Abstract
Hyper–immunoglobulin E syndrome (HIES) is a primary immune deficiency characterized by abnormal and devastating susceptibility to a narrow spectrum of infections, most commonly Staphylococcus aureus and Candida albicans. Recent investigations have identified mutations in STAT3 in the majority of HIES patients studied. Despite the identification of the genetic cause of HIES, the mechanisms underlying the pathological features of this disease remain to be elucidated. Here, we demonstrate a failure of CD4+ T cells harboring heterozygous STAT3 mutations to generate interleukin 17–secreting (i.e., T helper [Th]17) cells in vivo and in vitro due to a failure to express sufficient levels of the Th17-specific transcriptional regulator retinoid-related orphan receptor γt. Because Th17 cells are enriched for cells with specificities against fungal antigens, our results may explain the pattern of infection susceptibility characteristic of patients with HIES. Furthermore, they underscore the importance of Th17 responses in normal host defense against the common pathogens S. aureus and C. albicans.
Appropriate immune responses to particular pathogens are orchestrated by distinct T cell populations with relatively stable cytokine expression profiles that differentiate during antigen-specific clonal proliferation. Such differentiation of CD4+ T cells is determined by signals provided by components of the innate immune system and is subsequently stabilized by induction and repression of key transcriptional regulators (1). Recently, IL-17–producing CD4+ T cells (Th17) have been identified as the key mediators of inflammation and tissue damage in several animal models of human diseases (for review see references 1 and 2). The commitment of naive murine CD4+ T cells to the Th17 lineage requires exposure to TGF-β and either IL-6 (1, 3, 4) or IL-21 (2, 5, 6). These cytokines act in a STAT3-dependent manner to induce expression of retinoid-related orphan receptor γt (RORγt), the critical transcription factor necessary for the development of Th17 cells (7–10), which subsequently induces a set of effector molecules, including IL-17, IL-21, IL-22, and IL-23R plus several chemokines and other defense molecules (1, 2). In contrast to mice, much less is known about the requirements for generating human Th17 cells. Indeed, although there is consensus that TGF-β is not required for this process, and is actually inhibitory, controversy exists as to whether IL-6 or IL-23, in combination with IL-1β, is the predominant cue that mediates commitment of human naive CD4+ T cells to the Th17 lineage (11, 12). Given these differences, it is important to dissect further the molecular requirements for the development of human Th17 cells.
Although inappropriate Th17 differentiation helps explain autoimmune disease and inflammation, the physiological function of Th17 cells appears to lie in host protection against specific pathogens (1, 2). Based on the anatomical distribution of Th17 cells and the capacity of IL-17 to regulate granulopoiesis, neutrophil chemotaxis, and production of microbiocidal peptides by epithelium in mice, it is likely that Th17 cells confer immunity to pathogens on mucosal and possibly other epithelial surfaces (8, 13). For instance, Th17 cells appear to have a predominant role in antifungal immune responses in both humans and mice (14–16).
Hyper-IgE (HIES), or Job's, syndrome (OMIM 147060) is a primary immune deficiency characterized by abnormal susceptibility to a narrow spectrum of infections, most commonly Staphylococcus aureus and Candida albicans (17, 18). Patients with HIES often suffer from mucocutaneous candidiasis, whereas local and invasive S. aureus infections lead to persistent eczematoid eruptions, recurrent skin and joint abscesses, and lung damage. In addition to recurrent tissue-destructive infections, the HIES phenotype includes coarse facies, osteopenia resulting in fractures after relatively minor trauma, scoliosis and craniosynostosis, delayed shedding of the primary dentition, and short stature (18). Patients also exhibit dramatic increases in serum IgE (>10 times normal) (17, 18). Recently, heterozygous missense mutations and short deletions in STAT3 have been identified in the majority of HIES patients studied (19, 20). Despite the identification of the genetic cause of HIES, the mechanisms underlying the pathological features of this disease remain to be elucidated. Based on the observations that (a) defects in immunity to both Candida and S. aureus are prominent features of HIES, especially at epithelial surfaces (18), (b) human Th17 cells have specificities for fungal antigens, including C. albicans (14), (c) heterozygous STAT3 mutations are responsible for HIES in the majority of cases (19, 20), and (d) the development of murine Th17 cells is compromised in mice lacking expression of STAT3 in their CD4+ T cells (7, 9, 10), we hypothesized that STAT3 mutations conferred specific susceptibility in humans with HIES to infection via an inability to generate Th17 cells.
RESULTS AND DISCUSSION
Characteristics of HIES patients and STAT3 mutations
Five unrelated patients with HIES were identified according to their clinical phenotype (Table I). One patient (HIES4) had a family history of HIES (her mother had succumbed to infection but was not available for this analysis). All patients had a history of recurrent infection, including invasive S. aureus and mucocutaneous candidiasis. In addition, all patients had features typical of the autosomal-dominant form of HIES, with abnormalities of the skeleton, teeth, and soft tissue (Table I). Phenotype scores ranged from 26 to 71 according to the proposed HIES score (an age-sensitive scoring with HIES being most likely in patients with a score >40) (21).
Table I.
