Abstract
A pseudouridine-modified region of the U2 small nuclear (sn)RNA anneals with the intronic branchpoint sequence, and positions a bulged adenosine to serve as the nucleophile in the first chemical step of pre-mRNA splicing. We have determined three X-ray structures of RNA oligonucleotides containing the pseudouridylated U2 snRNA and the branchpoint consensus sequences. The expected adenosine branchpoint is extrahelical in a 1.65 Å resolution structure containing the mammalian consensus sequence variant, and in a 2.10 Å resolution structure containing a shortened Saccharomyces cerevisiae consensus sequence. The adenosine adjacent to the expected branchpoint is extrahelical in a third structure, which contains the intact yeast consensus sequence at 1.57 Å resolution. The hydration and base stacking interactions mediated by the U2 snRNA pseudouridines correlate with the identity of the unpaired adenosine. The expected adenosine bulge is associated with a well-stacked pseudouridine, which is linked via an ordered water molecule to a neighboring nucleotide. In contrast, the bulge of the adjacent adenosine shifts the base stacking and disrupts the water-mediated interactions of the pseudouridine. These structural differences may contribute to the ability of the pseudouridine modification to promote the bulged conformation of the branch site adenosine, and to enhance catalysis by snRNAs. Furthermore, iodide binding sites are identified adjacent the unconventional bulged adenosine, and the structure of the mammalian consensus sequence variant provides a high resolution view of a hydrated magnesium ion bound in a similar manner to a divalent cation binding site of the Group II intron.
Keywords: RNA splicing, RNA structure, pseudouridine, branchpoint sequence, U2 snRNA, iodide binding sites
The spliceosome, composed of more than 100 proteins and five small nuclear (sn)RNAs (U1, U2, U4, U5, and U6 snRNAs), is responsible for removing the intervening sequences from pre-mRNAs (reviewed in (1, 2)). The spliceosomal snRNAs are essential for recognition and pairing of the consensus sequences at the pre-mRNA splice sites, and are likely to participate directly in catalysis. One of the critical snRNA/pre-mRNA interactions involves a short duplex between the U2 snRNA and a consensus intron sequence, called the branchpoint sequence (BPS). In the first chemical step of splicing, the BPS forms a 2′-5′ branched intron lariat and releases the 5′ exon. During this reaction, the phosphate at the 5′ splice site undergoes nucleophilic attack by the 2′ hydroxyl group of a conserved adenosine in the BPS. In the second chemical step, a lariat intron and spliced exons are released following a second attack by the newly formed 3′ hydroxyl group of the upstream exon on the phosphate of the 3′ splice site. These phosphoryl transfer reactions of the spliceosome require divalent metal ions (3-5), which play structurally important roles and may activate the nucleophiles and leaving groups in an analogous manner to protein enzymes (6). Spliceosomal proteins are also essential for the pre-mRNA splicing process; the U2 snRNA/BPS duplex is contacted by proteins including the p14 subunit of the larger protein complex SF3b (7, 8), the arginine-serine rich domains of SR protein splicing factors (9), and the protein PRP8 at the catalytic center of the spliceosome (10, 11). Thus, the conformation and metal ion binding sites of the U2 snRNA/BPS duplex are important factors during the recognition and positioning of essential functional groups for the pre-mRNA splicing reaction.
The BPS is highly conserved in the yeast Saccharomyces cerevisiae (12) (UACUAAC, branchpoint adenosine underlined), but is more variable in mammals (YNCURAY, with Y pyrimidine, R purine, and N any nucleotide) (13, 14). A region near the 5′ end of the U2 snRNA (nucleotides 33-38 in humans) contains a sequence complementary to the BPS (15), with the exception of a bulged branchpoint adenosine. In addition to the major eukaryotic spliceosome, the minor class of nuclear spliceosomes and the self-splicing Group II introns are predicted to share an unpaired branchpoint adenosine, within an otherwise complementary region of an RNA duplex. In the minor spliceosome, the U2 snRNA is replaced by the U12 snRNA, which recognizes a slightly different branchpoint sequence of the pre-mRNA target (UCCAURAY). Self-splicing Group II introns, found in certain gene transcripts of bacteria and plant or fungal organelles, also employ an adenosine within a branch site region that otherwise diverges in sequence from the nuclear splicing complex (for example, CCUAU/GGGGG in the S. cerevisiae ai5γ intron) (16).
A highly conserved post-transcriptional pseudouridine (ψ) modification in the U2 snRNAs of eukaryotes ranging from yeast to vertebrates base pairs adjacent the branchpoint adenosine ((17, 18), reviewed in (19)). This pseudouridine neighboring the branchpoint adenosine is also conserved in the U12 snRNA of the minor spliceosome (20). The functional importance of pseudouridylation within the branch site region has been established using several systems. At the chemical level, branch site pseudouridylation enhances phosphotriester bond formation during catalysis by a minimal U2/U6 snRNA construct (21). Inhibition of pseudouridylation within the U2 branch site recognition positions, in conjunction with a secondary mutation in the region, inhibits splicing of most pre-mRNAs in S. cerevisiae at nonpermissive temperatures (22). Most strikingly, the conserved pseudouridylation site in the BPS is required for pre-mRNA splicing and active U2 ribonucleoprotein particle (RNP) assembly in vertebrates (23, 24).
Significant efforts have been devoted to understanding the conformations of the U2 snRNA/BPS duplex and the Group II intron counterpart. Previously, the X-ray structure was determined of a self-complementary RNA duplex designed to mimic the S. cerevisiae U2 snRNA/BPS site in the absence of pseudouridines (25). In this structure, the adenosines 5′ to the expected branchpoint site are extrahelical, possibly due to contacts with adjacent RNA molecules in the crystal. Subsequently, NMR structures of oligonucleotides containing the S. cerevisiae U2 snRNA/BPS consensus were determined in the presence and absence of the pseudouridine modification (26). Comparison of these structures, coupled with 2-aminopurine fluorescence titrations, indicates that the presence of the pseudouridine promotes an extrahelical conformation of the expected branchpoint adenosine. Similarly, an extrahelical conformation of the branch site adenosine was detected in NMR experiments with a domain of the Group II intron (27), although the X-ray structure of the catalytic Group II domains showed extrahelical conformations of both the expected branchpoint adenosine and the 3′ flanking uridine (28). Altogether, these different structural investigations paint a diverse view of possible conformations at the branch site region.
To investigate the conformation of the branch site region in complex with the pseudouridylated U2 snRNA at high resolution, the X-ray structures of three RNA duplexes containing variations of the pseudouridylated U2 snRNA/BPS consensus sequences are described here. Consistent with the previously established capacity of either BPS adenosine to serve as the branch site in the context of the intact spliceosome (29), the expected branchpoint adenosine is extrahelical in two of the structures, whereas the adjacent 5′ adenosine is extrahelical in the third structure. The unconventional bulged adenosine forms a pocket that accommodates iodide ions from the crystallization solution without requiring further changes in the overall structure. A structure containing a mammalian branchpoint variant reveals a hydrated magnesium ion bound in the major groove adjacent the bulged adenosine, a location similar to a divalent metal ion binding site bridging the Group II intron branchpoint-containing helix (28). The intramolecular base stacking and water-mediated interactions with the pseudouridines correlate with the extrahelical conformation of the expected branchpoint adenosine, which suggests possible means for the pseudouridine modification to influence the conformation of the branchpoint RNA.
MATERIALS AND METHODS
Crystallization and data collection
RNA oligonucleotides (including BPS-1: 5′-CGCUACUAACGCG; U2-1: 5′-GCGCGψAGUAGC; BPS-2: 5′-CGCACUAACCGCG; U2-2: 5′-GCGCGGψAGUGC; BPS-3: 5′-CGCUACUGACGCG; ψ: pseudouridine) were synthesized by Dharmacon Research, Inc. (Boulder, Colorado) and used without further purification. Deprotected RNAs were resuspended in buffer containing 100 mM sodium cacodylate pH 6.5, 10 mM MgCl2 and 2 mM spermidine. Complimentary strands (BPS1: BPS-1/U2-1; BPS2: BPS-2/U2-2; BPS3: BPS-3/U2-1) were mixed in equimolar amounts, heated at 65°C for 15 minutes, and annealed by gradual cooling to room temperature. Preliminary crystallization conditions were obtained from sparse matrix screens at 4°C and room temperature using the vapor diffusion hanging drop method, followed by additive screens to improve crystal quality. Seven duplexes were screened for crystallization in addition to the three whose structures are described here, without yielding diffraction quality crystals. Crystals were obtained from 1 mM RNA duplexes mixed in 1.2:1 ratio with well solution containing 1 M ammonium sulfate (BPS1 or BPS2) or 15% 2-methyl-2,4-pentanediol (MPD)/0.2 M magnesium acetate (BPS3) and then equilibrated against 0.7 ml of well solution at room temperature. For BPS1, the addition of 200 mM NaI to the crystallization stock was used to improve crystal quality. BPS1 and BPS2 crystals obtained maximum size within one week, whereas BPS3 crystals required one month.
