Abstract
The ribonucleic acid (RNA) helicases retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation–associated gene 5 (MDA5) recognize distinct viral and synthetic RNAs, leading to the production of interferons. Although 5′-triphosphate single-stranded RNA is a RIG-I ligand, the role of RIG-I and MDA5 in double-stranded (ds) RNA recognition remains to be characterized. In this study, we show that the length of dsRNA is important for differential recognition by RIG-I and MDA5. The MDA5 ligand, polyinosinic-polycytidylic acid, was converted to a RIG-I ligand after shortening of the dsRNA length. In addition, viral dsRNAs differentially activated RIG-I and MDA5, depending on their length. Vesicular stomatitis virus infection generated dsRNA, which is responsible for RIG-I–mediated recognition. Collectively, RIG-I detects dsRNAs without a 5′-triphosphate end, and RIG-I and MDA5 selectively recognize short and long dsRNAs, respectively.
The first line of defense against RNA virus infection relies on the innate immune system, which is initiated by the detection of viral components. Sensing of pathogens by innate immunity is mediated by host pattern recognition receptors (PRRs) that detect pathogen-specific molecular patterns (1). Three different classes of PRRs have been identified: Toll-like receptors (TLRs), retinoic acid–inducible gene-I (RIG-I)–like helicases (RLHs), and NOD-like receptors (2–4). The recognition of viruses by the PRRs leads to production of proinflammatory cytokines and type I IFNs. In particular, type I IFNs, comprised of multiple IFN-αs and -βs, are important for eliminating invading viruses by inducing death of infected cells, conferring resistance to viral infection on surrounding cells, and activating acquired immune responses.
Among PRRs, TLRs and RLHs are known to recognize viral infection. TLRs are transmembrane receptors with leucine-rich repeats and a cytoplasmic Toll/IL-1 receptor homology (TIR) domain. TLR3, 7, and 9 are located on endosome/ER membranes, and detect double-stranded (ds) RNA, single-stranded (ss) RNA, and DNA with a CpG motif, respectively (5–8). Upon encountering their cognate ligands, TLRs activate intracellular signaling cascades by recruiting the TIR domain containing adaptor molecules, such as MyD88 and TRIF (9, 10). Ultimately, the signaling leads to the expression of type I IFNs and proinflammatory cytokine genes. TLR7 and 9 are highly expressed on plasmacytoid DCs (pDCs), a cell type known to produce vast amounts of type I IFNs in response to viral infection (11). The importance of TLRs in the production of type I IFNs in pDCs have been shown by using mice deficient in MyD88, which is responsible for TLR7 and 9 signaling.
However, TLRs are dispensable for virus-induced IFN production in cell types other than pDCs. Instead, RIG-I family cytoplasmic RNA helicases play a key role in sensing RNA virus invasion. The RIG-I family consists of three helicases, RIG-I, melanoma-differentiation–associated gene 5 (MDA5), and LGP2 (12–14). RIG-I and MDA5 contain two caspase-recruitment domains (CARDs) and a DExD/H-box helicase domain. The helicase domains of RIG-I and MDA5 recognize viral RNAs, and their CARDs are responsible for signaling through interacting with a CARD-containing adaptor, IFN-β promoter stimulator-1 (IPS-1) (15, 16), also called MAVS, CARDIF, and VISA (17–20), which is located in the outer mitochondrial membrane. This interaction finally activates several transcriptional factors, IFN regulator factor 3, IFN regulator factor 7, and NF-κB for the induction of type I IFNs and proinflammatory cytokines (21, 22). In contrast, LGP2 does not possess a CARD, but only a DExD/H-box helicase domain, and has been reported to function as a negative regulator (14, 23), especially of the RIG-I–mediated pathway (24, 25).
Recent studies have demonstrated that RIG-I and MDA5 are differentially involved in antiviral responses. Picornaviruses, such as encephalomyocarditis virus (EMCV), are specifically recognized by MDA5, whereas RIG-I recognizes a wide variety of RNA viruses belonging to the paramyxovirus and rhabdovirus families, as well as Japanese encephalitis virus (26). Some viruses such as Dengue virus and reovirus were shown to be recognized by both RIG-I and MDA5 (27). These two helicases have also been shown to recognize distinct types of RNAs. Single-stranded (ss) RNA with 5′-triphosphate has been identified as a RIG-I ligand (26, 28–30). The cellar 5′-triphosphate ssRNA does not exist in the cytoplasm, but in the nucleus, and ssRNAs in the cytoplasm are normally capped or processed. Thus, RIG-I can distinguish viral RNAs from vast amounts of cellar RNAs. On the other hand, small dsRNAs (ranging from 21 to 27 nucleotides) have also been reported to induce IFN-inducible genes via both ATPase and helicase activities of RIG-I (31).
However, whether or not dsRNA is also a RIG-I ligand, in addition to 5′-triphosphate ssRNA, is still controversial. Although polyinosinic-polycytidylic acid (poly I:C), an artificial double-stranded (ds) RNA, was found to be a ligand for MDA5 (26, 28), the motif recognized by MDA5 should be characterized, and a natural ligand for MDA5 is also yet to be discovered.