Characteristics of HIES patients
| HIES 1 | HIES 2 | HIES 3 | HIES 4 | HIES 5 | |
|---|---|---|---|---|---|
| Age | 18 | 18 | 15 | 7 | 42 |
| Sex | M | M | M | F | F |
| Infection | |||||
| Candidiasis | Yes | No | Yes | Yes | Yes |
| S. aureus pneumonia | Yes | Yes | No | No | Yes |
| S. aureus abscesses | Yes | Yes | Yes | Yes | Yes |
| Other serious infection | No | No | No | No | No |
| Parenchymal lung damage | Yes | Yes | No | No | Yes |
| Characteristic face | Yes | Yes | Yes | Yes | Yes |
| Retained primary teeth | Yes | Yes | Yes | — | Yes |
| Fractures | Yes | Yes | Yes | No | No |
| Phenotype score (21) | 71 | 50 | 54 | 26 | 63 |
| IgE (kIU/L) | 18,314 | 37,620 | 2,545 | 8,075 | 7,491 |
| Genotype | V432M | R382Q | R382Q | H437P | Q644P |
| Affected STAT3 domain | DNABD | DNABD | DNABD | DNABD | SH2 |
DNABD, DNA binding domain; SH2, src homology 2 domain.
Genomic DNA was isolated from each patient, and exons 13–21 of STAT3, which encompass the DNA binding and SH2 domains, were sequenced. Heterozygous missense mutations were identified in all five patients. Four harbored mutations in the DNA binding domain (R382Q [n = 2], V432M, and H437P) and one had a mutation in the SH2 domain (Q644P; Table I). The R382 residue is a common site of mutation, accounting for ∼30% (18/58) of reported HIES genotypes, with R382Q in 8 of those 18 (19, 20). Most of the mutations we identified have previously been demonstrated to impair the ability of STAT3 to translocate to the nucleus and activate transcription of STAT3 target genes (19, 20, 22).
Heterozygous STAT3 mutations abrogate the generation of Th17 cells in vivo
To test the hypothesis that missense mutations in STAT3 would confer a defect in human Th17 cell differentiation, CD4+ T cells from HIES patients were examined and compared with those from matched normal controls. Flow cytometric analysis revealed normal development of CD4+ T cells, with comparable proportions of naive (CD45RA+ CD27+), central memory (CD45RA− CD27+) and effector memory (CD45RA− CD27−) cells in HIES patients and healthy controls (not depicted).
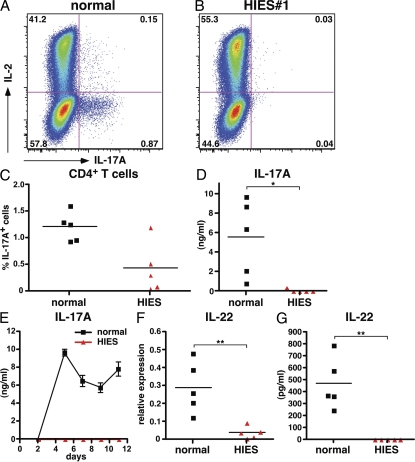
To investigate whether Th17 cells were present within the circulating T cell population of HIES patients in vivo, total CD4+ T cells were stimulated in vitro with anti-CD3 mAb, anti-CD28 mAb, and IL-2 for 4–5 d. The proportion of Th17 cells within the population was assessed by intracellular staining for IL-17 after restimulation with PMA and ionomycin. A small but consistent population of IL-17–expressing cells was detected in the CD4+ T cells examined for all healthy donors (1.2%; n = 5; Fig. 1, A and C). In contrast, there was a significant reduction in IL-17+ cells from the cultures of stimulated STAT3 mutant HIES CD4+ T cells (0.44%; n = 5; P < 0.05; Fig. 1, B and C). Indeed, no IL-17+ cells could be detected after stimulation of CD4+ T cells from two HIES patients (Fig. 1, B and C). These findings were corroborated by examination of IL-17 secretion by stimulated CD4+ T cells. Control CD4+ T cells secreted abundant quantities of IL-17, whereas IL-17 was undetectable in cultures of CD4+ T cells from four of five HIES patients (Fig. 1 D). In the fifth patient, the concentration of IL-17 was >20-fold lower than the average amount produced by control CD4+ T cells (Fig. 1 D).
Figure 1.
HIES patients have a defect in the generation of IL-17–producing CD4+ T cells in vivo. Intracellular expression of IL-17A and IL-2 in purified CD4+ T cells from a healthy donor (A) and a HIES patient (B) activated for 5 d (see Materials and methods). (C) The frequency of IL-17A+ cells, determined by intracellular cytokine expression, and (D) IL-17A secretion by CD4+ T cells from five healthy donors and five HIES patients that had been stimulated with anti-CD3 mAb, anti-CD28 mAb, and IL-2 for 4–5 d, as determined by ELISA. Each symbol represents the value from an individual donor or patient, and the line represents the mean. (E) Time course of IL-17A secretion by CD4+ T cells isolated from a normal donor and HIES patient. Similar results were obtained when CD4+ T cells from all HIES patients were examined over a culture period that continued for up to 11 d. (F and G) Expression of IL-22 mRNA (relative to housekeeping genes; F) and secreted protein (G) by normal and HIES CD4+ T cells was determined after 2 and 5 d, respectively, of stimulation with CD3/CD28/IL-2. *, P < 0.05; **, P < 0.01.