Prior to data collection at 100K, BPS1 or BPS2 crystals were cryoprotected by coating with 50% mineral oil/50% paratone-N mixture, and BPS3 crystals were briefly exposed to crystallization conditions with the MPD concentration increased to 35%. The BPS1 data set was collected on beamline X8C of the National Synchrotron Light Source (NSLS) at the Brookhaven National Laboratory using the ADSC Quantum-4R detector. The BPS2 data set was collected using the in-house Bruker-AXS X8 Proteum CCD detector and MICROSTAR generator at the Johns Hopkins School of Public Health. The BPS3 data set was collected at beamline A1 of the Cornell High Energy Synchrotron Source (CHESS) using the ADSC Quantum-210 CCD detector. For synchrotron data collection, a low resolution pass with minimum exposure times was included to measure the strong reflections. Data were integrated and scaled using HKL2000 (30). Crystallographic data collection statistics and unit cell parameters are summarized in Table 1 for BPS1 (Iodide), BPS2, and BPS3, and Supplementary Table 1 for BPS1 determined in the absence of iodide additive.
Table 1.
Crystallographic data and refinement statistics
| Sequence | BPS1 5′-GCGCG ψ AGUAGCGCGCAAUCAUCGC-5′ | BPS2 5′-GCGCGG ψ AGUGCGCGCCAAUCACGC-5′ | BPS3 5′-GCGCG ψ AGUAGCGCGCAGUCAUCGC-5′ |
|---|---|---|---|
| A: Data collection statistics a | |||
| Wavelength (Å) | 1.10 | 1.54 | 0.98 |
| Space group | C2 | C2 | P21 |
| Unit cell a (Å) | 68.30 | 67.98 | 31.19 |
| b (Å) | 40.80 | 40.47 | 38.28 |
| c (Å) | 32.55 | 32.21 | 32.08 |
| β (°) | 94.93 | 95.21 | 108.23 |
| Resolution limit (Å) | 1.57 | 2.10 | 1.65 |
| Observed reflections | 40,066 | 25,940 | 56,359 |
| Unique reflections | 12,181 | 4,759 | 8,754 |
| Completeness (%) | 96.5 (92.8) | 93.1 (83.2) | 99.5 (97.5) |
| I/σ(I) | 24.3 (9.7) | 12.0 (2.5) | 35.2 (9.1) |
| Redundancy | 3.3 (3.0) | 5.5 (1.7) | 6.4 (4.6) |
| Rsymb(%) | 4.3 (12.2) | 11.9 (48.6) | 5.6 (24.3) |
| B: Refinement statistics | |||
| Rcrystc(%) | 20.1 | 23.8 | 22.7 |
| Rfreec (%) | 22.8 | 24.7 | 25.2 |
| RMSD bond lengths (Å) | 0.003 | 0.005 | 0.004 |
| RMSD bond angles ° | 0.85 | 0.96 | 1.02 |
| Coordinate error from cross-validated Luzzati plot (Å) | 0.20 | 0.36 | 0.29 |
| Coordinate error from cross-validated σA plot (Å) | 0.08 | 0.52 | 0.27 |
| No. RNA atoms | 528 | 529 | 529 |
| No. water molecules | 158 | 81 | 125 |
| No. sulfate ions | 1 | 1 | 0 |
| No. iodide ions | 3 | 0 | 0 |
| No. magnesium ions | 0 | 0 | 1 |
| Average B-factor | |||
| all atoms (Å2) | 21.4 | 26.8 | 33.2 |
| RNA atoms (Å2) | 17.7 | 25.7 | 28.1 |
| water molecules (Å2) | 32.1 | 29.9 | 42.9 |
Values in parenthesis are for the highest resolution shell: 1.63-1.57 Å for BPS1, 2.18-2.10 Å for BPS2, 1.71-1.65 Å for BPS3.
, where Ii is an intersity I for the ith measurement of a reflection with indices hkl and 〈I〉 is the weighted mean of all measurements of I.
for the working set of reflections; Rfree is Rcryst for 7% of the reflections excluded from the refinement
Structure solution and crystallographic refinement
The BPS1 structure were solved by molecular replacement using a combination of PDB ID: 1NUV (31) and PDB ID: 1I9X (25) as the search model. A direct rotation search (RF-function=0.41 for the top rotation solution) followed by translation search and PC-refinement provided the molecular replacement solution (TR-function MONITOR=0.56/packing=0.34) using the program Crystallography & NMR System (CNS) with data in the 20-3.0Å resolution range (32). Initial rigid body refinement resulted in a working R factor of 40% with a slightly lower free R factor of 37%, possibly due to statistical fluctuations of the relatively small test set available for this resolution rage (100 reflections, 7% of data from 20-3.0Å resolution) (33), or correlation of the randomly-chosen test set and working set of reflections related to the pseudo-twofold symmetry of an RNA helix (34). The BPS1 structure was then used in turn as a model for the BPS2 and BPS3 structures. The register and bulged nucleotide conformations were modified based on composite omit electron density maps. Composite omit electron density maps calculated using the refined coordinates are shown for the bulged adenosines in Supplementary Fig. 1.
Atomic coordinates were refined using simulated annealing, energy minimization and restrained individual temperature factor refinement against maximum likelihood targets. The bulged adenosines were located by inspecting |Fo|-|Fc| omit electron density maps using the program O (35). The Molprobity validation server reported high quality scores for all-atom contacts of the structures compared with structures of comparable resolution (≥95th percentile) (36). Global helical parameters were calculated using the program Curves v5.3 (37), figures were made using the Pymol molecular graphics system (http://www.pymol.org), buried surface areas were calculated using NACCESS (38), and other manipulations made use of the Uppsala Software Factory (39) and the CCP4 suite of programs (40).
RESULTS
Overview of the structures
To obtain a high resolution view of the conformation and metal ion binding sites of the pseudouridylated U2 snRNA/BPS duplex, three structures were determined of 12mer/13mer RNA oligonucleotides containing complementary U2 snRNA/BPS sequences (Fig. 1). All of the structures contain a single bulged adenosine within the context of an otherwise A-form double helix, characterized by an average helical twist of 31°, rise per base pair of 3.25 Å, and C3′-endo sugar pucker. The bases on either side of the extrahelical adenosine stack against one another as if uninterrupted. Excluding the bulged nucleotide, the helical parameters relating the base pairs bordering the extrahelical adenosine are slightly overwound compared with A- or B-form double helices (38° between Psu6-Ade21 and Ade7-Uri19 of BPS1; 48° between Gua5-Cyt22 and Psu6-Gua20 of BPS3). All of the sequences terminate with Gua/Cyt overhangs, which promote the asymmetric, pseudocontinuous helical packing of the crystal through base pairs between symmetry-related duplexes. Despite similar overhangs, the crystal-packing interactions between consecutive duplexes differ between the BPS1/BPS2 and BPS3 structures; the BPS1 and BPS2 overhangs are engaged in an unusual Hoogsteen base pair (Supplementary Fig. 2) with an overwound helical twist of ∼60°, whereas the BPS3 termini are coupled by Watson-Crick base pairing interaction with an underwound helical twist of ∼15°.
