the surface of PIG-A/GPI-AP deficient AR1-c1 hES cells that are defective post-translationally and lack all the GPI-APs, we made a novel DRAGON cell-surface protein by fusing the mature DRAGON domain with a transmebrane domain. As predicted, the over-expression of the engineered cell surface DRAGON protein (DRAGON™) restored both high-level BMP activation and trophoblast differentiation (Fig 7), as did an activated type I BMP receptor (ALK3 or ALK6) gene vectors (Fig 6). Conversely, knockdown DRAGON or RGMa gene expression in H9 hES cells (Fig S8) pheno-copied the observation with Ar1-c1 (H1) hES cells. Based on these data, we suggest that cell surface forms of DRAGON/RGM protein family play a critical role for high-level activation of BMP signaling in hES cells, which is in turn critical for trophoblast differentiation. How the members of DRAGON/RGM GPI-AP subfamily modulate or oppose related signaling such as those mediated by Activin/Nodal remains to be determined (Besser, 2004; James et al., 2005; Xia et al., 2007). Our approach of constructing a transmembrane form of GPI-APs such as DRAGON™ and reconstituting in AR1-c1 hES cells will likely help to delineate the first step of requirement for other GPI-APs during development using hES cell initiated developmental systems.
It is generally believed based on mouse and other lower organism models that mesodermal induction is dependent on BMP signaling (Hogan, 1996). However, mesodermal differentiation in AR1-c1 cells lacking GPI-APs appears normal (e.g., CD34 gene expression in Fig S3 and CD34+ cell production in Fig S4), whereas trophoblast differentiation fails due to the reduced level of BMP signaling. One hypothesis is that GPI-anchored co-receptors such as DRAGON are only critical to the full activation of BMP signaling and in turn trophoblast or TE formation. This hypothesis then easily explains the fact that mesoderm induction was not affected in GPI-AP deficient AR1-c1 hES cells. This hypothesis is also supported by gene array data that BMP ligands (such as BMP4) and downstream target genes (such as human ID2 and eHAND/Hand1) are expressed significantly higher in human TE tissues vs. inner cell mass (Adjaye et al., 2005), and also higher (such as Id2) in mouse TS cells vs. mES cells (Tanaka et al., 2002). The generality of this novel hypothesis, such as in mice and other placental-bearing mammals, however, remains to be fully investigated. The present study highlights the excitement that hES cells provide an unprecedented and much needed research tool for understanding the development and function of human cells and tissues in normal and pathological processes.
Experimental procedures
Human ES cell culture
The H1 line was obtained from WiCell Research Institute, Inc. (Wisconsin, MI), and propagated as previously described (Zhan et al., 2004; Dravid et al., 2005; Zhou et al., 2007). For the feeder-free culture and pro-aerolysin selection, undifferentiated hES cell clumps were passaged onto Matrigel (BD Biosciences, 1/30 dilution) coated tissue plates and cultured in primary embryonic fibroblast (pMEF)-conditioned medium (Dravid et al., 2005). An H1 hES cell preparation (G-GFP, non-clonal) that stably expresses GFP after lentivial vector transduction (Dravid et al., 2005; Zhou et al., 2007) was used as the starting cell population to select GPI-AP deficient cells by pro-aerolysin selection. Cells were incubated with 500 nM pro-aerolysin (Protox Biotech, Victoria, Canada) at 37°C for 30′, and cultured under the feeder-free condition for one passage (5-7 days). The (pro-)aerolysin-resistant (AR) hES cells were expanded and monitored for the presence or absence of GPI-APs such as Thy-1/CD90 on cell surface. A detailed procedure of how we established two independent clones (AR1-c1 and AR2-c1) is described in Supplement Table S2. Gene transduction of AR hES cells by an inducible lentiviral vector (iDuet101/PIG-A) was previously described (Zhou et al., 2007). The karyotyping was conducted (300-400 bands, 20 mitotic spreads) as previous described (Zhan et al., 2004).
Induced hES cell differentiation
Various hES cell lines cultured under the feeder-free condition were used for embryoid formation (in suspension, with 20% fetal bovine serum) as previously described (Zhan et al., 2004). For BMP4 induced differentiation, hES cells were cultured under the feeder-free condition (on Matrigel, serum-free and with pMEF-conditioned medium) in the presence of exogenous BMP4 (Xu et al., 2002). Unless otherwise indicated, 50 ng/ml BMP-4 (R & D Systems) was used. The supernatant was collected at day 10 for a human chorionic gonadotrophin (hCG) ELISA (with an antibody specific to the hCGβ) performed by Johns Hopkins Hospital. The BMP4-treated cells were analyzed subsequently by either RT-PCR or immuno-staining.