To exclude a difference in the rate of IL-17 secretion by HIES CD4+ T cells, production of IL-17 by activated CD4+ T cells from healthy donors and HIES patients was measured over an 11-d time course. In cultures of healthy CD4+ T cells, IL-17 was first detected after 4 d, peaked on days 5–6, and remained stable or declined at later times (Fig. 1 E). In contrast, no IL-17 was detected in the supernatants of HIES CD4+ T cells at any of the time points examined (Fig. 1 E). We also assessed expression of IL-22, a Th17-derived cytokine thought to be important in epithelial immunity (23, 24). IL-22 mRNA (Fig. 1 F) was significantly reduced in activated CD4+ T cells from HIES patients compared with healthy controls (P < 0.01). Furthermore, there was a complete absence of IL-22 secretion in cultures of activated CD4+ T cells (Fig. 1 G). This finding demonstrates that production of both IL-17 and IL-22 by in vivo–generated Th17 cells is dependent on functional STAT3 protein and confirm that IL-22 is a signature Th17 cytokine.
HIES CD4+ T cells fail to up-regulate expression of RORγt, the Th17 lineage–specific transcriptional regulator
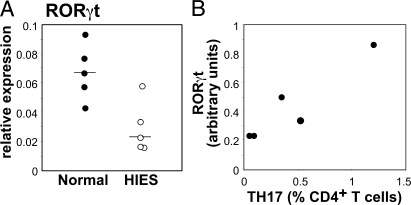
Differentiation of Th17 cells is determined by RORγt (8). Because induction of this key transcription factor in mice is regulated by STAT3 (7, 9, 10), we tested for a possible defect in the expression of RORγt mRNA in HIES CD4+ T cells. We found a fourfold reduction of RORγt message in stimulated HIES CD4+ T cells compared with control CD4+ T cells (Fig. 2 A).
Figure 2.
Deficiency of RORγt mRNA in HIES patients. (A) RORγt mRNA expression in CD4+ cells activated for 2 d with CD3/CD28/IL-2 in five HIES patients and five normal controls. (B) RORγt mRNA expression after 2 d of culture is proportional to the number of Th17 cells as determined by intracellular cytokine expression after 4–5 d.
Given that STAT3 monomers must undergo homodimerization to be functional (25), a fourfold reduction in expression of RORγt is consistent with the prediction that a single mutant allele of STAT3 would reduce functional STAT3 homodimers by 75%. In our cohort of patients, the quantity of RORγt mRNA correlated with the number of Th17 cells induced after stimulation in vitro, which would be expected if RORγt specifies Th17 differentiation (Fig. 2 B).
HIES CD4+ T cells can proliferate, produce Th1- and Th2-type cytokines, and undergo in vivo differentiation to T regulatory (T reg) and T follicular helper (TFH) cell phenotypes
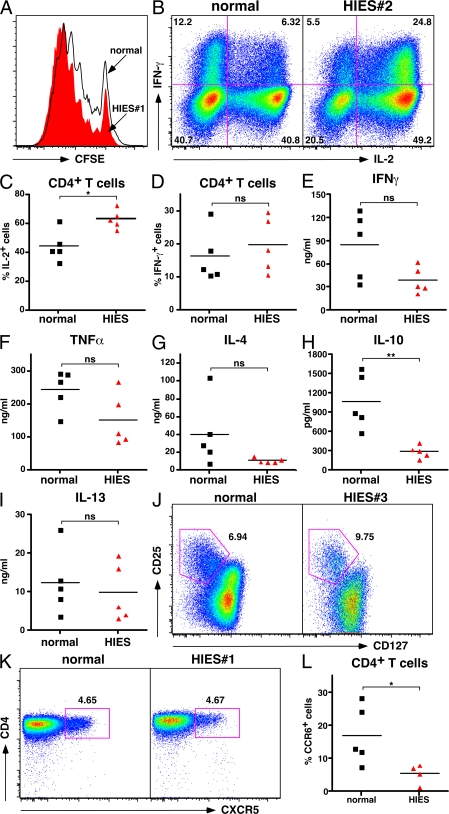
The defect in IL-17 secretion and Th17 cell differentiation by HIES CD4+ T cells did not appear to result from a general defect in lymphocyte activation. HIES CD4+ T cells proliferated to a similar extent as control counterparts (Fig. 3 A). Furthermore, there was no significant defect in production of IFN-γ (Fig. 3, B–E), TNF-α (Fig. 3 F), IL-4, or IL-13 (Fig. 3, G and I). IFN-γ secretion by activated HIES CD4+ T cells was reduced compared with controls (Fig. 3 E), but this reduction did not reach statistical significance, and there was no deficiency of IFN-γ–producing cells when assessed by flow cytometry after restimulation with PMA/ionomycin. Interestingly, there was a consistent and significant reduction (∼3.5-fold; P < 0.01) in IL-10 production by CD4+ T cells from all HIES patients examined relative to production by control CD4+ T cells (Fig. 3 H). This probably reflects a specific intrinsic requirement for STAT3 in IL-10 production by activated CD4+ T cells, as recently demonstrated in mice that specifically lack STAT3 in their CD4+ T cells (26). Despite reductions in secretion of IFN-γ and IL-10, measurable amounts of these cytokines were clearly present in the cultures of activated CD4+ T cells from HIES patients (Fig. 3 I), which contrasts the complete deficit in IL-17 production by these cells (Fig. 1, D and E). Interestingly, HIES CD4+ T cells consistently gave rise to an increased frequency of cells expressing IL-2 compared with normal controls (Fig. 3, B–D). Furthermore, T reg cells (defined as CD4+CD25hiCD127lo cells) (27) and TFH cells (defined as CD4+CXCR5+ cells) (28) were found to be present in normal numbers ex vivo in HIES patients (Fig. 3, J and K).
