Abstract
Huntington’s disease is caused by polyglutamine expansion in the huntingtin protein. Huntingtin directly interacts with profilin, a major actin monomer sequestering protein and a key integrator of signals leading to actin polymerization. We observed a progressive loss of profilin in the cerebral cortex of Huntington’s disease patients, and in cell culture and Drosophila models of polyglutamine disease. This loss of profilin is likely due to increased degradation through the ubiquitin-proteasome system. Profilin loss reduces the F/G actin ratio, indicating a shift in actin polymerization. Overexpression of profilin abolishes mutant huntingtin toxicity in cells and partially ameliorates the morphological and functional eye phenotype and extends lifespan in a transgenic polyglutamine Drosophila model. These results indicate a link between huntingtin and profilin and implicate profilin in Huntington’s disease pathogenesis.
Keywords: profilin, Huntington’s disease, polyglutamine, actin, ubiquitin proteasome system
Introduction
Huntington’s disease (HD) is an autosomal dominantly inherited progressive neurological disease that belongs to a group of neurodegenerative disorders caused by expansion of polyglutamine tracts in otherwise unrelated proteins (Zoghbi and Orr, 2000). Expansion of a (CAG)n trinucleotide repeat within the first exon of the IT15 gene leads to an abnormally long polyglutamine tract in the huntingtin (htt) protein (The Huntington's Disease Collaborative Research Group, 1993). Polyglutamine expansion increases the propensity of htt to aggregate and causes neuronal degeneration, particularly in the caudate nucleus of the basal ganglia and in the cerebral cortex. Htt is a 350 kDa, ubiquitously expressed protein containing a proline-rich region adjacent to the polyglutamine tract in its N-terminus as well as numerous HEAT repeats throughout the protein. Although its cellular function remains unclear, htt has been implicated in several processes, including cell signalling, endocytosis, and vesicle transport, based on the known functions of its interacting partners (Harjes and Wanker, 2003). The underlying molecular mechanism of HD pathogenesis is yet to be fully elucidated, but it is thought that mutant htt gains novel toxic properties, possibly through polyglutamine dependent conformational changes and aberrant protein interactions.
Various htt interactors have been identified through yeast two-hybrid screening, co-immunoprecipitation, and in vitro binding assays, including proteins that are regulators of cytoskeletal organization and participants in endocytic pathways (Wanker et al., 1997; Singaraja et al., 2002; Goehler et al., 2004). One of the proteins thus identified is profilin-2a (Goehler et al., 2004). Profilins are eukaryotic actin monomer (G-actin) binding proteins that interact with proline-rich regions in a variety of proteins (reviewed by (Witke, 2004)) and regulate actin dynamics. Profilin was originally thought to be a G-actin sequestration protein that inhibits actin polymerization, but subsequent studies have shown that, depending on the conditions, profilin may either sequester G-actin or promote actin polymerization (Pantaloni and Carlier, 1993).
In mammals, there are four profilin genes, with tissue-specific expression. Profilin-1 is the major isoform in most tissues; profilin-2 is predominantly expressed in the brain (Kwiatkowski and Bruns, 1988); and profilins 3 and 4 are almost exclusively expressed in the testis (Hu et al., 2001; Obermann et al., 2005). Profilin-2 has two splice variants, 2a and 2b (Di Nardo et al., 2000; Lambrechts et al., 2000), and shares approximately 60% sequence identity with profilin-1. Neurons express profilin-1 and -2a (Lambrechts et al., 2000; Neuhoff et al., 2005), and biochemical studies have suggested that both isoforms are closely related with respect to actin binding and their affinity for poly(L-proline). Ablation of profilin-1 expression in knockout mice results in early embryonic lethality (Witke et al., 2001). Mice lacking profilin-2, although viable, show abnormalities in neurotransmitter release and presynaptic excitability, highlighting the importance of profilin-2 in the central nervous system (Pilo Boyl et al., 2007).
Here we report that profilin levels are progressively reduced in cortical tissue from HD patients, in cultured cells expressing mutant htt, and in a Drosophila polyglutamine model. We further demonstrate that restoration of profilin mitigates polyglutamine-induced toxicity in cells and in Drosophila, indicating that profilin may be involved in the HD pathogenesis.
Materials and Methods
Antibodies
Polyclonal antibodies against profilin-1 and -2a were generated by Open Biosystems (Huntsville, AL) by injecting into rabbits the linear peptides KCYEMASHLRRSQY and KAYSMAKYLRDSGF conjugated to KLH. Following immunization, blood was collected, and crude serum was purified with an affinity column containing the respective profilin peptide. Other antibodies used include a monoclonal anti-chickadee, chi1J (Developmental Studies Hybridoma Bank, University of Iowa), monoclonal anti-actin (Chemicon), monoclonal anti-tubulin (Chemicon), monoclonal anti-actin (AC-40; Sigma-Aldrich), monoclonal anti-GFP (BD Biosciences), and rabbit anti-profilin (Alexis Biochemicals).
Human Tissues
Frozen blocks of human cortical tissue from Brodmann area 9 were obtained for western blotting from the Harvard Brain Tissue Resource Center and Vanderbilt University Medical Center in accordance with institutional guidelines (NIH, Office of Human Subjects Research). Specimens were matched for postmortem interval (PMI) and divided into groups based on supplied information: (a) controls (n = 3, age = 65 ± 16 years, PMI = 10 ± 2 hours); (b) presymptomatic HD gene carriers (n = 3, age = 46 ± 31 years, PMI = 13 ± 6 hours); (c) grade 1 HD patients (n = 3, age = 60 ± 10 years, PMI = 13 ± 7 hours); (d), grade 4 HD patients (n = 3, age = 67 ± 6 years, PMI = 16 ± 7 hours). Neuropathological classification was based on previously established guidelines (Vonsattel et al., 1985). The mutant alleles in the HD patients ranged from 45 to 50 CAGs.