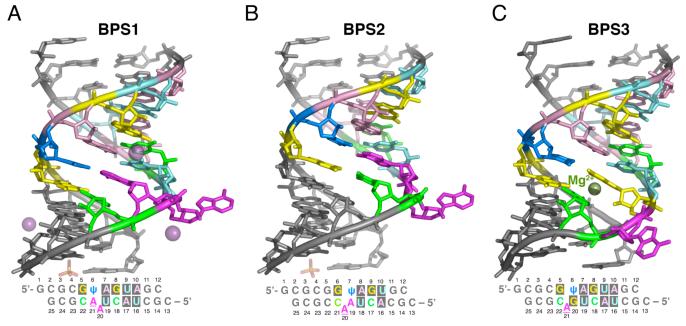
Figure 1.
Overall structures of the U2 snRNA/BPS-containing duplexes. (A) BPS1, (B) BPS2, (C) BPS3. The consensus sequences are colored by residue here and throughout: Gua, yellow; Cyt, green, Ade, magenta; Uri, cyan; Psu, marine. Oligonucleotide sequences and numbering used in the text are given below. The top strand contains the U2 snRNA consensus sequence; the bottom strand contains the BPS consensus sequence. Iodide ions bound to BPS1 are shown as space-filling spheres.
The initial structure of the S. cerevisiae U2 snRNA/BPS consensus sequence includes the functionally important U2 snRNA pseudouridine nearest the branch site, flanked on either side by nearly equivalent numbers of Gua-Cyt base pairs to promote duplex stability (5′-CGCUACUAACGCG/5′-GCGCGψAGUAGC, consensus in bold)(BPS1, Fig. 1A). Crystals of BPS1 diffracted beyond 1.6 Å resolution in the presence of an iodide additive (Table 1), and the structure was determined by molecular replacement using a search model composed of A-form RNA and bulged nucleotide fragments. For comparison, a lower, 2.5Å resolution structure of BPS1 was determined in the absence of added iodide (Supplementary Table 1). The BPS1 structures are nearly identical in the presence and absence of iodide (0.4 Å RMSD between 526 matching atoms), so that the higher resolution BPS1 structure is used for the detailed analysis presented below.
Formally, it was possible for either of the tandem adenosines in the BPS consensus sequence (Ade20 and Ade21) to base pair with the adjacent pseudouridine (Psu6) of the U2 snRNA. Although NMR studies indicate that the presence of the pseudouridine promotes an extrahelical conformation of the branchpoint adenosine (26), this adenosine (Ade21) was base paired with the pseudouridine within the BPS1 duplex. Instead, the neighboring adenosine (BPS1-Ade20) was extrahelical, a base pairing arrangement shared by the previously-determined structure of a self-complementary duplex containing the S. cerevisiae U2 snRNA/BPS consensus sequences, despite the absence of pseudouridines in this previous structure (25) (0.4 Å RMSD between 131 matching atoms of the consensus sequences).
To further investigate the conformation of the BPS1 branch site, the S. cerevisiae U2 snRNA/BPS consensus sequences were truncated by one nucleotide and rearranged within the context of the flanking Gua-Cyt base pairs (5′-CGCACUAACCGCG/5′-GCGCGGψAGUGC, consensus in bold) (BPS2, Fig. 1B). The BPS2 crystal form was isomorphous with that of BPS1, and the BPS2 structure was determined at 2.10 Å resolution starting from the BPS1 coordinates (Table 1). Accordingly, the overall conformation of the BPS2 structure is similar to that of BPS1 (1.2 Å RMSD between 249 matching atoms of U2 snRNA/BPS consensus, with register adjusted to match bulged nucleotides), but slipped in register by one nucleotide to place the consensus branchpoint adenosine in an extrahelical position. Thus, the BPS2 structure provides a high resolution view of the pseudouridylated U2 snRNA/BPS consensus sequences with the expected adenosine bulge.
Independently, a third structure (BPS3) was determined in a distinct crystal form at 1.65 Å resolution (Fig. 1C), for comparison of the BPS1 and BPS2 structures containing the S. cerevisiae consensus sequences with the conformation of a branchpoint variant more frequently observed in higher eukaryotes (5′-CGCUACUGACGCG/5′-GCGCGGψAGUAGC, consensus in bold). The composite BPS consensus sequences of plants, rats, humans, chickens, and fruit flies display a guanosine preceding the branch site with slightly higher frequency than adenosine (42% compared with 39%, respectively) (14). This distinct BPS consensus in higher eukaryotes is further developed in mammals, as indicated by the 60% frequency of guanosines at this position of mammalian splice sites (41). In contrast, the tandem adenosines within the S. cerevisiae BPS are strictly conserved (100%) (12). Accordingly, the BPS3 sequence containing the mammalian consensus was identical to BPS1, with the exception of an adenosine to guanosine substitution preceding the branchpoint adenosine.
Although the distinctive BPS3 guanosine preceding the branchpoint (Gua20) lacks a Watson-Crick base pairing partner, this guanosine engages in a wobble base pair with the neighboring pseudouridine (Psu6) of the U2 snRNA consensus (Fig. 1C). Despite the similarity of the BPS3 sequence register with the BPS1 rather than the BPS2 oligonucleotides, the expected branchpoint adenosine of BPS3 is extrahelical. The RMSD between the BPS3 and BPS1 or BPS2 structures is higher than that between the BPS1 and BPS2 structures (RMSD of 1.9 Å for BPS3 with 246 matching atoms of BPS1, comparing consensus sequences adjusted for the register of bulged nucleotide). More dramatically, the conformation of the BPS3 bulged adenine has rotated by 60° compared with the BPS2 or BPS1 bulged adenine bases, as described in more detail below.
Conformations of branchpoint sites
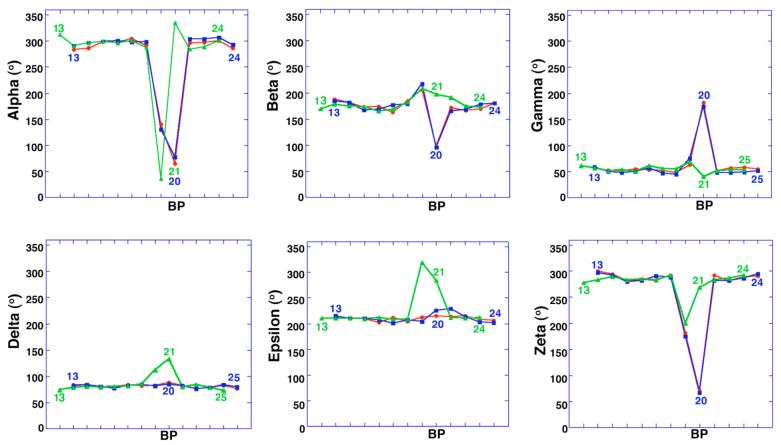
The torsion angles of the nucleic acid backbone near the bulged adenosine deviate from the nearly ideal A-form values of the flanking helical regions (Fig. 2)., The conformations of the bulged adenosines (Ade20) are nearly identical between the BPS1 and BPS2 structures. In contrast, the bulged adenosine (Ade21) of BPS3 adopts a distinct conformation (Fig. 3A), characterized by distinct backbone torsion angles. The phosphodiester linkages of the BPS1 and BPS2 bulged adenosines display large deviations in the alpha, beta, gamma, and zeta angles. To compensate for the bulged nucleotide, the alpha and zeta angles of the BPS1 or BPS2 nucleotide preceding Ade20 are also distorted (BPS1-Uri19 or BPS2-Ade19). For BPS3, the alpha and zeta angles of the bulged Ade21 remain close to A-form. Instead, the BPS3-Gua20 nucleotide is marked by a large deviation in the alpha torsion angle, and a lesser deviation in the zeta torsion angle. The delta and epsilon torsion angles are also slightly distorted for BPS3-Gua20 and Ade21, unlike the A-form delta and epsilon torsion angles of BPS1 and BPS2. Altogether, these conformational differences generate the 60° rotation about an axis perpendicular to the base plane, that relates the bulged adenine of BPS3 with those of BPS1 or BPS2.
Figure 2.
Torsion angles of the sugar-phosphate backbone versus nucleotide position. Alpha, O5′-P; beta, O5′-C5′; gamma, C5′-C4′; delta, C4′-C3′; epsilon, C3′-O3′; zeta, O3′-P. BP, branchpoint. The register of BPS3 (green) is shifted by one nucleotide to align the bulged nucleotide with those of BPS1 (red) and BPS2 (blue).