Figure 3.
CD4+ T cells from HIES patients undergo normal proliferation and differentiation into Th1-, Th2-, T reg–, and TFH-type effector cells. (A) CFSE profiles of normal (outline black histogram) and HIES (solid red histogram) CD4+ T cells after stimulation with CD3/CD28 mAb and IL-2 for 5 d. (B) Intracellular expression of IFN-γ and IL-2 in CD4+ T cells purified from normal donors and HIES patients after activation for 4–5 d with CD3/CD28/IL-2. The FACS plots in B are representative of one normal control and one HIES patient. (C and D) Frequency of CD4+ T cells from normal controls and HIES patients that expressed IL-2 (C) or IFN-γ (D). (E–I) Secretion of IFN-γ (E), TNF-α (F), IL-4 (G), IL-10 (H), and IL-13 (I) by normal and HIES CD4+ T cells stimulated with anti-CD3/CD28 mAb and IL-2 for 5 d. (J and K) CD4+ T cells were labeled with either (J) CD25 and CD127 to identify T reg cells (CD25hiCD127lo) or (K) CXCR5 to identify TFH cells, as indicated by the gated populations. The values represent the frequency of gated cells and are from one healthy donor and one HIES patient and are representative of at least four patient and control samples. (L) PBMCs were labeled with anti-CD4 and CCR6 mAb, and the frequency of CD4+ CCR6+ cells in healthy donors and HIES patients was enumerated. For all graphs (E–I and L), each symbol represents the value from an individual donor or patient, and the line represents the mean. *, P < 0.05; **, P < 0.01; ns, not significant.
It has been recently reported that Th17 cells express the chemokine receptor CCR6 (14, 29). Because IL-17–producing cells were absent from the CD4+ T cell compartment of HIES patients, we enumerated CD4+ CCR6+ T cells in peripheral blood. In healthy donors, ∼20% of CD4+ T cells expressed CCR6 (Fig. 3 L). In contrast, <10% of CD4+ T cells expressed CCR6 in HIES patients (Fig. 3 L), consistent with the suggestion that CCR6 identifies Th17 cells. Furthermore, because human CCR6+ CD4+ T cells have been shown to secrete both IL-17 and IFN-γ under some circumstances (29), deficiency of this subset might account for the reduction in IFN-γ sometimes observed in HIES. It is important to emphasize that previous assessments of IFN-γ production in HIES are inconsistent, which could also be due to heterogeneity of HIES cohorts before genotyping for STAT3 became available. Indeed, recent evidence indicates that some HIES-like patients with normal STAT3 exhibit deficiencies of IFN-γ production, whereas STAT3-deficient patients have no IFN-γ defect (30).
Naive CD4+ T cells fail to differentiate into Th17 cells in vitro
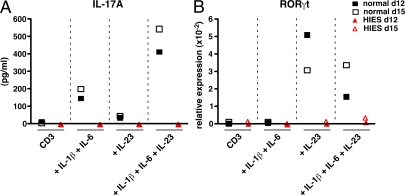
Human naive CD4+ T cells do not produce IL-17 when stimulated through CD3 and CD28 (11, 12). Thus, the detection of IL-17 in the cultures of total CD4+ T cells activated with anti-CD3/CD28 mAb reflects production by memory CD4+ T cells that have differentiated to the Th17 lineage in vivo. The requirement of STAT3 in CD4+ T cell differentiation in vivo appears to be specific to the Th17 lineage because CD4+ T cells from HIES patients could give rise to effector cells with features of Th1-, Th2-, T reg–, and TFH-type cells in vivo. Because STAT3 is ubiquitously expressed and functions downstream of numerous cytokine receptors (25), it was possible that the inability to generate Th17 cells in vivo was a consequence of a requirement for STAT3 function in non-CD4+ T cells, for instance antigen-presenting cells, that instruct the differentiation of CD4+ T cells. It was therefore important to demonstrate an intrinsic requirement of STAT3 in naive CD4+ T cells for their commitment to the Th17 lineage. Thus, naive CD4+ T cells were sort-purified and then cultured with beads coated with anti-CD2, anti-CD3, and anti-CD28 together with various combinations of IL-1β, IL-6, and IL-23. Secretion of IL-17A by normal CD4+ T cells was not detected until the cells had been subjected to three rounds of stimulation in vitro with anti-CD2, anti-CD3 plus anti-CD28 mAb-coated beads, and either IL-1β plus IL-6 or IL-23, with each stimulation period lasting 4–5 d (Fig. 4 A). The combination of IL-1β plus IL-6 was approximately fivefold more effective at inducing production of IL-17A by activated naive CD4+ T cells than IL-23, and there was a further two- to threefold increase in the presence of all three cytokines (Fig. 4 A). In complete contrast, when naive HIES CD4+ T cells were cultured under the same optimized conditions they failed to produce IL-17A (Fig. 4 A). Although these culture conditions induced IL-17A, we were unable to detect IL-17A–expressing cells by intracellular staining (not depicted). Because only ∼1% of total activated CD4+ T cells expressed IL-17A, yet secreted up to 10,000 pg/ml IL-17A (see Fig. 1, A–E), our inability to detect IL-17A+ cells in cultures of naive CD4+ T cells most likely reflects the fact that these cells secreted 10–20-fold less IL-17A than memory CD4+ T cells.