Cell Culture
The cell model used for these experiments, a stably transfected, inducible PC12 cell line expressing exon 1 of the HD gene with either 23 or 74 CAGs under the control of a tetracycline promoter, has been previously described (PC12–23Q and PC12–74Q, (Wyttenbach et al., 2001)). The cells were maintained in high glucose Dulbecco’s Modified Eagle Medium (DMEM) with 100 U/ml penicillin/streptamycin, 2 mM L-glutamine, 10% horse serum (HS), 5% Tet-free fetal bovine serum (FBS), and 100 µg/ml G418 at 37° C and 10% CO2. Induction of htt expression was done by incubating the cells in media containing 1 µg/ml of doxycycline for the specified periods of time. For differentiation, the cells were incubated with 100 ng/ml of nerve growth factor (NGF) in media containing 1% HS and no FBS.
F-actin Sedimentation
The htt-inducible PC12 cells were incubated in media containing 1 µg/ml doxycycline for 48 hr. For the specified experiments, cells were transfected with empty vector (mock) or with profilin-1 cDNA using Fugene HD as per manufacturer’s instructions (Roche). Following induction cells were scraped, washed in phosphate-buffered saline (PBS), and then lysed in 0.75 ml of actin lysis buffer (50 mM NaCl, 5 mM MgCl2, 1 mM ATP, 5% glycerol, 5 mM EGTA, 0.1% Triton X-100, 0.1% Nonidet P-40, 0.1% Tween 20, 0.1% β-mercaptoethanol, 50 mM piperazine-N,N'-bis(2-ethanesulfonic acid), pH 6.9). The F-actin and G-actin pools were separated by ultracentrifugation at 100,000 × g at 30 °C. The supernatant was diluted 1:2 with Laemmli buffer, and the pellet was resuspended in cold distilled H2O with 1 µM cytochalasin D and sonicated for 10 s. The pellet fraction was kept on ice for 45 min and then diluted 1:4 with Laemmli buffer. Both fractions were then boiled for 5 min and centrifuged at 14,000 × g for 10 min at 4 °C to remove remaining connective tissue. The same relative amounts of supernatant and pellet fractions (2:1) were loaded on 10% polyacrylamide gels and analyzed by Western blot using an anti-actin antibody (AC-40; Sigma-Aldrich). The F/G-actin ratio was determined by scanning densitometry. The values are presented as the means ± standard error. Statistical comparisons were done using Student’s t-test (p values < 0.05 were considered statistically significant).
Cell Death
The htt-inducible PC12 cells were seeded in 35 mm dishes at 4 × 105 cells / well. On day 1, the cells were transfected with empty vector (mock) or with profilin-1 cDNA using Fugene HD as per manufacturer’s instructions (Roche). The following day, cells were induced with 1 µg/ml doxycycline. 72 h following induction, the cells were trypsinized and centrifuged at 800 × g for 5 min at room temperature. The cell pellets were washed and re-suspended in 400 ul PBS. Propidium iodide (PI) was added to the cell suspension at 1 µg/ml. A total of 50,000 cells were collected and analyzed by flow cytometry (Becton Dickinson FACS Calibur). PI positive cells were detected using a 530 nm excitation and 617 nm emission filter. Cell populations were gated on size (forward scatter) and granularity (side scatter) to exclude debris and cell clumps. An uninduced, unstained negative control sample and a positive control sample stained with PI were used to establish reference regions. Cell death was calculated as the percent cells that were PI positive and reported as the mean ± standard error of three independent experiments. Statistical comparisons were performed using Student’s t-test (P values < 0.05 were considered statistically significant).
Western Blotting
For human specimens, pieces of frozen cortical tissue were immersed in liquid nitrogen and pulverized with a BioPulverizer™ (Biospec Products, Bartlesville, OK). Powdered tissue was homogenized in a buffer consisting of 25 mM Tris-HCl (pH 7.5), 5 mM EDTA, 1% SDS, and protease inhibitors (Complete Mini, Roche) at 1:10 w/v. The samples were sonicated on ice for 10 seconds three times and then centrifuged at 14,000 g for 10 minutes at 4° C to remove any particulate matter. For cells, at specified time points, the samples were washed with PBS, scraped, collected, and lysed in ice-cold RIPA buffer (50 mM Tris-HCl, 1% NP40, 0.5% Na-DOC, 150 mM NaCl, 0.1% SDS, and 2 mM EDTA) containing protease inhibitors. The cell lysate was sonicated on ice and incubated for 20 minutes on ice. For Drosophila, flies were frozen on dry ice and the heads were collected on a frozen plate and transferred to an eppendorf tube with RIPA buffer (1:5 w/v). The heads were then homogenized with a pestle and sonicated on ice. The lysates were centrifuged at 15,000 g for 20 minutes at 4° C for 20 minutes. Afterwards, for all samples the supernatant were collected and kept on ice. Protein concentrations for all samples were determined using the Pierce BCA protein assay (Rockford, IL). Laemmli buffer (BioRad, Hercules, CA) was added to each supernatant, and the samples were heated to 95° C for 5 minutes. Equal protein amounts of cell extract (20 or 60 µg total protein) were separated by SDS-PAGE and then transferred to nitrocellulose. The blots were then incubated in 5% non-fat dry milk in phosphate-buffered saline (PBS) containing 0.1% Tween-20 (PBS-T) for 1 hour at room temperature. The blocking solution was then removed, and each blot was incubated overnight at 4° C in a solution of primary antibody diluted as indicated in PBS-T (anti-profilin, 1:1000; anti-profilin-1, 1:1000; anti-profilin-2a 1:1000; anti-chickadee 1:10; anti-actin 1:5000; anti-tubulin 1:3000). The blots were then washed three times in PBS-T and incubated with either anti-mouse IgG or anti-rabbit IgG horseradish peroxidase-conjugated secondary antibodies (Amersham, Buckinghamshire, England) diluted 1:5000 in PBS-T for 1 hour at room temperature. The membranes were washed, incubated with chemiluminescent detection reagents (ECL-Plus, Amersham, Buckinghamshire, England) for 5 minutes at room temperature, and exposed to autoradiography film (X-OMAT, Kodak, Rochester, NY) to visualize the labeled proteins. Exposed films were scanned at 600 dots per inch, and relative levels of protein expression were determined by analyzing the pixel intensity of the bands using NIH Image software (Version 1.62) and normalized to actin or tubulin. For each data set, the experimental conditions were then normalized to control samples so that control was always set to 100%. The results from three independent sets of tissues (each set consisting of a control, presymptomatic, grade 1, and a grade 4 specimen) were pooled and assessed by one-way ANOVA for statistical significance followed by Fisher’s probable least-squares difference post-hoc test. For cells and Drosophila, protein levels were analyzed by Student’s t-test for statistical significance.