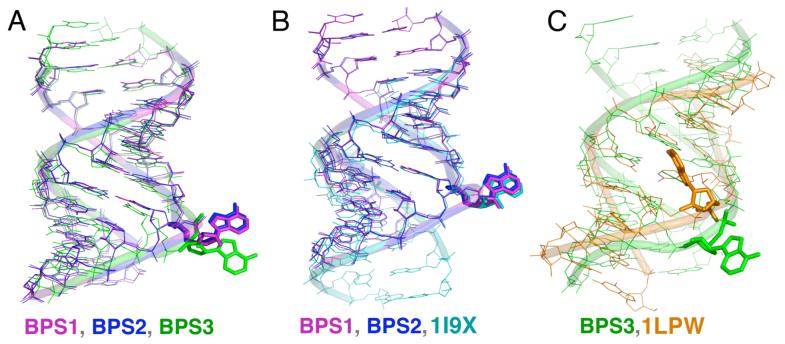
Figure 3.
Superposition of U2 snRNA/BPS-containing structures. (A) BPS1, BPS2, and BPS3. (B) BPS1, BPS2, and unmodified counterpart from (25), PDB ID: 1I9X. (C) BPS3 and NMR structure from (26), PDB ID: 1LPW. Structures were superimposed using matching atoms in consensus sequence regions.
The conformations of the BPS1 and BPS2 bulged adenosines are remarkably similar to the palindromic X-ray structure determined in the absence of pseudouridines (PDB ID: 1I9X) (Fig. 3B) (25). Despite the approximation of pseudouridine with uridine, this similarity is reflected by a closely matched overlay of the key nucleotides of the consensus regions including the extrahelical Ade20, the expected branchpoint Ade21, and the Psu6 target of pseudouridylation (BPS1 numbering). Although the detailed nature of the interactions differ, the bulged BPS1, BPS2, and 1I9X adenosines all share a similar mode of crystal packing via an A-minor motif in the minor groove of a symmetry-related duplex (Fig. 4A). In the BPS1 and BPS2 structures, the Ade20 base is centered over the cytidine of the Cyt22-Gua5 base pair, but also forms hydrogen bonds with the exocyclic guanine amine as well as the cytidine 2′ hydroxyl group. The 2′ hydroxyl of Ade20 is deeply buried in the minor groove, engaged by hydrogen bonds with the Gua5 base and ribose 2′ hydroxyl group. Accordingly, the solvent accessible surface areas of the bulged BPS1 or BPS2 adenosines are largely buried by these symmetry-related interactions of the crystal (41% of the solvent accessible adenosine surface area buried by intermolecular contacts).
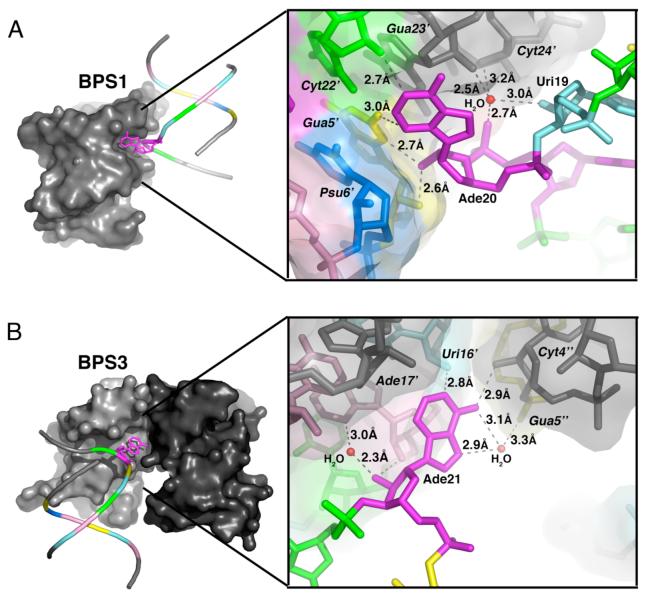
Figure 4.
Interactions mediated by bulged adenosines. (A) BPS1-Ade20 interacts with minor groove of a symmetry-related duplex. The interactions mediated by BPS2-Ade20 are similar. (B) BPS3-Ade21 of BPS3 interacts with the phosphodiester backbones of two symmetry-related duplexes. The overall structure of one duplex is represented by a coil for the phosphodiester backbone, and a ball-and-stick diagram for the bulged adenosine. The solvent accessible surface is shown for interacting, symmetry-related duplex(es). An expanded view of the detailed interactions is given in the right panel. Residue names of symmetry-related molecules are distinguished by primed, italicized font.
In contrast with BPS1 or BPS2, the BPS3 bulged adenosine is located in an exposed environment that diverges from the 1I9X structure (60° rotation of bulged adenine base relative to 1I9X). The conformation of the BPS3 bulge also differs from an NMR structure of oligonucleotides containing the S. cerevisiae U2 snRNA/BPS consensus sequences (PDB ID: 1LPW) (Fig. 3C) (3.6 Å RMSD for 272 matching consensus sequence atoms). The branchpoint adenosine of the NMR structure containing the S. cerevisiae consensus sequences interacts with the minor groove, at the cost of a >5 Å gap introduced between the flanking base pairs. This gap may be the result of overfitting during the NMR refinement rather than a structural feature, given an ∼15 kcal/mol energetic penalty for disrupted base stacking (42). In the X-ray structure, the branchpoint adenosine of BPS3 is rotated away from the minor or major grooves in a manner that allows the flanking nucleotides (Gua20 and Cyt22) to engage one another in energetically-favorable base stacking interactions.
The bulged adenosine of BPS3 docks loosely on the ribose backbone of one symmetry- related duplex, and interacts with another via a single hydrogen bond (Fig. 4B). Two direct hydrogen bonds connect the bulged Ade21 base with neighboring helices; one from the Ade21 N1 imine to a symmetry-related Uri16′ 2′ hydroxyl group (where primed italics indicate a symmetry-related molecule), and the other from the Ade21 exocyclic N6 amine to the Cyt4”; 2′ hydroxyl group of the distinct, symmetry-related molecule. The six-member ring of the bulged Ade21 stacks on the O4′ atom of a symmetry-related Ade17′ ribose, in a sugar-base mode of nucleic acid interactions commonly observed in Z-DNA and aromatic drug-DNA complexes (43). In addition to the interactions with the adeninebase, the 2′ hydroxyl group of the Ade21 ribose forms hydrogen bonds with the 2′ hydroxyl group of Ade17′, as well as with a bound water molecule. Otherwise, this 2′ hydroxyl group of the bulged Ade21, which would serve as a nucleophile in the context of an activated spliceosome, is solvent accessible.
Pseudouridine conformation
When uridine is converted to pseudouridine, the glycosidic linkage isomerizes to a carbon-carbon bond between the uracil and the ribose, positioning the N1-H imino group in the major groove (Fig. 5A). All of the pseudouridines of the U2 snRNA consensus oligonucleotides are marked by a single water molecule that forms hydrogen bonds with the unique N1-H group of the base and a nonbridging oxygen of the 5′ phosphate (Fig. 5). Similarly located water molecules are observed bound to the pseudouridines in tRNA and ribosomal rRNA structures ((44), personal observation from PDB ID: 1S72 (45)). A second theme shared among the different structures is offset base stacking that exposes the 3′ face of the pseudouracil; in all three cases, the 3′ flanking adenosine (respectively Ade7, Ade8, or Ade7 of BPS1, BPS2, and BPS3) buries only the Watson-Crick edge of the base.
Figure 5.