Figure 4.
Naive CD4+ T cells from HIES patients fail to differentiate to the Th17 lineage in vitro in response to the STAT3-activating cytokines IL-6 and IL-23. Naive CD4+ T cells isolated from a normal donor (▪, □) and a HIES patient (▴, ▵) were cultured with anti-CD2, anti-CD3, and anti-CD28 mAb-coated beads either alone (CD3) or in the presence of IL-1β plus IL-6 (IL-1β+IL-6), IL-23 (IL-23), or all three cytokines (IL-1β +IL-6+IL-23). The cells were harvested after 5 d and then recultured under the same conditions for another two rounds of stimulation. (A) IL-17A secretion was determined after either 3 d (▪, ▴; total time of culture, 12 d) or 6 d of the third round of restimulation (□, ▵; total time of culture, 15 d). (B) Expression of RORγt mRNA in cells stimulated for 5 d (▪, ▴) or 15 d (□, ▵) was determined by quantitative PCR.
Consistent with the defect in differentiation of naive HIES CD4+ T cells into Th17 cells in the presence of IL-1β, IL-6, and IL-23, we demonstrated a defect in induction of RORγt in T cells in response to each of these stimuli. The induction of RORγt mRNA was >10-fold greater in IL-23–containing culture conditions in normal CD4+ T cells compared with those from HIES patients (Fig. 4 B). Interestingly, induction of RORγt expression appeared to be absolutely dependent on IL-23, whereas there was a lesser effect of IL-6 (Fig. 4 B). These findings recapitulate the in vivo Th17 differentiation defect in HIES and are consistent with the ability of IL-6 (25) and IL-23 (31) to both activate STAT3. Furthermore, although IL-23 can also activate STAT-1, -4, and -5 (31), it would appear that IL-23–induced activation of STAT3 plays a predominant role in amplifying the generation of Th17 cells from human naive CD4+ T cells induced by IL-6 (Fig. 2).
Our findings identify a fundamental defect in Th17 differentiation in HIES. Thus, enumeration of IL-17 production by activated CD4+ T cells, coupled with measuring the frequency of CCR6+CD4+ T cells, emerges as a relatively quick and simple procedure that could be used to screen patients suspected of having HIES due to inactivating mutations in STAT3. In addition, our findings also point to a crucial role for Th17 cells in the normal host response. Infections with Candida spp. and S. aureus, the most prevalent etiological agents in the infections encountered in patients with HIES, are also important pathogens in other clinical settings. Mechanisms for defense against Candida and other fungal infections have been contentious. Although human Th17 cells respond selectively to Candida (14), other evidence suggests that production of IL-17 and IL-23 increases susceptibility to fungal infection in vivo (32). Furthermore, a role for T reg cells has been proposed, whereby down-regulation of Th17-mediated inflammation is necessary to prevent fungal-induced pathology (33). However, we did not detect a deficiency of T reg cells, at least by cell surface phenotype, in HIES. Our observations using CD4+ T cells from HIES patients supports the data of Acosta-Rodriguez et al. (14) that Th17 cells play a crucial role in coordinating the host defense against Candida. This most likely reflects a direct function of IL-17 produced by Th17 cells, as demonstrated by the requirement of the IL-17R in protective immune responses in mice to infection with C. albicans (15). Recent evidence from Milner et al. (30) demonstrates that CD4+ T cells from HIES patients fail to differentiate in vitro into IL-17–producing effector cells in response to stimulation with C. albicans and S. aureus. Deficiency of IL-17 is not necessarily the only mechanism of immunodeficiency in HIES because IL-22 has been shown to contribute to protection against Klebsiella pneumoniae in mice (23), and we show that IL-22 production is also reduced significantly in HIES.
Although our findings help to explain the spectrum of infections in HIES, other aspects of the complex phenotype require elucidation. A similar cell-intrinsic effect of STAT3 acting within chondrocytes or osteoblasts may explain some of the skeletal defects as evidenced in mice with a conditional hematopoietic cell-specific disruption of the STAT3 gene (34). Craniosynostosis and midline abnormalities, for example, can result from defects in the transcription factor TWIST, which is induced by STAT3 (35, 36). The mechanism of the HIES component of the phenotype is also likely to be complex because STAT3 operates intrinsically in B cells and is activated in response to many regulatory cytokines, including IL-6, IL-10, and IL-21, and, as shown here, T helper signals are also disrupted.
Finally, illumination of the aspects of the HIES phenotype that arise as a consequence of Th17 deficiency may sound a note of caution when considering IL-17 antagonism in autoimmune and inflammatory diseases (9, 10). On the other hand, administration of IL-17 or IL-22 may represent a therapeutic approach to controlling specific pathogen infections in HIES patients, as well as in other patient groups who exhibit susceptibility to Candida.
MATERIALS AND METHODS
Reagents.
Allophycocyanin-conjugated anti–IFN-γ mAb was purchased from Invitrogen, and FITC–anti–IL-17A was purchased from eBioscience. Alexa 647–anti-CXCR5, allophycocyanin–anti-CD25, FITC–anti-CD45RA, PE-conjugated anti–IL-2 and anti-CD27, PE-Cy7–anti-CD4, and PE–anti-CCR6 were purchased from BD Biosciences. ECD-conjugated anti-CD4 and PE–anti-CD127 were from Beckman Coulter. Recombinant human IL-2 (rIL-2) was purchased from Endogen, recombinant human IL-1β and IL-6 were from PeproTech, and recombinant human IL-23 was purchased from eBioscience. Anti-CD28 mAb was purchased from BD Biosciences, anti-CD3 mAb (Spv-T3b) was provided by H. Spits (NKI, Amsterdam, Netherlands), and anti-CD2–, anti-CD3–, and anti-CD28–coated beads were purchased from Miltenyi Biotec (T cell activation/expansion kit). PMA, calcium ionophore (ionomycin), brefeldin A, and saponin were all purchased from Sigma-Aldrich. CFSE was purchased from Invitrogen.