Real-Time Quantitative PCR
Following induction for 1, 3, and 5 days, PC12-74Q cells were harvested from 6 cm dishes and centrifuged at 12,000 g for 10 minutes. The pellets were resuspended in 1 ml of Trizol (Invitrogen, Carlsbad, CA), and RNA was extracted. The RNA was then purified according to the company’s protocol using the RNAeasy Mini kit (Qiagen Valencia, CA, USA). The total RNA concentration of each sample was then quantified by absorbance at 260 nm using an Ultraspec 3100pro (GE Healthcare, Piscataway, NJ). A total of 1 µg of RNA was converted to cDNA using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). PCR reactions were run in triplicate on an ABI Prism 7100 Sequence Detection System using 1.25 ng of RNA and 12.5 µl of Taqman Universal PCR MasterMix (Roche Diagnostics, Indianapolis, IN) per reaction. PCR primer pairs specific to rat profilin-1, rat profilin-2a, and rat ribosomal protein, S18 were obtained from Applied Biosystems. S18 was used as an endogenous control, and profilin transcript levels were calculated using the ΔΔCt method as per Applied Biosystems specifications. Four independent RNA extractions were tested.
Pulse-Chase Protein Labeling
Following two days of induction, PC12-74Q cells were incubated in DMEM lacking cysteine and methionine for 2 h at 37 °C with 10% CO2. The cells were then pulse-labeled in cysteine/methionine-free media containing 100 µCi/µl 35S-labeled cysteine/methionine ProMix (GE Healthcare, Piscataway, NJ, USA) for 30 minutes. After labeling, the cells were washed once with culture medium containing a 10-fold excess of unlabeled methionine and cysteine (2 mM each) and incubated further for the indicated times in media alone or media containing MG132 (10 µM) or rapamycin (2 µg/ml). Cells were collected at the different time points, and lysed in 200 µl RIPA buffer (50 mM Tris—HCl, 1% (v/v) Nonidet P-40, 0.1% (w/v) deoxycholate, 0.1% (w/v) SDS, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1 mM PMSF and 1 × proteinase inhibitor, Complete Mini (Roche), and equal amounts of protein from the cleared lysates were immunoprecipitated with anti-profilin-1 and protein G-sepharose overnight at 4°C. The immune complexes were washed 3 times in RIPA buffer and separated on 16% SDS-PAGE. The gels were dried and exposed to a phosphorimager screen for visualization on a STORM phosphorimager system (Molecular Dynamics, Piscataway, NJ, USA).
Drosophila Stocks
The UAS-pros127Q flies used for these experiments have been previously described (Kazemi-Esfarjani and Benzer, 2002). The UAS-chic line was the generous gift of Lynn Cooley (Yale University) and has also been previously described (Hopmann and Miller, 2003). The GMR-gal4 and Elav-gal4 lines were supplied by K-T Min (NINDS). The flies were maintained on a standard mixture of corn meal, yeast, and agar at room temperature unless otherwise noted.
Lifespan Analysis
Newly eclosed flies were collected and reared on standard corn meal agar medium in a 25°C incubator. The flies were transferred to fresh media every 4 days, and the number of dead flies was counted daily. Differences in survival between the groups were examined using the log-rank test.
Behavioral Assay
Phototaxis measurement was performed as described previously(Min and Benzer, 1999). Shortly after eclosion, flies were placed in a 25°C incubator in the dark. On day 3, the flies were brought into a dark room and placed into a counter-current tube system. The flies were tapped gently to the bottom of the first tube and then the tube rack was placed horizontally with a light source at one end. The flies were allowed to move toward the light for 30 seconds, and those that did were then tapped down to the bottom of the next tube; this process was repeated six times. The number of flies in each tube was calculated as a percentage of the overall population tested. The mean ± standard error of five independent experiments was calculated for each fly strain; a total of 200 flies were tested per strain. Results between the groups were assessed by one-way ANOVA for statistical significance followed by Fisher’s probable least-squares difference post-hoc test for statistical difference between tubes for each fly strain.