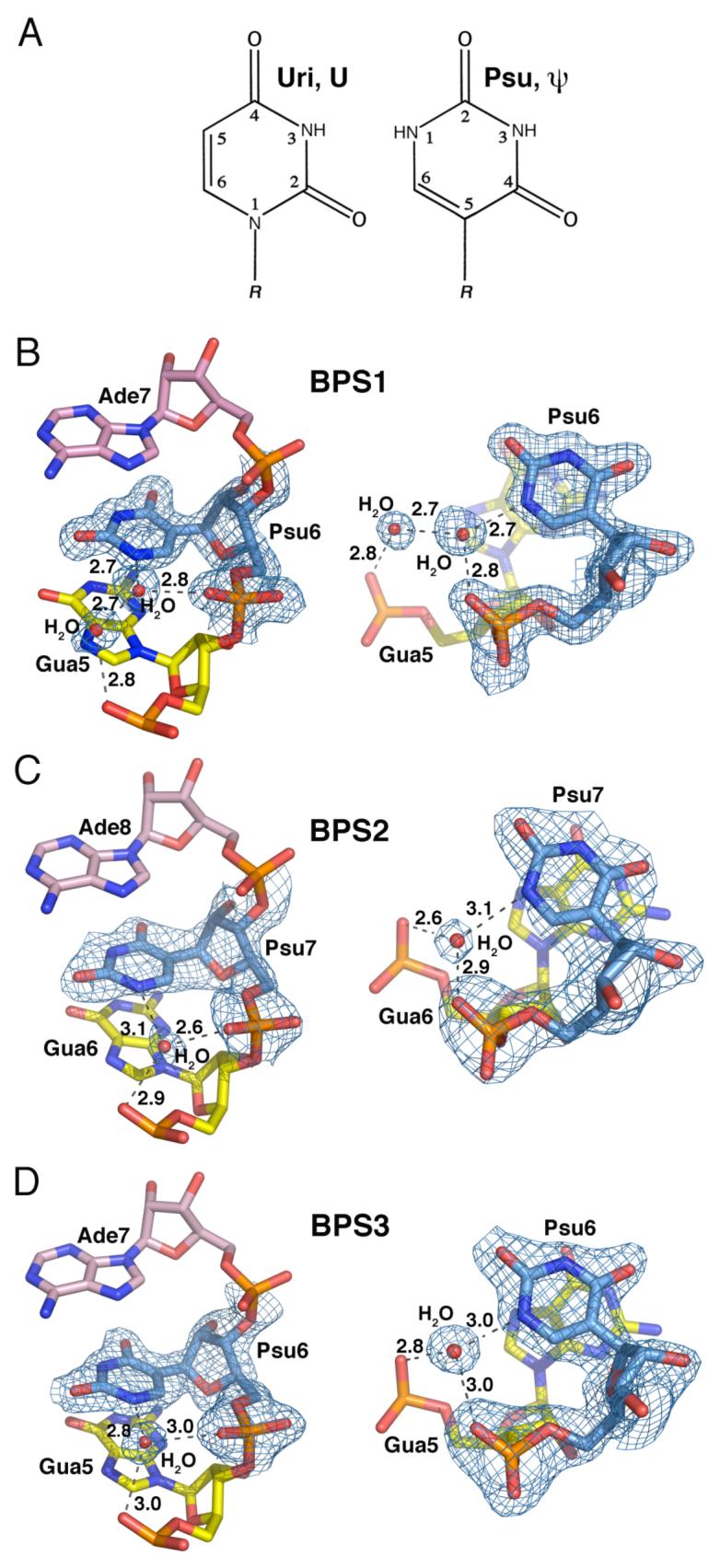
Pseudouridine base stacking interactions and hydration. (A) Chemical structures, nomenclature, and numbering convention of uridine compared with pseudouridine. R, 5′-phosphate-ribose. (B) BPS1-Psu6. (C) BPS2-Psu7. (D) BPS3-Psu6. Composite 2|Fo|-|Fc| omit electron density maps are contoured around the pseudouridines at 1σ level. Views on the right are rotated 90° about the x-axis compared with views on the left.
Despite these similarities, the interactions mediated by the pseudouridine-bound water molecule, and the nature of the pseudouracil stacking with the 5′ base, vary depending on the identity of the branchpoint adenosine (Fig. 5). The BPS2 and BPS3 pseudouridines (BPS2-Psu7 and BPS3-Psu6) are sufficiently near one of the nonbridging phosphate oxygens of the 5′ flanking guanosine to form a hydrogen bond via the pseudouridine-bound water molecule. However, the BPS1 structure includes a second water molecule in a lengthened, two-water bridge between the guanosine phosphate and pseudouridine (Psu6). Moreover, the unique N1-H of the pseudouridine in the BPS2 and BPS3 structures centers over the endocyclic N7 imino group of the 5′ guanosine (BPS2-Gua6 and BPS3-Gua5). This stacking register is expected to be energetically favorable, given the proximity of the electropositive potential of the N1-H to the electronegative potential of the N7. In contrast, the N1-H group of the BPS1 pseudouridine stacks against the center of the larger aromatic ring in the guanine (BPS1-Gua5), which leaves the five-member aromatic ring of the guanine exposed to solvent.
Anion binding sites of BPS1 and BPS2 structures
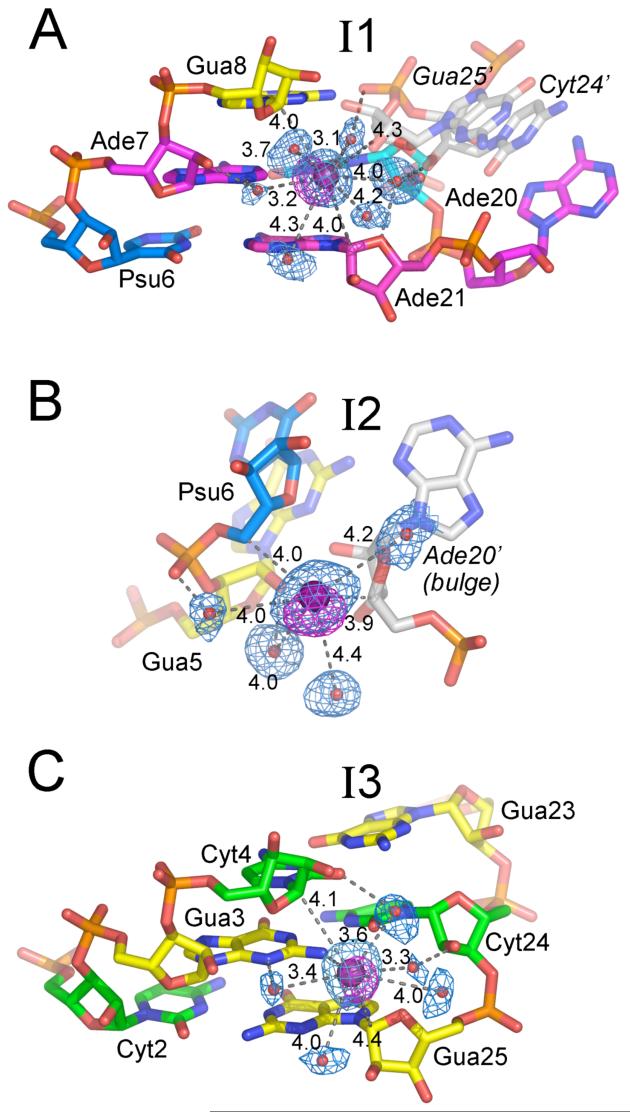
Iodide ions (200 mM NaI) promoted crystallization of the BPS1 oligonucleotides, whereas addition of iodide had little effect on the BPS2 or BPS3 crystal quality. Three iodide binding sites were identified in the BPS1 structure based on significant r.m.s. electron density values of 24, 19, or 11σ in anomalous difference Fourier maps (Fig. 6), consistent with f”; of 3.9 electrons for iodine at γ = 1.10 Å. Further, the packing interactions with the iodide ions (∼3.3-4.4 Å) were compatible with water-iodide coordination distances (∼3.7 Å O—I-) (46,47) and the van der Waals radius of iodine (1.98 Å), as opposed to the typical 2.6-3.0 Å hydrogen bond distances of bound water molecules (48). Only one of the iodides (I3) directly interacts with the polar nucleotide atoms in a manner previously described for chloride or sulfate ions (49) (Fig. 6C). This I3 iodide contacts the Gua3 exocyclic amine and is relatively solvent-exposed in the minor groove of the Gua3-Cyt24 base pair, consistent with the lowest electron density and highest temperature factor among the bound iodides (11σ, B=44 Å2). Water molecules coordinated to the I3 iodide mediate further contacts with the imino or hydroxyl groups of the Gua3, Cyt24 and Cyt4 nucleotides. Nevertheless, the I3 iodide ion also engages in hydrophobic interactions that would be uncharacteristic of chloride ions, packing between the C1′ carbons of the Gua25 and Cyt4 riboses.
Figure 6.