Patients.
Five normal donors and five patients with the clinical diagnosis of HIES were recruited from Immunology Clinics in Canberra and Sydney, Australia. The cited HIES score (21) is age sensitive; therefore, the low score in the 7-yr-old patient did not argue against testing her for mutations in STAT3. All studies were approved by the institutional human research ethics committees (Canberra and Westmead hospitals).
Identification of STAT3 mutations in HIES patients.
Genomic DNA was isolated from saliva samples, and exons 13–21 encompassing the DNA binding and SH2 domains were amplified by PCR and sequenced (ABI Prism 7700). The following primers were used: exons 12–14, TAGTTTAAAGAAATGCCCAGGAGCACAGAG and TTGGCCTAAGTGACTTTTTGGAATAACTACAGC; exon 15, TGCTGTGCTGCTTAGACTGG and CCCCTGTACGTAGCCTCTCCA; exon 16, GAGATGCGGGTGAAGAGATT and CTTGTTTAGATGAGGGATGGTG; exon 17, AGTGCCCCCTCCTTTTAGTT and CCCAAATGAACAGCCCTATG; exons 18 and 19, TGCACACACACAACAGTGC and TCTGTCCAACCTACCCTTCG; exon 20, GGACAGAGTGTGCACAGATGTAA and CACTGGAGCAAGCAAAACAA; and exon 21, GCTTAAGTCTTTTCCCCTTCG and GCATTTGCCTATCTATCCTCCA.
CD4+ T cell isolation, phenotyping, and cell culture.
Whole blood was collected from normal donors and HIES patients, and PBMCs were prepared by centrifugation using Ficoll-Paque. CD4+ T cells were then isolated by positive selection using the CD4-DYNA beads. Purified CD4+ T cells were cultured in 24-well plates (2 × 105 cells/ml) with immobilized 5 μg/ml anti-CD3 mAb, 750 ng/ml anti-CD28 mAb, and 20 U/ml rIL-2 (37). CD4+ T cells were also labeled with CFSE (37), and their proliferation was determined after 5 d of in vitro culture. For in vitro differentiation to the Th17 lineage, naive CD4+ T cells were isolated by sorting CD45RA+CD27+ cells (37) and then culturing them with anti-CD2–, anti-CD3–, and anti-CD28–coated beads (one bead per five cells), and 20 U/ml rIL-2 alone or in the presence of 20 ng/ml IL-23 only, 10 ng/ml IL-1β plus 50 ng/ml IL-6, or IL-1β, IL-6, and IL-23. The cells were harvested every 4–5 d before being washed and recultured under the same conditions at 2 × 105 cells/ml (11, 12).
The frequencies of CD4+ T cells that were of a naive, conventional memory, or effector memory phenotype, were determined by enumerating CD4+ T cells that were either CD45RA+CD27+, CD45RA−CD27+, or CD45RA−CD27−, respectively. T reg and TFH cells were quantitated by determining the frequency of CD4+ T cells that had a CD25hiCD127lo (27) or CXCR5+ (28) phenotype, respectively.
Intracellular cytokine staining and ELISA.
After 5 or 6 d of activation with the conditions mentioned above, CD4+ T cells were harvested and restimulated for 6 h with 100 ng/ml PMA and 750 ng/ml ionomycin, with 10 μg/ml brefeldin A added after 2 h. After this time, cells were fixed in 2% formaldehyde (Sigma-Aldrich) and stained for expression of intracellular cytokines (IL-2, IL-17A, and IFN-γ). All antibody staining and wash steps were performed in 0.5% saponin solution (37). Data were acquired using a FACSCanto or LSRII (BD Biosciences) and analyzed using FlowJo software (Tree Star). IL-17A in culture supernatants was measured using the human IL-17A ELISA Ready-SET-Go kit according to the manufacturer's instructions (eBioscience). IL-22 secretion was measured using the Human IL-22 ELISA Development kit from PeproTech. Secretion of IL-4, IL-10, IL-13, IFN-γ, and TNF-α was determined by ELISA using purified and biotinylated cytokine-specific mAb (IL-4: 8D4-8 and MP4-25D2; IL-10: JES3-12G8 and JES3-9D7; IL-13: PVM13-1 and polyclonal; IFN-γ: NIB42 and 4S.B3; TNF-α: MAb1 and MAb11; from BD Biosciences and eBioscience) as the capture and detection mAbs, respectively, and visualization with streptavidin–horseradish peroxidase and TMB substrate.
mRNA quantitation.