Results
Levels of profilin-1 and -2a are reduced in HD brain and polyglutamine disease models
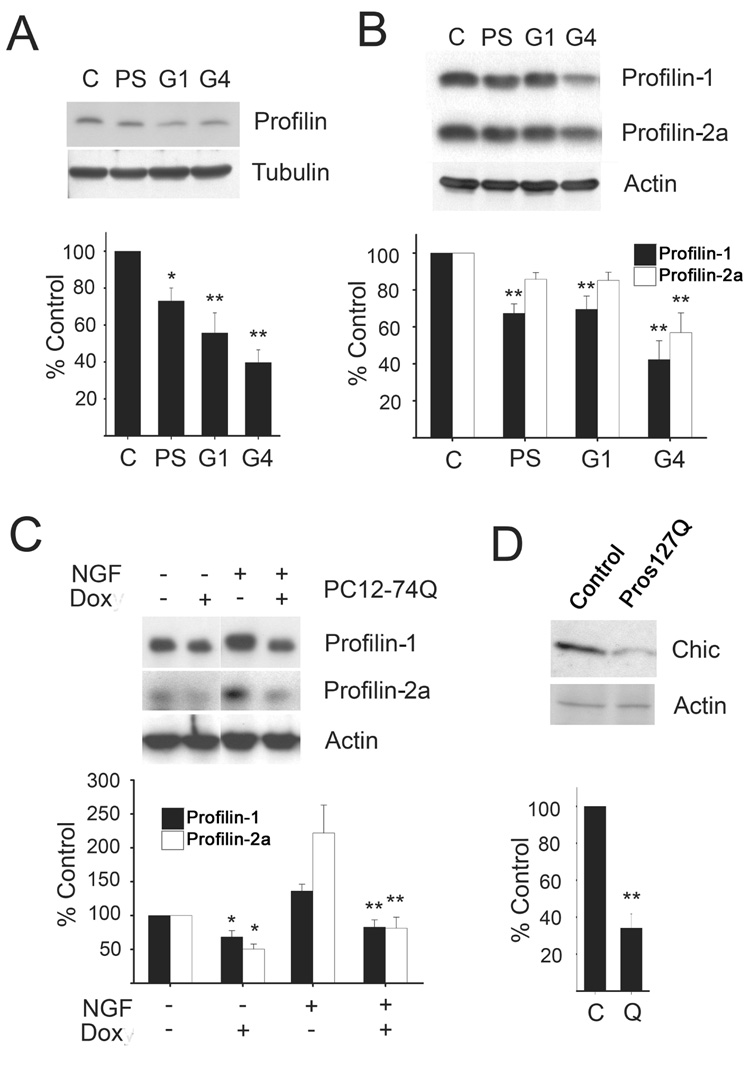
Since cytoskeletal alteration is believed to contribute to cellular toxicity in HD, we investigated the levels of profilin in HD brain tissue. For the quantification of profilin levels in brain tissue, antibodies that specifically recognize profilin-1, -2a, and total profilin were used (Fig. 1A–C). Protein lysates prepared from postmortem human cortical brain tissue (Brodmann area 9) of different HD patients and age matched controls were analyzed by immunoblotting. Fig. 1A shows that in comparison to control tissue (C) the total profilin levels in presymptomatic (PS), early stage (G1), and late stage (G4) HD brains were significantly reduced. This indicates that the total amount of profilin in HD patient brain decreases progressively with disease severity and histological grade. Presymptomatic and early stage HD patients (G1) had profilin levels of 73 ± 7% and 56 ± 11% of controls, respectively, while in late stage HD patients (G4) the total amount of profilin was further decreased to 40 ± 7% of controls.
Fig. 1. Decreased profilin in HD brain and polyglutamine models.
(A and B) Equal amounts of homogenates from presymptomatic (PS), grade 1 (G1) and grade 4 (G4) HD cortical brain tissue were resolved by SDS-PAGE, transferred to nitrocellulose and probed for total profilin, profilin-1, and profilin-2a; the membranes were stripped and reprobed for actin or tubulin. Following densitometry, relative intensities were normalized to actin or tubulin and calculated as percentage of control for three independent experiments. Profilin levels decreased with disease progression, beginning in presymptomatic samples. (C) Quantitation of profilin-1 and 2a levels in PC12 cells overexpressing mutant htt. Expression of EGFP-HD74Q was induced by the addition of doxycycline (Dox) to the culture medium. Treatment of the cells with nerve growth factor (NGF; 100 ng/ml) under low serum conditions (1%) induced differentiation and an increase in the expression of profilin-1 and -2a. Doxycycline-induced mutant htt expression led to significantly reduced profilin levels in both undifferentiated and differentiated cells. (D) Quantitation of the levels of chickadee (Chic), the Drosophila ortholog of mammalian profilin, in heads of 3 day old transgenic flies overexpressing the protein pros127Q (GMR-gal4/+; UAS-pros127Q/UAS-LacZ) and controls (+/+; UAS-pros127Q/UAS-pros127Q). *p<0.05; **p<0.01.
A similar result was obtained when the antibodies that detect profilin-1 and -2a were used for the quantification of the protein levels in the HD brains using immunoblots. In late stage HD brain tissue (G4) the amount of profilin-1 and -2a was reduced to 42 ± 10% and 57 ± 11% of controls, respectively (Fig. 1B).
Next, we investigated the levels of profilin-1 and -2a in a doxycycline-inducible PC12 cell model of HD (Wyttenbach et al., 2001). The polyglutamine-containing htt protein EGFP-HD74Q was overexpressed for 3 days, and protein extracts of induced and uninduced cells were analyzed by SDS-PAGE and immunoblotting. In mutant htt overexpressing PC12 cells, the levels of profilin-1 and -2a were significantly reduced (55 ± 22% and 45 ± 17% of control levels, respectively), consistent with the observations in HD brain tissue (Fig. 1C). A similar result was also obtained when the expression of mutant htt protein was induced for 3 days in PC12 cells differentiated with nerve growth factor (NGF). Differentiation of the cells with NGF under low serum conditions significantly elevated the basal levels of profilin-1 and -2a which would be expected in a neuronal-like cells compared to the undifferentiated state (Fig. 1C).
Finally, we examined whether the profilin levels are altered in a transgenic Drosophila model of polyglutamine disease (Kazemi-Esfarjani and Benzer, 2002). This model is similar to other polyglutamine models in that it contains a small protein fragment with an expansion of its normal polyglutamine tract; in this case 127 glutamines in the prospero protein. This prospero protein fragment is similar in size to the huntingtin fragments used in other Drosophila models, and like the others contains a polyproline domain close to the polyglutamine repeat. This model manifests toxicity similar to other polyglutamine models (reviewed in (Bonini and Fortini, 2003)) and offers the advantage of an easily monitored external phenotype. Overexpression of pros127Q in cells of the retina with the glass multimer repeat (GMR) promoter results in a progressive eye degeneration phenotype. Protein lysates prepared from the heads of 3 day old transgenic and control flies were analyzed by Western blotting. As shown in Fig. 1D, the levels of the Drosophila profilin ortholog, chickadee (Chic), were significantly reduced in the transgenic flies (34 ± 8%) compared to controls. Thus, profilin loss occurs in HD patient tissues, a cell model of HD, and a fly model of polyglutamine toxicity.
Mutant htt enhances profilin degradation
Analysis of SDS-insoluble aggregates with a filter trap assay showed the presence of huntingtin but not profilin after three days induction of mutant htt in PC12 cells (Supplemental Fig. 2). This suggests that loss of profilin is probably not due to incorporation into SDS-resistant htt aggregates. To investigate whether the reduction of profilin-1 and -2a protein levels in PC12 cells overexpressing EGFP-HD74Q is due to a decrease in transcription, we measured profilin-1 and -2a mRNA levels by quantitative real-time PCR (Fig. 2A). Following 1, 3, and 5 days of mutant htt induction, mRNA levels of profilin-1 did not change from control, uninduced levels. Profilin-2a mRNA levels were increased rather than decreased, possibly as a compensatory mechanism, in cells expressing mutant htt. This indicates that the reduction of profilin protein is not caused by decreased transcription.