Immuno-staining
Adherent hES cells before or after differentiation were fixed with PBS containing 4% paraformaldehyde for 20′ at RT. Following blocking of non-specific binding with 4% goat serum, the cells were incubated with mouse monoclonal (IgG) antibodies recognizing alkaline phosphatase (TRA-2-49), SSEA-4 (MC-813-70) and TROMA-1 (Developmental Studies Hybridoma Bank, Iowa City, IA), or Cripto and placenta-Cadherin/CDH3 (both from R&D Systems). The stained cells were visualized by anti-mouse goat IgG conjugated with Alexa 594 or Alexa 555 (Invitrogen). For intracellular staining of TROMA-I and Cripto, fixed cells were first permeablized by treatment with 0.1% Triton X-100 and 1% BSA for 45′. Flow cytometric (FACS) analysis was used as previously described (Zhou et al., 2007).
Analysis of gene expression by RT-PCR
Total RNA isolated from various hES cell populations before and after induced differentiation was analyzed as before (Dravid et al. 2005). Primers used for semi-quantitative PCR are listed in Table S3. For quantitative (real-time) RT-PCR, specific primers and probes were purchased from Applied Biosystems (Foster City, CA), and used as described before (Dravid et al., 2005). Total RNA isolated from normal hES cell-derived teratoma (consisting of mixed cell population) was used to normalize between various hES cell samples, before or after differentiation. After normalization to the level of the ß-actin gene, the expression level of each gene is plotted in a log scale relative to the level of the given gene expression in teratoma defined as 100.
Northern blot
Ten microgram total RNA of undifferentiated and BMP4 induced hES cells were denatured, separated by electrophoresis, and transferred onto Hybond-N filter (Amersham Biosciences, Piscataway, NJ). After crosslinking RNA by UV irradiation, the filter was hybridized with α-32P-dCTP (Amersham Biosciences) labeled PIG-A gene probe in ULTRAhyb Hybridization Buffer (Ambion, CA) overnight. After striping, the filter was re-probed with the β-actin gene probe.
Western blot
The protein extracts from hES cells +/- BMP4 induction (50 ng/ml for 6 hours) were made and separated by 8-16% SDS-PAGE (Bio-RAD Laboratories, Hercules, CA) were transferred onto Hybond™-P membrane (Amersham Biosciences). Purified antibodies recognizing phosphorylated SMAD1/5/8 (Cell Signaling Technology Inc.) were used (James et al. 2005). The purified rabbit antibody recognizes phosphorylated SMAD1/5/8 at SMAD1 (S463/465), SMAD5 (S463/465) and SMAD8 (S426/428). After striping, the filter was re-probed with an antibody recognizing GAPDH, a housekeeping protein used as an internal control.
Construction and use of a vector expressing a transmembrane form of DRAGON protein
To express the DRGON/RGMb molecule on the cell surface of AR1-c1 hES cells deficient of PIG-A and all the GPI-APs, we had to engineer a chimeric protein in which the mature form of DRAGON peptide (without the C-terminal region that should be cleaved by the GPI anchoring process) is fused to a transmembrane (TM) domain. The cDNA encoding the mature form of human DRAGON/RGMB (NP_001012779, from the 87th to 452th amino acids, lacking the 26 amino acids at the C-terminus) is cloned and inserted into the expression vector called pDisplay (Invitrogen) at the Bgl II and Sac II sites (Fig S7b). This allows in-frame fusion of DRAGON mature peptide to the TM domain of a PDGF receptor, resulting in a TM called DRAGON™. The cell-surface expression of DRAGON™ from the pDisplay-based vector is shown in Fig. S7).
Transient transfection of hES cells to measure BMP4 induced signaling or restoration
We used the reporter plasmid in which the luciferase gene is controlled by the BMP-responsive element from the Id1 promoter (Id120-Luc) (Lopez-Rovira et al., 2002), or the Id2 promoter (from Dr. Xiao-Hong Sun, Oklahoma Medical Research Foundation). Human ES cells plated on Matrigel-coated 24-well plates were transfected by lipofectamine 2000 as previously described (Dravid et al., 2005). In short, 0.6 μg Id120-Luc plasmid and 0.2 μg EF.lacZ plasmid were added into transfection mixture (0.1 ml), and added to 0.5 ml culture medium. Six hours after transfection, hES cells were treated with various concentrations of BMP4. After additional 24 hours, transfected cells were lysed and normalized lucferase activities were measured (Dravid et al., 2005).