After 2 d of culture, CD4+ T cells were harvested and washed with PBS. Total RNA was isolated using the QIAGEN RNeasy kit according to the manufacturer's instructions. For quantitative RT-PCR, total RNA was reverse transcribed using oligo-dT according to the Invitrogen protocol. cDNA expression was determined using the ABI Prism 7900 sequence detection system and Syber-green reagents (Applied Biosystems). The following primers were used: RORγt, TCAGTCATGAGAACACAAATTGA and GCACCCCTCACAGGTGATAA; UBE2D2, TGAAGAGAATCCACAAGGAATTGA and CAACAGGACCTGCTGAACACTG; and IL22, AGGCTCAGCAACAGGCTAAG and TCCTTCAGCTTTTGCACATT. Fluorescence signals were measured over 40 PCR cycles, and the cycle (Ct) at which signals crossed a threshold set within the logarithmic phase was recorded. The Ct for the target gene was subtracted from the Ct for UBE2D2 (ΔCt). The relative amount of mRNA was calculated as 2ΔCt.
Acknowledgments
We thank Drs. Carola Vinuesa and Elissa Deenick for helpful discussions, Gill Tangye for establishing the cytokine ELISAs, and Karen Fenimore for providing reagents.
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia. C.S. Ma, G.Y.J. Chew, and S.G. Tangye are the recipients of Research Fellowships awarded by the NHMRC.
The authors have no conflicting financial interests.
S.G. Tangye and M.C. Cook contributed equally to this paper.
References
- 1.Weaver, C.T., R.D. Hatton, P.R. Mangan, and L.E. Harrington. 2007. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25:821–852. [DOI] [PubMed] [Google Scholar]
- 2.Bettelli, E., T. Korn, and V.K. Kuchroo. 2007. Th17: the third member of the effector T cell trilogy. Curr. Opin. Immunol. 19:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veldhoen, M., R.J. Hocking, C.J. Atkins, R.M. Locksley, and B. Stockinger. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. [DOI] [PubMed] [Google Scholar]
- 4.Mangan, P.R., L.E. Harrington, D.B. O'Quinn, W.S. Helms, D.C. Bullard, C.O. Elson, R.D. Hatton, S.M. Wahl, T.R. Schoeb, and C.T. Weaver. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234. [DOI] [PubMed] [Google Scholar]
- 5.Nurieva, R., X.O. Yang, G. Martinez, Y. Zhang, A.D. Panopoulos, L. Ma, K. Schluns, Q. Tian, S.S. Watowich, A.M. Jetten, and C. Dong. 2007. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 448:480–483. [DOI] [PubMed] [Google Scholar]
- 6.Korn, T., E. Bettelli, W. Gao, A. Awasthi, A. Jager, T.B. Strom, M. Oukka, and V.K. Kuchroo. 2007. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 448:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laurence, A., C.M. Tato, T.S. Davidson, Y. Kanno, Z. Chen, Z. Yao, R.B. Blank, F. Meylan, R. Siegel, L. Hennighausen, E.M. Shevach, and J. O'Shea. 2007. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 26:371–381. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov, I.I., B.S. McKenzie, L. Zhou, C.E. Tadokoro, A. Lepelley, J.J. Lafaille, D.J. Cua, and D.R. Littman. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. [DOI] [PubMed] [Google Scholar]
- 9.Yang, X.O., A.D. Panopoulos, R. Nurieva, S.H. Chang, D. Wang, S.S. Watowich, and C. Dong. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282:9358–9363. [DOI] [PubMed] [Google Scholar]
- 10.Harris, T.J., J.F. Grosso, H.R. Yen, H. Xin, M. Kortylewski, E. Albesiano, E.L. Hipkiss, D. Getnet, M.V. Goldberg, C.H. Maris, et al. 2007. Cutting edge: an in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J. Immunol. 179:4313–4317. [DOI] [PubMed] [Google Scholar]
- 11.Acosta-Rodriguez, E.V., G. Napolitani, A. Lanzavecchia, and F. Sallusto. 2007. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat. Immunol. 8:942–949. [DOI] [PubMed] [Google Scholar]
- 12.Wilson, N.J., K. Boniface, J.R. Chan, B.S. McKenzie, W.M. Blumenschein, J.D. Mattson, B. Basham, K. Smith, T. Chen, F. Morel, et al. 2007. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 8:950–957. [DOI] [PubMed] [Google Scholar]
- 13.Aujla, S.J., P.J. Dubin, and J.K. Kolls. 2007. Th17 cells and mucosal host defense. Semin. Immunol. 19:377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acosta-Rodriguez, E.V., L. Rivino, J. Geginat, D. Jarrossay, M. Gattorno, A. Lanzavecchia, F. Sallusto, and G. Napolitani. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646. [DOI] [PubMed] [Google Scholar]
- 15.Huang, W., L. Na, P.L. Fidel, and P. Schwarzenberger. 2004. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J. Infect. Dis. 190:624–631. [DOI] [PubMed] [Google Scholar]
- 16.LeibundGut-Landmann, S., O. Gross, M.J. Robinson, F. Osorio, E.C. Slack, S.V. Tsoni, E. Schweighoffer, V. Tybulewicz, G.D. Brown, J. Ruland, and C. Reis e Sousa. 2007. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol. 8:630–638. [DOI] [PubMed] [Google Scholar]