Fig. 2. Mutant htt enhances profilin degradation.
(A) Quantitation of profilin-1 and-2a transcript levels in PC12 cells overexpressing EGFP-HD74Q (induced cells) and in uninduced control cells. The ribosomal protein S18 was used as an internal control. The profilin-1 and -2a mRNA expression levels were not significantly different between cells expressing mutant htt and controls. (B) Pulse-chase experiments for assessing profilin-1 turnover in PC12 cells overexpressing the protein EGFP-HD74Q two days after induction (74Q+Dox). Uninduced cells were used as a control (74Q). Induced and uninduced PC12 cells treated with the proteasome inhibitor MG132 were also analyzed. Doxycycline-induced mutant htt led to a rapid decrease in profilin levels, which was blocked by MG132.
We then used pulse-chase experiments to determine whether profilin turnover is altered by mutant htt expression in PC12 cells (Fig. 2B). In uninduced PC12 cells, profilin-1 remained stable for up to 24 hours. In cells expressing EGFP-HD74Q, however, degradation of radiolabelled profilin-1 increased markedly. Two hours after profilin-1 pulse labeling, the protein was barely detectable. This indicates that mutant htt strongly stimulates profilin-1 clearance. To test whether proteasome activity is necessary for the enhanced degradation of profilin-1, the pulse-chase experiments were repeated after addition of the proteasome inhibitor MG132. As shown in Fig. 2B, clearance of profilin-1 was completely blocked in MG132 treated cells overexpressing mutant htt, suggesting that proteasome activity is indeed responsible for enhanced profilin-1 turnover. No such effect was seen after treatment with rapamycin, an inhibitor of autophagy (data not shown).
Mutant htt reduces F/G actin ratio
Since profilin levels are reduced after induction of mutant htt, we sought to determine whether actin dynamics are also affected. This was done by analyzing the relative levels of polymerized (F) and monomeric (G) actin in the HD PC12 cell model. Using actin sedimentation analysis we found that in comparison to uninduced cells the F/G actin ratio was markedly reduced 48 hours following induction of mutant htt. In contrast, the F/G actin ratio was unchanged after induction of wild-type htt (Fig. 3A, B). One possible explanation for this shift in F/G actin ratios could be the initiation of apoptosis by mutant htt. However, induction of mutant htt in these cell lines is not reported to cause significant cell death at this time point (48 hrs.), which we confirmed using propidium iodide staining and flow cytometry analysis (data not shown and (Wyttenbach et al., 2001)). This data indicates that mutant htt expression alters actin dynamics. We next examined whether the observed change in F/G actin ratio is attributable to the loss of profilin. We found that replacement of profilin by transfection with exogenous profilin-1 following induction of htt resulted in partial restoration of the F/G actin ratio (Fig. 3C). Together, these data indicate an effect of mutant htt on actin dynamics, likely through the increased degradation of profilin.
Fig. 3. Mutant Htt expression reduces F/G actin ratio.
(A) Representative Western blot of F-actin and G-actin levels in uninduced PC12 cells and cells induced to express wild-type or mutant htt. Induction of mutant htt resulted in a decrease in the F/G actin ratio (B) Quantitation of data in (A); Data represent mean ± standard error from four independent experiments. (C) Overexpression of profilin by transfecting exogenous profilin-1 after induction of mutant htt partially restores the F/G actin ratio; n = 3. *P < 0.05. Data represent mean ± standard error. A representative Western blot of endogenous and transfected profilin levels is shown in inset. (D) Induction of mutant htt results in a 10 increase in cell death by PI staining and flow cytometry. Overexpression of profilin-1 blocks mutant htt toxicity. Data represent mean ± standard error; n = 3. *P < 0.05.
Profilin reduces mutant htt toxicity in HD model cells
Given that restoration of profilin reversed the shift in F/G actin ratios brought about by expression of mutant htt, we sought to determine whether mutant htt toxicity was also affected by restoring profilin. We observed a 10% increase in cell death (at 72 hrs.) in cells expressing mutant htt compared to uninduced cells, as measured by propidium Iodide staining and flow cytometry (Fig. 3D). Overexpressing profilin-1 did not affect baseline cell death in uninduced cells but significantly mitigated mutant htt toxicity 3 days following induction (p<0.05). These findings are consistent with profilin rescue of mutant htt toxicity.
Restoration of profilin reduces polyglutamine toxicity in Drosophila
Since profilin loss occurs early in HD tissues and models and restoration abrogates toxicity in vitro, we investigated whether increased profilin expression ameliorates polyglutamine toxicity in flies. When the pros127Q fly line was crossed with a line carrying a transgene for UAS-chickadee, the chickadee levels were restored from 34 ± 8% to 86 ± 9% of control levels (Fig. 4A). We then asked whether this restoration of chickadee levels could reduce polyglutamine-induced degeneration. One day after eclosion, the eyes of pros127Q flies showed mild depigmentation, which became more severe by day 10 (Fig. 4B). In contrast, eyes of pros127Q flies overexpressing chickadee were nearly normal at day 1 and had much less depigmentation by day 10. Flies expressing LacZ or chickadee alone were normal at days 1 and 10. To test whether the preservation of eye pigmentation is associated with functional restoration, we performed a counter-current phototaxis test that assesses the number of flies that repeatedly move toward a light source when placed in glass tubes in successive trials (Min and Benzer, 1999). Flies with intact vision move toward light in each trial, whereas flies that do not see light distributed randomly. Three days after eclosion, the majority of control flies (GMR-gal4) responded to light and completed the test by reaching the final tube (Fig. 4C). In contrast, flies expressing pros127Q showed random distribution across the tubes, indicating visual impairment. Polyglutamine-expressing flies rescued with chickadee demonstrated improved eye function, with significantly more flies in the final tube. To further assess whether profilin mitigates polyglutamine toxicity, the fly lines were crossed with flies expressing the neuron-specific driver Elav-gal4. Control flies expressing LacZ in the central and peripheral nervous system under Elav-gal4 regulation remained alive throughout the 30 day observation period (Fig. 4D). However, expression of pros127Q using Elav-gal4 regulation resulted in premature death with an average lifespan of 19.5 days. Elav-gal4 driven co-expression of chickadee with pros127Q significantly extended survival to an average lifespan of 21.5 days (p<0.05). Similar life span extension was observed in another HD fly model crossed with the chickadee overexpressing transgenic flies (M. Diamond, personal communication). Thus, restoration of chickadee partially rescues polyglutamine-induced toxicity in vivo.