Iodide binding sites of BPS1, arranged from highest to lowest anomalous electron density.(A) I1. (B) I2. (C) I3. Composite 2|Fo|-|Fc| omit electron density maps are shown in blue at 1σ contour level, and anomalous difference Fourier maps are shown in magenta at 5σ contour level surrounding the iodide ions and coordinated water molecules. Distances of direct interactions with the iodide (<4.5 Å) are given (Å). Dashed lines indicate hydrogen bonds of coordinated water molecules with the RNA (<3.0 Å). Symmetry-related moleculesare indicated by gray color and labeled with italicized, primed font.
Iodide ions with stronger anomalous electron density and lower temperature factors occupy nonpolar niches created by the extrahelical conformation of adenine preceding the designated branchpoint site combined with the intermolecular crystal packing. The apparently most occupied iodide binding site (I1, 24σ, B=28 Å2) is located in a hydrophobic pocket created by the extrusion of the Ade20 preceding the branchpoint site (Fig. 6A). This I1 iodide packs in the minor groove of the Ade7-Uri19 base pair, between the C1′ carbons of Gua8 and Ade21 across the bulged site. A C5′ methylene carbon donated by a symmetry-related, Gua25′ ribose completes the hydrophobic pocket. As for I3, bound water molecules mediate further interactions between I1 and the polar functional groups, including the edges of the bases, 2′ hydroxyl atoms of the ribose sugars, and phosphodiester backbone. A site with apparently intermediate occupancy (I2, 19σ, B=33 Å2) also interacts with the extrahelical Ade20 conformation (Fig. 6B). In this case, the I2 iodide mediates nonpolar contacts among the C5′ and C4′ methylenes of the bulged Ade20 ribose of one duplex, and Psu6′ of the next. Additional contacts between I2 and the phosphodiester backbone of the preceding, symmetry-related Gua5′ nucleotide are mediated by coordinated water molecules.
In addition to the iodide ions, a sulfate ion is observed in both the BPS1 and BPS2 structures, bound to the junction of two symmetry-related helices. The 3.6-3.9 Å distance between the sulfur and the base nitrogen atoms is consistent with the average sulfate-water (S—O ∼3.8 Å) coordination distances derived from X-ray and neutron diffraction studies of aqueous ionic solutions (46, 47). At the 1.5 Å resolution of the BPS1 structure, the tetrahedral sulfate shape is clearly visible (Supplementary Fig. 2). The sulfate binds to a previously documented sulfate-binding site available in the Watson-Crick edge of guanines (49), which is left exposed by the Hoogsteen base-pairing conformation of Gua1 overhang with Cyt13′ of the neighboring BPS1 or BPS2 duplex. Two oxygen atoms of the sulfate are engaged in hydrogen bonds with the exocyclic amine and amide nitrogen groups of the guanine base. The absence of bound sulfate in the BPS3 structure is consistent with lack of sulfate ions in the crystallization solution, and the burial of the Gua1 base edge in Watson-Crick rather than Hoogsteen base-pairing.
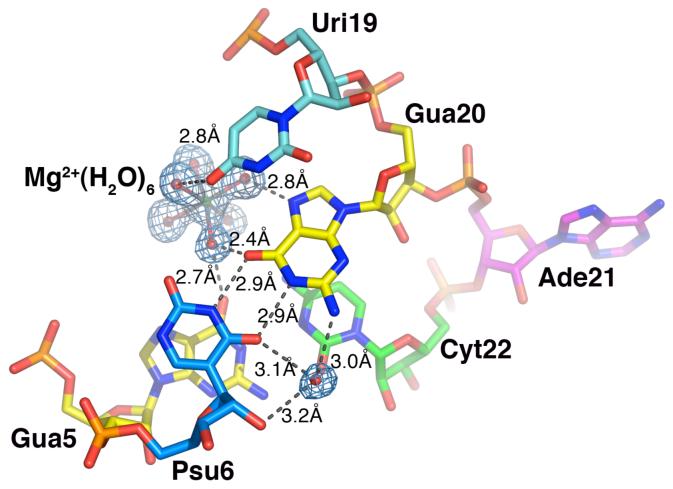
Magnesium ion binding site of BPS3 structure
A hydrated magnesium ion [Mg2+(H2O)6] was identified in the major groove of the Psu6-Gua20 and Gua5-Cyt22 base pairs flanking the bulged Ade21 of the BPS3 structure (Fig. 7). The magnesium ion is coordinated by an octahedral arrangement of six water molecules, which in turn interact with the edges of the bases. One of the coordinated water molecules mediates an interstrand hydrogen bond between the O6 carbonyl groups of the Gua20 and Gua5 bases, whereas another donates a hydrogen bond to the endocyclic N7 imine of Gua20. An additional hydrogen bond is observed between a third coordinated water molecule and the O4 carbonyl group of Uri19. Thus, three of the four hydrogen bonds in the [Mg2+(H2O)6]-binding site depend on the presence of Gua20 in the BPS3 sequence, which is distinct from the Ade20 counterpart of the BPS1 and BPS2 sequences. Given that the exocyclic N7 amine of adenine replaces the guanine O6 carbonyl, the consensus branchpoint sequence of the BPS1 and BPS2 oligonucleotides lacks the [Mg2+(H2O)6]-binding site offered by the BPS3 oligonucleotide. Similar interactions between hydrated magnesium ions and the O6 and N7 atoms of the guanine base edges are observed for other A-form RNA and DNA duplexes in general (50-52). Accordingly, soaking experiments and difference Fourier analysis of the X-ray structure of the Group II intron (28), in combination with further NMR experiments and fluorescence titrations (27, 53) established an analogous metal binding site in the major groove of the BPS site.
Figure 7.
Magnesium ion coordination and water-mediated interactions with major groove of BPS3 branchpoint sequence. An |Fo|-|Fc| omit electron density map is drawn at 3σ contour level around the hydrated magnesium ion and water molecule in the major groove.
DISCUSSION
Variable extrahelical adenosine
Although pseudouridine was included in all of the U2 snRNA sequences studied here, the BPS1 structure showed an extrahelical conformation for the preceding adenosine rather than the expected branchpoint site. A high concentration of ammonium sulfate (>1 M) was required for crystallization of the BPS1 and BPS2 oligonucleotides, and may have influenced the conformations captured for view by X-ray crystallography. In a similar manner, thestructure of a construct containing the fifth and sixth domains of the Group II intron revealed an unanticipated extrahelical conformation of two nucleotides in the branch site region in the presence of ∼1 M ammonium sulfate (28). Previous NMR and fluorescence studies demonstrate that the pseudouridine modification promotes an extrahelical conformation of the branch site adenosine in RNA oligonucleotides containing the S. cerevisiae branchpoint consensus sequence, whereas both of the consecutive adenosines remain within the double helix when paired opposite an unmodified uridine (26). Given that this bulged adenosine is engaged by crystal contacts, and can be reverted to the branchpoint adenosine by simply changing the register of the oligonucleotide sequence, crystal packing forces are likely to influence the conformations of the BPS1 and BPS2 bulges. Nevertheless, use of the 5′ nucleotide as the branch site can occur at low frequency in pre- mRNA splicing assays, and experiments with nucleotide analogues suggest that either of the two extrahelical nucleotides observed here may alternatively bulge from the U2 snRNA/BPS duplex in the context of the intact spliceosome (29).
The expected branchpoint adenosine of BPS3 was extrahelical, despite the ability to form a Watson-Crick base pair with Psu6 in lieu of the observed wobble Gua20-Psu6 base pair. In contrast with BPS1 and BPS2, the BPS3 structure was determined under relatively low salt conditions. The symmetry-related contacts with the bulged adenosine are fewer, and are nonspecific for the base identity. Thus, the preference for the bulged Ade21 of BPS3, as opposed to an alternative nucleotide such as Gua20, is independent of the arrangement of molecules in the crystal. Instead, inherent thermodynamic and electrostatic properties of the sequences are likely to assist the preference for the bulged adenosine over guanosine of BPS3. First, the energetic penalty for the observed 5′-GAC-3′ bulge is 1.2 kcal/mol lower than the alternative 5′-UGA-3′ bulge (54). Second, in vitro selection experiments show a strong inherent preference for use of an adenosine as the branch site nucleophile for catalytic formation of a 2′-5′ phosphodiester linkage (55). Third, a survey of the single nucleotide bulges in the ribosomal RNAs show that more than half involve bulged adenosines (56). Group II introns, which unlike the spliceosome have the capacity to operate in the absence of accessory proteins, also make use of a branchpoint adenosine (97% contain Ade as the predicted branchpoint nucleotide) (16). Hence, the BPS3 structure reflects a role for the branchpoint adenosine and the flanking sequences in facilitating choice of the appropriate nucleophile for the splicing reaction.