We also co-transfected AR1-c1 and G-GFP hES cells by a plasmid vector expressing an activated form of ALK3 (Q233D) or ALK6 (Q203D) gene (Chen et al., 1998), or from the pDisplay-based DRAGON™ vector. A mixture of 0.3 μg of an expression vector and 0.3 μg Id120-Luc reporter, plus 0.2 μg EF.lacZ plasmid, was used (Σ=0.8 μg per well). Human ES cells under the feeder-free condition were transfected by either ALK3 or ALK6 expression vector at day 0 for 6 hours and then treated with or without BMP4 (50 ng/ml). BMP4-induced Id1-Luc activities were measured 20 hours after. For reconstitution of trophoblast formation in AR1-c1 cells, 0.8 μg of the activated ALK3 or pDisplay-based DRAGON™ vector was used to transfect hES cells per well in 24-well plates. 48 hours later, we repeated the procedure. By day 5, cells were either harvested to RT-PCR analysis for elevated expression of trophoblast genes or fixed for staining by TROMA-I.
Statistical Analysis
Data plotted are typically expressed as mean ± standard deviation (SD). Statistical analysis was performed using GraphPad Prism version 4.0 (GraphPad Software, San Diego, CA). Significance of differences was examined using the Student t-test (two-sided, unequal variance), or non-parametric Mann-Whitney rank sum test when sample sizes were smaller. It is considered significant if p value is ≤0.05.
Supplementary Material
Acknowledgments
We thank Holly Hammond and Jeffrey Lin in the Cheng lab for teratoma formation and other technical assistance, Drs. C Cain and G Stetten of the Johns Hopkins Prenatal Cytogenetics Lab for karyotyping. We also thank Dr. L Li and J Ross for providing plasmids expressing activated forms of ALK3 and ALK6 genes, Dr. F Ventura for permission and Dr. R Xu for providing the Id1-luc, and Dr. XH Sun for the Id2-luc reporter plasmid. We thank Drs. J Gearhart and L Li, C Kerr for critical reading. Dr. G Chen was funded in part by NIH postdoctoral fellowships (T32HL007525 and F32HL086168). This work was supported in part by research grants from NIH (HL73781 and CA70970), Stem Cell Research Foundation (S2005-026) and the Johns Hopkins Institute for Cell Engineering to L Cheng.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, et al. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. 2005;23:1514–25. doi: 10.1634/stemcells.2005-0113. [DOI] [PubMed] [Google Scholar]
- Babitt JL, Zhang Y, Samad TA, Xia Y, Tang J, Campagna JA, Schneyer AL, Woolf CJ, Lin HY. Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280:29820–7. doi: 10.1074/jbc.M503511200. [DOI] [PubMed] [Google Scholar]
- Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–9. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J Biol Chem. 2004;279:45076–84. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- Brodsky RA, Mukhina GL, Nelson KL, Lawrence TS, Jones RJ, Buckley JT. Resistance of Paroxysmal Nocturnal Hemoglobinuria Cells to the Glycosylphosphatidylinositol-Binding Toxin Aerolysin. Blood. 1999;93:1749–56. [PubMed] [Google Scholar]
- Brulet P, Jacob F. Molecular cloning of a cDNA sequence encoding a trophectoderm-specific marker during mouse blastocyst formation. PNAS (USA) 1982;79:2328–32. doi: 10.1073/pnas.79.7.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ji X, Harris MA, Feng JQ, Karsenty G, Celeste AJ, Rosen V, Mundy GR, Harris SE. Differential roles for BMP receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J Cell Biol. 1998;142:295–305. doi: 10.1083/jcb.142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–9. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- Dravid G, Ye Z, Hammond H, Chen G, Pyle A, Donovan PJ, Yu X, Cheng L. Defining the role of Wnt/beta-catenin signaling in the survival, proliferation and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1498. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- Dunn DE, Tanawattanacharoen P, Boccuni P, Nagakura S, Green SW, Kirby MR, Kumar MS, Rosenfeld S, Young NS. Paroxysmal Nocturnal Hemoglobinuria Cells in Patients with Bone Marrow Failure Syndromes. Ann Intern Med. 1999;131:401–8. doi: 10.7326/0003-4819-131-6-199909210-00002. [DOI] [PubMed] [Google Scholar]
- Dunn DE, Yu J, Nagarajan S, Devetten M, Weichold FF, Medof ME, Young NS, Liu JM. A knock-out model of paroxysmal nocturnal hemoglobinuria: Pig-a(-) hematopoiesis is reconstituted following intercellular transfer of GPI-anchored proteins. PNAS (USA) 1996;93:7938–43. doi: 10.1073/pnas.93.15.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Naini B, Dovzhenko OV, Durning M, Wegner FH, Thomson JA, Golos TG. Trophoblast differentiation in embryoid bodies derived from human embryonic stem cells. Endocrinology. 2004;145:1517–24. doi: 10.1210/en.2003-1241. [DOI] [PubMed] [Google Scholar]
- Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J, et al. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10(13):1580–94. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–45. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- Hu R, Mukhina GL, Piantadosi S, Barber JP, Jones RJ, Brodsky RA. PIG-A mutations in normal hematopoiesis. Blood. 2005;105:3848–54. doi: 10.1182/blood-2004-04-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–82. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Kaufman DS, Hanson ET, Lewis RL, Auerbach R, Thomson JA. Hematopoietic colony-forming cells derived from human embryonic stem cells. PNAS (USA) 2001;98:10716–21. doi: 10.1073/pnas.191362598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P, Payne JL, Tremml G, Greer PA, Gaboli M, Pandolfi PP, Bessler M. FES-Cre targets phosphatidylinositol glycan class A (PIGA) inactivation to hematopoietic stem cells in the bone marrow. J Exp Med. 2001;194:581–9. doi: 10.1084/jem.194.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Ohishi K, Takeda J. GPI-anchor synthesis in mammalian cells: genes, their products, and a deficiency. J Biochem. 1997;122:251–7. doi: 10.1093/oxfordjournals.jbchem.a021746. [DOI] [PubMed] [Google Scholar]
- Lensch MW, Daley GQ. Scientific and clinical opportunities for modeling blood disorders with embryonic stem cells. Blood. 2006;107:2605–12. doi: 10.1182/blood-2005-07-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rovira T, Chalaux E, Massague J, Rosa JL, Ventura F. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J Biol Chem. 2002;277:3176–85. doi: 10.1074/jbc.M106826200. [DOI] [PubMed] [Google Scholar]
- Luzzatto L, Bessler M, Rotoli B. Somatic mutations in paroxysmal nocturnal hemoglobinuria: a blessing in disguise? Cell. 1997;88:1–4. doi: 10.1016/s0092-8674(00)81850-4. [DOI] [PubMed] [Google Scholar]
- Minchiotti G, Parisi S, Liguori G, Signore M, Lania G, Adamson ED, Lago CT, Persico MG. Membrane-anchorage of Cripto protein by glycosylphosphatidylinositol and its distribution during early mouse development. Mech Dev. 2000;90:133–42. doi: 10.1016/s0925-4773(99)00235-x. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, Ward-van Oostwaard D, Mummery C, et al. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J Cell Sci. 2004;117:1269–80. doi: 10.1242/jcs.00970. [DOI] [PubMed] [Google Scholar]
- Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. PNAS(USA) 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti V, Tremml G, Soares V, Pandolfi PP, Luzzatto L, Bessler M. Murine embryonic stem cells without pig-a gene activity are competent for hematopoiesis with the PNH phenotype but not for clonal expansion. J Clin Invest. 1997;100:1028–36. doi: 10.1172/JCI119613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong SJ, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, et al. DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280:14122–9. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- Schenke-Layland K, Angelis E, Rhodes KE, Heydarkhan-Hagvall S, Mikkola HK, Maclellan WR. Collagen IV induces trophoectoderm differentiation of mouse embryonic stem cells. Stem Cells. 2007;25:1529–38. doi: 10.1634/stemcells.2006-0729. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Takeda J, Miyata T, Kawagoe K, Iida Y, Endo Y, Fujita T, Takahashi M, Kitani T, Kinoshita T. Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell. 1993;73:703–711. doi: 10.1016/0092-8674(93)90250-t. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Kunath T, Kimber WL, Jaradat SA, Stagg CA, Usuda M, Yokota T, Niwa H, Rossant J, Ko MS. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12:1921–8. doi: 10.1101/gr.670002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–41. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- Xia Y, Sidis Y, Mukherjee A, Samad TA, Brenner G, Woolf CJ, Lin HY, Schneyer A. Localization and action of Dragon (repulsive guidance molecule b), a novel bone morphogenetic protein coreceptor, throughout the reproductive axis. Endocrinology. 2005;146:3614–21. doi: 10.1210/en.2004-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Yu PB, Sidis Y, Beppu H, Bloch KD, Schneyer AL, Lin HY. Repulsive guidance molecule (RGMa) alters utilization of bone morphogenetic protein (BMP) type II receptors by BMP2 and BMP4. J Biol Chem. 2007;282:18129–40. doi: 10.1074/jbc.M701679200. [DOI] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–4. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2:185–90. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–92. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Zhan X, Dravid G, Ye Z, Hammond H, Shamblott M, Gearhart J, Cheng L. Functional antigen-presenting leucocytes derived from human embryonic stem cells in vitro. Lancet. 2004;364:163–71. doi: 10.1016/S0140-6736(04)16629-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li L. BMP signaling and stem cell regulation. Dev Biol. 2005;284:1–11. doi: 10.1016/j.ydbio.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Zhou BY, Ye Z, Gao PZ, Chen G, Zhang YA, Cheng L. Inducible and reversible transgene expression in human stem cells after efficient and stable gene transfer. Stem Cells. 2007;25:779–89. doi: 10.1634/stemcells.2006-0128. [DOI] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–21. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