Fig. 4. Expression of the profilin ortholog, chickadee, ameliorates olyglutamineinduced eye degeneration in Drosophila.
(A) Overexpression of pros127Q, but not LacZ, reduces the levels of chickadee (Chic) in the heads of 3 day old adult flies. Transgenic expression of Chic in pros127Q flies restores Chic to nearly normal levels: C, fly heads overexpressing LacZ, Q, fly heads overexpressing only prosQ127, and R, fly heads overexpressing prosQ127 and Chic. (B) Representative Drosophila eyes overexpressing Chic (GMR-gal4/+; UAS-chic/+), LacZ (GMR-gal4/+; UAS-LacZ/+), pros127Q+LacZ (GMR-gal4/+; UAS-pros127Q/UASLacZ) or pros127Q + Chic (GMR-gal4/+; UAS-pros127Q/UAS-chic). The eye degeneration was less pronounced at both time points in flies overexpressing both pros127Q and Chic, indicating that increased levels of Chic restore the eye phenotype. (C) Overexpression of Chic in pros127Q flies partially restores phototactic behavior. In the countercurrent distribution experiment, the population of flies is fractionated according to the number of positive responses of moving toward light in repeated trials. Pros127Q + Chic flies (GMR-gal4/+; UAS-pros127Q/UAS-chic) performed significantly better than those expressing pros127Q (GMR-gal4/+; UAS-pros127Q/UAS-LacZ) but not as well as controls (GMR-gal4/GMR-gal4;+/+). (D) Polyglutamine-induced decreased lifespan (Elav/x; UAS-pros127Q/UAS-LacZ) is significantly prolonged in pros127Q flies overexpressing Chic (Elav/x; UAS-pros127Q/UAS-chic). *p<0.05; **p<0.01. The mean ± standard error of five independent experiments was calculated for each fly strain; a total of 200 flies were tested per strain.
Discussion
This study provides evidence that mutant htt enhances degradation of profilin, a critical G-actin binding protein, and suggests a link between htt pathogenesis and actin dynamics. Profilin plays a crucial role in regulating actin polymerization by catalyzing the exchange of actin-bound ADP to ATP, priming the monomer for polymerization (Selden et al., 1999; Da Silva et al., 2003). As one of the major proteins in the cell, actin plays important structural and functional roles, such as maintaining cell morphology, cell motility, exo- and endocytosis, and cell division (Pollard and Cooper, 1986; Singer, 1992; Olson et al., 1995). Apart from its essential role in maintaining normal mammalian cell function and development (Witke et al., 2001), profilin plays vital roles in specialized neuronal functions such as trafficking of neurotransmitter vesicles (Wang et al., 1999), neurite outgrowth (Da Silva et al., 2003), and dendritic spine formation and stability (Ackermann and Matus, 2003).
Our finding of a progressive loss of profilin beginning in presymptomatic HD gene carriers coincides with early changes observed in HD mouse models, such as altered neurotransmitter release (Nicniocaill et al., 2001) and loss of dendritic spines (Guidetti et al., 2001), which are indicative of changes in actin dynamics. The effects of profilin loss have been reported in various systems. For example, deletion of both profilin-1 and profilin-2 disrupts motility and cytokinesis of Dictyostelium amoebae while actin cytoskeleton formation is substantially inhibited in profilin-deficient cells (Haugwitz et al., 1994; Ding et al., 2006). Loss of the Drosophila ortholog of profilin, chickadee, results in cell proliferation and migration defects followed by late embryonic lethality (Verheyen and Cooley, 1994), and loss of profilin-1 in mice is lethal at very early stages of development (Witke et al., 2001). This early stage embryonic lethality has made it difficult to ascertain in vivo the specific neurological role of profilin-1. In contrast, profilin-2 knockout mice are viable with normal brain development and anatomy (Pilo Boyl et al., 2007). In fact, synaptic plasticity and learning and memory are essentially spared. Loss of profilin-2, however, results in hyperactive synaptic vesicle release in the striatal spiny neurons, which indicates a role in regulating and restricting synaptic vesicle release. To fully understand the role of profilin in the central nervous system and possibly provide further insight into the connection between profilin loss and HD pathophysiology, conditional profilin-1 knockout animals would need to be generated to bypass its critical developmental requirement.
Although the mechanism by which mutant htt facilitates profilin degradation remains unclear, the data reported here supports involvement of the ubiquitin proteasome system (UPS). Profilin is normally a very stable protein, and the marked increase in its turnover in the presence of mutant htt is quite remarkable. It is likely that the increased profilin turnover we observed with mutant htt is a result of selective degradation related to htt-profilin interaction rather than a general increase in UPS activity. Although there is evidence of altered UPS function in HD (Ortega et al., 2007), previous studies of the levels of synaptic and cytoskeletal protein elements in HD patient tissue and cell models have reported that very few of these proteins are altered (Edwardson et al., 2003; DiProspero et al., 2004).
Profilins regulate actin polymerization. In cultured cells we show that expression of mutant htt reduces profilin levels and thus alters the ratio of polymerized to unpolymerized (F/G) actin. Abrogating profilin loss by transient transfection partially restores the F/G actin ratios indicating that the observed changes in F/G actin ratios are due, at least in part, to loss of profilin. The effect of profilin on actin dynamics is complex, given that it can either sequester actin or promote actin polymerization. In this study, profilin was returned to baseline levels when we overexpressed profilin-1 in cells expressing mutant htt. Given that actin and profilin interact at a 1:1 ratio and profilin levels are usually less than 10% of actin (Babcock and Rubenstein, 1993), the stoichiometry is consistent with increased actin polymerization rather than G-actin sequestration as the cause for the restoration of F/G actin ratios.