- 17.Buckley, R.H. 2002. Primary immunodeficiency diseases: dissectors of the immune system. Immunol. Rev. 185:206–219. [DOI] [PubMed] [Google Scholar]
- 18.Grimbacher, B., S.M. Holland, and J.M. Puck. 2005. Hyper-IgE syndromes. Immunol. Rev. 203:244–250. [DOI] [PubMed] [Google Scholar]
- 19.Holland, S.M., F.R. DeLeo, H.Z. Elloumi, A.P. Hsu, G. Uzel, N. Brodsky, A.F. Freeman, A. Demidowich, J. Davis, M.L. Turner, et al. 2007. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357:1608–1619. [DOI] [PubMed] [Google Scholar]
- 20.Minegishi, Y., M. Saito, S. Tsuchiya, I. Tsuge, H. Takada, T. Hara, N. Kawamura, T. Ariga, S. Pasic, O. Stojkovic, et al. 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 448:1058–1062. [DOI] [PubMed] [Google Scholar]
- 21.Grimbacher, B., A.A. Schaffer, S.M. Holland, J. Davis, J.I. Gallin, H.L. Malech, T.P. Atkinson, B.H. Belohradsky, R.H. Buckley, F. Cossu, et al. 1999. Genetic linkage of hyper-IgE syndrome to chromosome 4. Am. J. Hum. Genet. 65:735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath, C.M., Z. Wen, and J.E. Darnell Jr. 1995. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9:984–994. [DOI] [PubMed] [Google Scholar]
- 23.Aujla, S.J., Y.R. Chan, M. Zheng, M. Fei, D.J. Askew, D.A. Pociask, T.A. Reinhart, F. McAllister, J. Edeal, K. Gaus, et al. 2008. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolk, K., S. Kunz, E. Witte, M. Friedrich, K. Asadullah, and R. Sabat. 2004. IL-22 increases the innate immunity of tissues. Immunity. 21:241–254. [DOI] [PubMed] [Google Scholar]
- 25.Leonard, W.J., and J.J. O'Shea. 1998. Jaks and STATs: biological implications. Annu. Rev. Immunol. 16:293–322. [DOI] [PubMed] [Google Scholar]
- 26.Stumhofer, J.S., J.S. Silver, A. Laurence, P.M. Porrett, T.H. Harris, L.A. Turka, M. Ernst, C.J. Saris, J.J. O'Shea, and C.A. Hunter. 2007. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 8:1363–1371. [DOI] [PubMed] [Google Scholar]
- 27.Seddiki, N., B. Santner-Nanan, J. Martinson, J. Zaunders, S. Sasson, A. Landay, M. Solomon, W. Selby, S.I. Alexander, R. Nanan, et al. 2006. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J. Exp. Med. 203:1693–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinuesa, C.G., S.G. Tangye, B. Moser, and C.R. Mackay. 2005. Follicular B helper T cells in antibody responses and autoimmunity. Nat. Rev. Immunol. 5:853–865. [DOI] [PubMed] [Google Scholar]
- 29.Annunziato, F., L. Cosmi, V. Santarlasci, L. Maggi, F. Liotta, B. Mazzinghi, E. Parente, L. Fili, S. Ferri, F. Frosali, et al. 2007. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 204:1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milner, J.D., J.M. Brenchley, A. Laurence, A.F. Freeman, B.J. Hill, K.M. Elias, Y. Kanno, C. Spalding, H.Z. Elloumi, M.L. Paulson, et al. 2008. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 452:773–776. [DOI] [PMC free article] [PubMed]
- 31.Parham, C., M. Chirica, J. Timans, E. Vaisberg, M. Travis, J. Cheung, S. Pflanz, R. Zhang, K.P. Singh, F. Vega, et al. 2002. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 168:5699–5708. [DOI] [PubMed] [Google Scholar]
- 32.Zelante, T., A. De Luca, P. Bonifazi, C. Montagnoli, S. Bozza, S. Moretti, M.L. Belladonna, C. Vacca, C. Conte, P. Mosci, et al. 2007. IL-23 and the Th17 pathway promote inflammation and impair antifungal immune resistance. Eur. J. Immunol. 37:2695–2706. [DOI] [PubMed] [Google Scholar]
- 33.De Luca, A., C. Montagnoli, T. Zelante, P. Bonifazi, S. Bozza, S. Moretti, C. D'Angelo, C. Vacca, L. Boon, F. Bistoni, et al. 2007. Functional yet balanced reactivity to Candida albicans requires TRIF, MyD88, and IDO-dependent inhibition of Rorc. J. Immunol. 179:5999–6008. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Z., T. Welte, N. Troiano, S.E. Maher, X.Y. Fu, and A.L. Bothwell. 2005. Osteoporosis with increased osteoclastogenesis in hematopoietic cell-specific STAT3-deficient mice. Biochem. Biophys. Res. Commun. 328:800–807. [DOI] [PubMed] [Google Scholar]
- 35.Boeck, A., C. Kosan, P. Ciznar, and J. Kunz. 2001. Saethre-Chotzen syndrome and hyper IgE syndrome in a patient with a novel 11 bp deletion of the TWIST gene. Am. J. Med. Genet. 104:53–56. [DOI] [PubMed] [Google Scholar]
- 36.Howard, T.D., W.A. Paznekas, E.D. Green, L.C. Chiang, N. Ma, R.I. Ortiz de Luna, C. Garcia Delgado, M. Gonzalez-Ramos, A.D. Kline, and E.W. Jabs. 1997. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat. Genet. 15:36–41. [DOI] [PubMed] [Google Scholar]
- 37.Ma, C.S., N.J. Hare, K.E. Nichols, L. Dupre, G. Andolfi, M.G. Roncarolo, S. Adelstein, P.D. Hodgkin, and S.G. Tangye. 2005. Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J. Clin. Invest. 115:1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]