The bulged adenosines of all the structures presented here adopt extrahelical conformations, whereas the NMR structure shows the bulged adenine base inserted in the minor groove of the pseudouridylated duplex (26). This minor groove-bound conformation buries the 2′ hydroxyl group, the proposed nucleophile of the splicing reaction, via intramolecular interactions with the phosphodiester backbone and base of the 3′ flanking nucleotide. Moreover, the conformation of the branchpoint adenosine in the NMR structure creates an energetically unlikely cavity that interrupts the base stacking of the flanking base pairs. In contrast, the 2′ hydroxyl group of the extrahelical BPS3 conformation observed here is prominently exposed, and the flanking base pairs are favorably stacked. For comparison, the ribosomal rRNAs contain only two intrahelical bases out of thirty single nucleotide bulges, and no intramolecular interactions of the bulged nucleotides with the adjacent minor groove (56). Instead, the bulged nucleotides of rRNAs are most commonly extrahelical, with more than 50% of these nucleotides interacting with other rRNA nucleotides. The spliceosome is a ribonucleoprotein machine like the ribosome, and the molecular environments of the spliceosome and ribosome are likely to share similar themes (57, 58). The proximity of the U2 snRNA/BPS duplex to the U2/U6 snRNA duplex and possibly other RNAs in the active spliceosome (1, 59), coupled with the need for the 2′ hydroxyl group of the bulged adenosine to access the substrate, suggests that an extrahelical conformation of the branchpoint adenosine such as described here may be functionally relevant for catalysis.
Context of pseudouridine interactions
Details of the pseudouridine hydration and base stacking interactions are influenced by the location of the single nucleotide bulge in the U2 snRNA/BPS consensus oligonucleotides. In both of the structures with the expected location of the single nucleotide bulge (BPS2 and BPS3), the water molecule that bridges the unique pseudouracil N1-H and 5′ phosphate also mediates an interaction with the phosphate of the 5′ nucleotide. This type of water-mediated interactions with pseudouridine stabilizes base stacking interactions by increasing the rigidity of the phosphodiester backbone located 5′ to the site of pseudouridylation (60). Accordingly, the BPS2 or BPS3 pseudouracil exhibits a favorable stacking arrangement, with the unique N1-H packed against the N7 imine of the 5′ guanine. In contrast, the BPS1 structure with the alternative extrahelical adenosine differs in the base stacking and water-mediated links with the pseudouridine. It is possible that these coupled hydration and base stacking characteristics of the pseudouridine serve to stabilize the expected adenosine bulge. Moreover, the altered hydration and exposed pseudouridine surfaces associated with proximity to the bulged adenosine would provide distinctive features for recognition by proteins and RNAs within the spliceosome.
Specific iodide binding sites at the branch site
The presence of three iodide ions in the BPS1 structure provides insights concerning the preferred iodide binding sites of nucleic acids, and further suggests that the possibility of successful extrapolation of the established use of iodide anions to phase protein structures to RNA (61). Despite the established binding of chloride and sulfate ions to nucleic acids (49), little is known concerning the nature of potential iodide binding sites. The BPS1 structure shows that iodide ions may occupy similar sites observed for chloride and sulfate ions near the electropositive base edges, but in the context of nonpolar environments provided by the ribose sugar carbons. Similar nonpolar binding sites have been previously observed for halide binding to proteins (61). Moreover, it is possible that similar anion binding sites have been overlooked for other nucleic acid structures (49), given the inherent difficulties of distinguishing atoms by 2|Fo|-|Fc| electron density alone at the high resolution limits of most macromolecular X-ray structures, coupled with a general predisposition towards considering nucleic acids as cation, rather than anion, binders. Although the native structure shows that iodide is not essential for the observed BPS1 conformation, the diffraction limit is improved, and accordingly two iodide binding sites are created by the extrusion of the Ade20 preceding the expected branch site Ade21. This raises the question of whether iodide ions may stabilize an extrahelical conformation for the adenine preceding the conventional branch site.
Significance of magnesium ion binding site
Both the spliceosome and Group II introns share a requirement for the presence of magnesium ions for folding and catalysis of RNA splicing (reviewed in (57)). Several lines of evidence demonstrate that group II self-splicing introns bind magnesium in a site comparable to the major groove of the guanine-containing consensus sequence described here (27, 28, 53, 62). The X-ray structure of the catalytically essential Group II intron domains shows a cobalt hexammine mimic of magnesium ions in the major groove of the branch site (28), and titration of the Group II intron branch site domain with magnesium ions induces chemical shifts of the nucleotides flanking the branch site (27, 53). Comparison of the Group II intron and BPS3 oligonucleotides demonstrates that in both cases, magnesium ions are similarly located in the major groove of a Gua-Uri wobble base pair. The BPS3 structure provides a high resolution example of this class of magnesium binding site, showing that a hydrated magnesium ion interacts via coordinated water molecules with the edges of the base pairs flanking the extrahelical branch site.
The position of the magnesium ion bridging the base pairs adjacent the expected branchpoint suggests that the bound ion may facilitate base closure and stacking surrounding the bulged nucleotide. Accordingly, 2-aminopurine fluorescence experiments with the Group II intron domain indicate that the addition of magnesium ions increases the extrahelical conformation of the branch site adenosine (27). A structural role for the magnesium ion in indirectly positioning the branch site nucleophile for the initial step of the splicing reaction, rather than directly activating the nucleophile or leaving group, is consistent with its location more than 15 Å from the 2′ hydroxyl group of the BPS3 branch site adenosine. Although magnesium ions are required to directly coordinate the leaving group during chemical catalysis of RNA splicing (4, 57), other metal binding sites, including a site of the U6 snRNA intramolecular stem loop/Group II domain 5 (reviewed in (59)), are likely candidates for this function.
As reinforced by the [Mg2+(H2O)6]-binding sites of the BPS3 structure presented here and numerous other nucleic acid structures (50-52), the major groove of guanosine-containing base steps offers a favorable binding site for hydrated magnesium ions in the major groove. Furthermore, a Gua-Uri(Psu) wobble pair combined with an adjacent Gua-Cyt base pair increases the electronegative surface potential of the major groove to promote association of magnesium ions (63). Accordingly, favorable [Mg2+(H2O)6]-binding sites would be provided by the Gua-Psu or Gua-Cyt base pairs adjacent the branchpoint adenosine of mammalian splice sites (42% and 45% sequence conservation, respectively (14)). Likewise, Gua-Uri or Gua-Cyt base pairs bordering the branchpoint adenosines of Group II introns (47% and 63% sequence conservation, respectively (16)) are likely to bind magnesium ions, as supported by the location of a cobalt hexammine ion in the Group II intron structure (28). However, an adenosine is frequently substituted for the guanosine within mammalian BPS (14), and this Ade-Psu base pair is stringently conserved in yeast BPS (12). The adenosine substitution would alter the key O6 functional group that coordinates the hydrated magnesium ion. This observation, coupled with the established association of guanosine-containing sites with magnesium ions (28, 50-52), suggests that these adenosines within the yeast BPS consensus sequence and a subset of mammalian BPS may weaken a specific [Mg2+(H2O)6]-binding site.
Coordinates
Structure factor data and coordinates for the refined structures are deposited in the RCSB Protein Data Bank (BPS1 iodide PDB ID: 3CGP; BPS1 native PDB ID: 3CGQ; BPS2 PDB ID: 3CGR; BPS3 PDB ID: 3CGS)
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01 GM070503 and an award from the Faculty Research Initiatives Fund (Johns Hopkins School of Public Health) to C.L.K. CHESS is supported by the National Science Foundation under award DMR 0225180. The Macromolecular Diffraction at CHESS (MacCHESS) facility is supported by award RR-01646 from the National Institutes of Health, through its National Center for Research Resources. Financial support for NSLS comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. We are grateful to Dr. Alexei Soares for assistance with data collection at NSLS.