There is growing evidence implicating the cytoskeleton in neurodegenerative diseases. For example, rod-like inclusions containing actin-depolymerization factor (ADF) and cofilin were found in hippocampal and cortical neurites of the post-mortem brains of Alzheimer's patients (AD) (Minamide et al., 2000). In addition hyperphosphorylated forms of the microtubule-associated protein tau accumulate in AD and are associated with accumulation of F-actin and the formation of actin-rich rods in models of tauopathy (Fulga et al., 2007)). Tau directly interacts with actin and this interaction is thought to alter the actin cytoskeleton and mediate tau-induced neurotoxicity in AD. The Drosophila ortholog of ataxin-2, a polyglutamine disease protein mutated in spinocerebellar ataxia type 2 (SCA2), appears to regulate actin formation (Satterfield et al., 2002). As in HD, an early event in SCA2 pathogenesis is a change in dendritic architecture (Huynh et al., 2000). Early, progressive loss of profilin may alter actin dynamics and disrupt neuronal function in SCA2 as well as in HD.
Here we report a marked reduction of profilin in patient tissue, an HD cell model and a fly model of polyglutamine disease. In cultured cells profilin is more rapidly degraded in the presence of mutant htt, resulting in a shift in the F/G actin ratio. Restoring profilin not only reverses the changes in F/G actin ratios but also mitigates the cellular toxicity of mutant htt. Furthermore, in a fly model of polyglutamine disease we show that returning profilin to almost baseline levels can reduce the toxicity of the mutant polyglutamine protein. The amelioration of polyglutamine toxicity may be due to the restoration of a depleted protein that mediates a broad range of cellular activities on which neurons are particularly dependent. Additionally, changes in actin dynamics may affect aggregation and toxicity of polyglutamine proteins. (Pollitt et al., 2003). Our data may provide further insight into HD pathogenesis, and highlights an alternative pathway for therapeutic intervention.
Supplementary Material
Fig. S1. Specificity of isoform-directed anti-profilin antibodies. SDS-PAGE gels of recombinant human profilins-1 and -2a were transferred onto PVDF-membranes which were then probed with polyclonal antibodies raised against profilin-1 and 2a-derived peptides. The anti-profilin-1 antibody recognized only recombinant profilin-1, and the anti-profilin-2a antibody recognized only recombinant profilin-2a.
Fig. S2. Profilin is not detected in htt aggregates. Cell lysates containing 150 µg of total protein from PC12 cells induced to express EGFP-HD23Q and EGFP-HD74Q were used for filtration on cellulose acetate membranes (Scherzinger et al., 1997). SDS-resistant htt aggregates were detected with anti-GFP and profilin detected with a profilin-1 specific antibody.
Acknowledgements
The authors wish to thank Erich Wanker and Herwig Schüler (Max Delbrueck Center for Molecular Medicine (MDC)) for characterizing the selectivity of the profilin antibodies and Dan Tagle (NINDS) for helpful guidance and suggestions. We also would like to thank David Rubinsztein (MRC Cambridge) for the kind gift of the inducible PC12 cell line and Lynn Cooley (Yale University) for the generous gift of the transgenic chickadee Drosophila line. Elav-gal4, GMR-gal4, and the UAS-LacZ fly lines were kindly provided by Kyung-Tai Min (NINDS). The monoclonal antibody to Drosophila profilin (chic1J) developed by Lynn Cooley was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This project was supported by intramural funds from the NINDS, an NINDS Competitive Fellowship (BGB) and by ClinPRAT support from NIGMS (to NAD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6:1194–1200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- Babcock G, Rubenstein PA. Control of profilin and actin expression in muscle and nonmuscle cells. Cell Motil Cytoskeleton. 1993;24:179–188. doi: 10.1002/cm.970240305. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Fortini ME. Human neurodegenerative disease modeling using Drosophila. Annu Rev Neurosci. 2003;26:627–656. doi: 10.1146/annurev.neuro.26.041002.131425. [DOI] [PubMed] [Google Scholar]
- Da Silva JS, Medina M, Zuliani C, Di Nardo A, Witke W, Dotti CG. RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol. 2003;162:1267–1279. doi: 10.1083/jcb.200304021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Gareus R, Kwiatkowski D, Witke W. Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J Cell Sci. 2000;113(Pt 21):3795–3803. doi: 10.1242/jcs.113.21.3795. [DOI] [PubMed] [Google Scholar]
- Ding Z, Lambrechts A, Parepally M, Roy P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J Cell Sci. 2006;119:4127–4137. doi: 10.1242/jcs.03178. [DOI] [PubMed] [Google Scholar]
- DiProspero NA, Chen EY, Charles V, Plomann M, Kordower JH, Tagle DA. Early changes in Huntington's disease patient brains involve alterations in cytoskeletal and synaptic elements. J Neurocytol. 2004;33:517–533. doi: 10.1007/s11068-004-0514-8. [DOI] [PubMed] [Google Scholar]
- Edwardson JM, Wang CT, Gong B, Wyttenbach A, Bai J, Jackson MB, Chapman ER, Morton AJ. Expression of mutant huntingtin blocks exocytosis in PC12 cells by depletion of complexin II. J Biol Chem. 2003;278:30849–30853. doi: 10.1074/jbc.M304615200. [DOI] [PubMed] [Google Scholar]
- Fulga TA, Elson-Schwab I, Khurana V, Steinhilb ML, Spires TL, Hyman BT, Feany MB. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat Cell Biol. 2007;9:139–148. doi: 10.1038/ncb1528. [DOI] [PubMed] [Google Scholar]
- Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Guidetti P, Charles V, Chen EY, Reddy PH, Kordower JH, Whetsell WO, Jr, Schwarcz R, Tagle DA. Early degenerative changes in transgenic mice expressing mutant huntingtin involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp Neurol. 2001;169:340–350. doi: 10.1006/exnr.2000.7626. [DOI] [PubMed] [Google Scholar]
- Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- Haugwitz M, Noegel AA, Karakesisoglou J, Schleicher M. Dictyostelium amoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell. 1994;79:303–314. doi: 10.1016/0092-8674(94)90199-6. [DOI] [PubMed] [Google Scholar]
- Hopmann R, Miller KG. A balance of capping protein and profilin functions is required to regulate actin polymerization in Drosophila bristle. Mol Biol Cell. 2003;14:118–128. doi: 10.1091/mbc.E02-05-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu E, Chen Z, Fredrickson T, Zhu Y. Molecular cloning and characterization of profilin-3: a novel cytoskeleton-associated gene expressed in rat kidney and testes. Exp Nephrol. 2001;9:265–274. doi: 10.1159/000052621. [DOI] [PubMed] [Google Scholar]
- Huynh DP, Figueroa K, Hoang N, Pulst SM. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat Genet. 2000;26:44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Benzer S. Suppression of polyglutamine toxicity by a Drosophila homolog of myeloid leukemia factor 1. Hum Mol Genet. 2002;11:2657–2672. doi: 10.1093/hmg/11.21.2657. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Bruns GA. Human profilin. Molecular cloning, sequence comparison, and chromosomal analysis. J Biol Chem. 1988;263:5910–5915. [PubMed] [Google Scholar]
- Lambrechts A, Braun A, Jonckheere V, Aszodi A, Lanier LM, Robbens J, Van Colen I, Vandekerckhove J, Fassler R, Ampe C. Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol Cell Biol. 2000;20:8209–8219. doi: 10.1128/mcb.20.21.8209-8219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min KT, Benzer S. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science. 1999;284:1985–1988. doi: 10.1126/science.284.5422.1985. [DOI] [PubMed] [Google Scholar]
- Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol. 2000;2:628–636. doi: 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- Neuhoff H, Sassoe-Pognetto M, Panzanelli P, Maas C, Witke W, Kneussel M. The actin-binding protein profilin I is localized at synaptic sites in an activity regulated manner. Eur J Neurosci. 2005;21:15–25. doi: 10.1111/j.1460-9568.2004.03814.x. [DOI] [PubMed] [Google Scholar]
- Nicniocaill B, Haraldsson B, Hansson O, O'Connor WT, Brundin P. Altered striatal amino acid neurotransmitter release monitored using microdialysis in R6/1 Huntington transgenic mice. Eur J Neurosci. 2001;13:206–210. doi: 10.1046/j.0953-816x.2000.01379.x. [DOI] [PubMed] [Google Scholar]
- Obermann H, Raabe I, Balvers M, Brunswig B, Schulze W, Kirchhoff C. Novel testis-expressed profilin IV associated with acrosome biogenesis and spermatid elongation. Mol Hum Reprod. 2005;11:53–64. doi: 10.1093/molehr/gah132. [DOI] [PubMed] [Google Scholar]
- Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- Ortega Z, Diaz-Hernandez M, Lucas JJ. Is the ubiquitin-proteasome system impaired in Huntington's disease? Cell Mol Life Sci. 2007;64:2245–2257. doi: 10.1007/s00018-007-7222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Pilo Boyl P, Di Nardo A, Mulle C, Sassoe-Pognetto M, Panzanelli P, Mele A, Kneussel M, Costantini V, Perlas E, Massimi M, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. Embo J. 2007;26:2991–3012. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- Pollitt SK, Pallos J, Shao J, Desai UA, Ma AA, Thompson LM, Marsh JL, Diamond MI. A rapid cellular FRET assay of polyglutamine aggregation identifies a novel inhibitor. Neuron. 2003;40:685–694. doi: 10.1016/s0896-6273(03)00697-4. [DOI] [PubMed] [Google Scholar]
- Satterfield TF, Jackson SM, Pallanck LJ. A Drosophila homolog of the polyglutamine disease gene SCA2 is a dosage-sensitive regulator of actin filament formation. Genetics. 2002;162:1687–1702. doi: 10.1093/genetics/162.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Selden LA, Kinosian HJ, Estes JE, Gershman LC. Impact of profilin on actin-bound nucleotide exchange and actin polymerization dynamics. Biochemistry. 1999;38:2769–2778. doi: 10.1021/bi981543c. [DOI] [PubMed] [Google Scholar]
- Singaraja RR, Hadano S, Metzler M, Givan S, Wellington CL, Warby S, Yanai A, Gutekunst CA, Leavitt BR, Yi H, et al. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum Mol Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- Singer SJ. Intercellular communication and cell-cell adhesion. Science. 1992;255:1671–1677. doi: 10.1126/science.1313187. [DOI] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group, H. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120:717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Wang X, Kibschull M, Laue MM, Lichte B, Petrasch-Parwez E, Kilimann MW. Aczonin, a 550-kD putative scaffolding protein of presynaptic active zones, shares homology regions with Rim and Bassoon and binds profilin. J Cell Biol. 1999;147:151–162. doi: 10.1083/jcb.147.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanker EE, Rovira C, Scherzinger E, Hasenbank R, Walter S, Tait D, Colicelli J, Lehrach H. HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum Mol Genet. 1997;6:487–495. doi: 10.1093/hmg/6.3.487. [DOI] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci U S A. 2001;98:3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach A, Swartz J, Kita H, Thykjaer T, Carmichael J, Bradley J, Brown R, Maxwell M, Schapira A, Orntoft TF, et al. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington's disease. Hum Mol Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Specificity of isoform-directed anti-profilin antibodies. SDS-PAGE gels of recombinant human profilins-1 and -2a were transferred onto PVDF-membranes which were then probed with polyclonal antibodies raised against profilin-1 and 2a-derived peptides. The anti-profilin-1 antibody recognized only recombinant profilin-1, and the anti-profilin-2a antibody recognized only recombinant profilin-2a.
Fig. S2. Profilin is not detected in htt aggregates. Cell lysates containing 150 µg of total protein from PC12 cells induced to express EGFP-HD23Q and EGFP-HD74Q were used for filtration on cellulose acetate membranes (Scherzinger et al., 1997). SDS-resistant htt aggregates were detected with anti-GFP and profilin detected with a profilin-1 specific antibody.