REFERENCES
- 1.Valadkhan S. The spliceosome: a ribozyme at heart? Biol Chem. 2007;388:693–697. doi: 10.1515/BC.2007.080. [DOI] [PubMed] [Google Scholar]
- 2.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 3.Sontheimer EJ, Gordon PM, Piccirilli JA. Metal ion catalysis during group II intron self-splicing: parallels with the spliceosome. Genes Dev. 1999;13:1729–1741. doi: 10.1101/gad.13.13.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon PM, Sontheimer EJ, Piccirilli JA. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: extending the parallels between the spliceosome and group II introns. RNA. 2000;6:199–205. doi: 10.1017/s1355838200992069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Luhrmann R, Query CC. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;20:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Query CC, Strobel SA, Sharp PA. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- 9.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 10.Turner IA, Norman CM, Churcher MJ, Newman AJ. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA. 2006;12:375–386. doi: 10.1261/rna.2229706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spingola M, Grate L, Haussler D, Ares M., Jr. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. Rna. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruskin B, Greene JM, Green MR. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985;41:833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- 14.Harris NL, Senapathy P. Distribution and consensus of branch point signals in eukaryotic genes: a computerized statistical analysis. Nucleic Acids Res. 1990;18:3015–3019. doi: 10.1093/nar/18.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tycowski KT, Kolev NG, Conrad NK, Fok V, Steitz JA. The ever-growing world of small nuclear ribonucleoproteins. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World III. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2006. pp. 327–368. [Google Scholar]
- 16.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns--a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 17.Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol. 1999;19:2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Yu YT. Detection and quantitation of RNA base modifications. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massenet S, Mougin A, Branlant C. Posttranscriptional modifications in the U small nuclear RNAs. In: Grosjean H, Benne R, editors. The Modification and Editing of RNA. ASM Press; Washington, DC: 1998. pp. 201–228. [Google Scholar]
- 20.Massenet S, Branlant C. A limited number of pseudouridine residues in the human atac spliceosomal UsnRNAs as compared to human major spliceosomal UsnRNAs. RNA. 1999;5:1495–1503. doi: 10.1017/s1355838299991537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valadkhan S, Manley JL. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 2005;280:6655–6662. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- 23.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Yu YT. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA. 2004;10:681–690. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berglund JA, Rosbash M, Schultz SC. Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines. RNA. 2001;7:682–691. doi: 10.1017/s1355838201002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol. 2002;9:958–965. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- 27.Schlatterer JC, Crayton SH, Greenbaum NL. Conformation of the Group II intron branch site in solution. J Am Chem Soc. 2006;128:3866–3867. doi: 10.1021/ja0578754. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Doudna JA. Structural insights into group II intron catalysis and branch-site selection. Science. 2002;295:2084–2088. doi: 10.1126/science.1069268. [DOI] [PubMed] [Google Scholar]
- 29.Query CC, Moore MJ, Sharp PA. Branch nucleophile selection in pre- mRNA splicing: evidence for the bulged duplex model. Genes Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Wedekind JE, McKay DB. Crystal structure of the leadzyme at 1.8 A resolution: metal ion binding and the implications for catalytic mechanism and allo site ion regulation. Biochemistry. 2003;42:9554–9563. doi: 10.1021/bi0300783. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 34.Fabiola F, Korostelev A, Chapman MS. Bias in cross-validated free R factors: mitigation of the effects of non-crystallographic symmetry. Acta Crystallogr D Biol Crystallogr. 2006;62:227–238. doi: 10.1107/S0907444905040680. [DOI] [PubMed] [Google Scholar]
- 35.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 38.Hubbard SJ, Thornton JM, NACCESS, Dept. Biochem. & Mol. Biol. University College of London; 1993. [Google Scholar]
- 39.Klegwegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN - programs for reformatting, analysis, and manipulation of biomacromoleular electron-density maps and reflection data sets. Acta Crystallogr. 1996;D52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 40.CCP The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Keller EB, Noon WA. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci USA. 1984;81:7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sponer J, Jurecka P, Marchan I, Luque FJ, Orozco M, Hobza P. Nature of base stacking: reference quantum-chemical stacking energies in ten unique B- DNA base-pair steps. Chemistry. 2006;12:2854–2865. doi: 10.1002/chem.200501239. [DOI] [PubMed] [Google Scholar]
- 43.Egli M, Gessner RV. Stereoelectronic effects of deoxyribose O4′ on DNA conformation. Proc Natl Acad Sci USA. 1995;92:180–184. doi: 10.1073/pnas.92.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudouridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 45.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 46.Ohtaki H. Ionic solvation in aqueous and nonaqueous solutions. Monatsh Chem. 2001;132:1237–1268. [Google Scholar]
- 47.Ohtaki H, Radnai T. Structure and dynamics of hydrated ions. Chem. Rev. 1993;93:1157–1204. [Google Scholar]
- 48.Jeffrey GA. An introduction to hydrogen bonding. Oxford University Press; 1997. [Google Scholar]
- 49.Auffinger P, Bielecki L, Westhof E. Anion binding to nucleic acids. Structure. 2004;12:379–388. doi: 10.1016/j.str.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Correll CC, Freeborn B, Moore PB, Steitz TA. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 51.Jovine L, Djordjevic S, Rhodes D. The crystal structure of yeast phenylalanine tRNA at 2.0 A resolution: cleavage by Mg(2+) in 15-year old crystals. J Mol Biol. 2000;301:401–414. doi: 10.1006/jmbi.2000.3950. [DOI] [PubMed] [Google Scholar]
- 52.Egli M. DNA-cation interactions: quo vadis? Chem Biol. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 53.Erat MC, Zerbe O, Fox T, Sigel RK. Solution Structure of Domain 6 from a Self-Splicing Group II Intron Ribozyme: A Mg(2+) Binding Site is Located Close to the Stacked Branch Adenosine. Chembiochem. 2007;8:306–314. doi: 10.1002/cbic.200600459. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka F, Kameda A, Yamamoto M, Ohuchi A. Thermodynamic parameters based on a nearest-neighbor model for DNA sequences with a single-bulge loop. Biochemistry. 2004;43:7143–7150. doi: 10.1021/bi036188r. [DOI] [PubMed] [Google Scholar]
- 55.Zelin E, Wang Y, Silverman SK. Adenosine is inherently favored as the branch-site RNA nucleotide in a structural context that resembles natural RNA splicing. Biochemistry. 2006;45:2767–2771. doi: 10.1021/bi052499l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Znosko BM, Silvestri SB, Volkman H, Boswell B, Serra MJ. Thermodynamic parameters for an expanded nearest-neighbor model for the formation of RNA duplexes with single nucleotide bulges. Biochemistry. 2002;41:10406–10417. doi: 10.1021/bi025781q. [DOI] [PubMed] [Google Scholar]
- 57.Villa T, Pleiss JA, Guthrie C. Spliceosomal snRNAs: Mg(2+)-dependent chemistry at the catalytic core? Cell. 2002;109:149–152. doi: 10.1016/s0092-8674(02)00726-2. [DOI] [PubMed] [Google Scholar]
- 58.Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- 59.Butcher SE, Brow DA. Towards understanding the catalytic core structure of the spliceosome. Biochem Soc Trans. 2005;33:447–449. doi: 10.1042/BST0330447. [DOI] [PubMed] [Google Scholar]
- 60.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;232:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dauter Z, Dauter M, Rajashankar KR. Novel approach to phasing proteins: derivatization by short cryo-soaking with halides. Acta Crystallogr D Biol Crystallogr. 2000;56:232–237. doi: 10.1107/s0907444999016352. [DOI] [PubMed] [Google Scholar]
- 62.Sigel RK, Vaidya A, Pyle AM. Metal ion binding sites in a group II intron core. Nat Struct Biol. 2000;7:1111–1116. doi: 10.1038/81958. [DOI] [PubMed] [Google Scholar]
- 63.Xu D, Landon T, Greenbaum NL, Fenley MO. The electrostatic characteristics of G.U wobble base pairs. Nucleic Acids Res. 2007;35:3836–3847. doi: 10.1093/nar/gkm274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.