In this study, we investigated how RIG-I and MDA5 differentially recognize RNAs by chemically modifying poly I:C. We found that the MDA5 ligand, poly I:C, was converted to a RIG-I ligand after shortening of the dsRNA length. RIG-I and MDA5 preferentially bind to short and long poly I:Cs, respectively. In addition, dsRNAs prepared from viruses differentially activated RIG-I and MDA5, depending on the length. Whereas influenza virus infection failed to generate dsRNA in the infected cells, vesicular stomatitis virus (VSV) infection generated dsRNA, which is responsible for RIG-I–mediated recognition. Collectively, RIG-I detects dsRNAs without a 5′-triphosphate end, and RIG-I and MDA5 selectively recognize short and long dsRNAs, respectively.
RESULTS
The switching of poly I:C from an MDA5 ligand to a RIG-I ligand
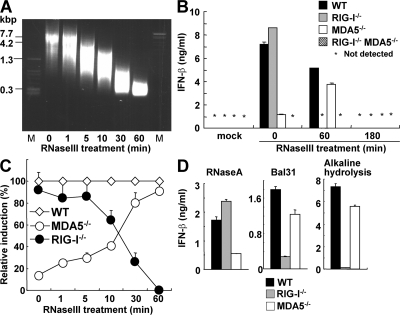
We first estimated the length of the MDA5 ligand poly I:C by agarose gel electrophoresis and found that untreated poly I:C migrated as a smeared band corresponding to the mobility of ∼4–8 kbp dsDNA fragments (Fig. 1 A). To analyze the importance of poly I:C length in IFN-inducing activity, we partially digested poly I:C with a dsRNA-specific endonuclease, RNase III. The size of the poly I:C became shorter in a digestion time-dependent manner, and the median size of 60 min–treated poly I:C corresponded to 300 bp dsDNA (Fig. 1 A). When WT, Rig-I−/−, Mda5−/−, Rig-I−/−Mda5−/− mouse embryonic fibroblasts (MEFs) were stimulated with untreated poly I:C, IFN-β was induced in an MDA5-dependent manner that was consistent with previous reports (26, 28). Surprisingly, IFN-β induction by poly I:C treated with RNase III for 60 min depended on RIG-I, but not on MDA5 (Fig. 1 B). However, complete digestion of poly I:C by 180-min treatment with RNase III produced 10–20 nt dsRNA that failed to activate even wild-type cells (Fig. 1 B and unpublished data). When cells were stimulated with poly I:C partially digested for different periods, Mda5−/− MEFs gained the ability to respond to RNase III–treated poly I:C as the digestion proceeded, whereas Rig-I−/− MEFs gradually lost the ability to produce IFN-β in response to shortened poly I:C (Fig. 1 C). In addition, conventional DCs derived from bone marrow or prepared from spleen from Rig-I−/− and Mda5−/− mice also failed to produce IFN-β in response to RNase III–digested and undigested poly I:C, respectively (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20080091/DC1). Furthermore, production of IFN-β in the sera in response to digested and undigested poly I:C conjugated with a transfection reagent was dependent on the presence of RIG-I and MDA5, respectively (Fig. S2). These results indicate the switching of poly I:C from being an MDA5 ligand to being a RIG-I ligand after partial digestion with RNase III.
Figure 1.
Preferential recognition of long and short poly I:C by MDA5 and RIG-I. (A) The indicated RNAs are shown on the ethidium bromide–stained agarose gel. M, DNA marker. (B) WT, Rig-I−/−, Mda5−/−, and Rig-I−/−Mda5−/− MEFs were treated with 1 μg/ml of untreated or RNase III–treated poly I:C for 16 h. The production of IFN-β in the supernatant was measured by ELISA. mock, no RNA. (C) Relative induction of IFN-β after stimulation with poly I:C treated with RNase III for the indicated periods. (D) The production level of IFN-β in the stimulation with RNase A–treated, Bal31-treated, or alkaline-hydrolyzed poly I:C. Error bars show the SDs between triplicates.
Poly I:C partially digested with human Dicer also induced IFN-β in a RIG-I–dependent manner (unpublished data), indicating that this phenomenon is not specific to bacterial RNase III. In contrast, digestion of poly I:C with a ssRNA-specific nuclease, RNase A, did not affect its recognition by MDA5 (Fig. 1 D), suggesting that the shortening of the dsRNA part of poly I:C is necessary for the conversion from an MDA5 ligand to a RIG-I ligand. Consistent with the aforementioned observations, treatment with Bal 31, which degrades ssRNA and linear duplex RNA progressively from both the 5′- and 3′-ends, as well as alkaline hydrolysis, also converted poly I:C to a RIG-I ligand (Fig. 1 D). Poly I:C is an artificial dsRNA generated by annealing poly I and poly C. Poly I and C are synthesized by a bacterial enzyme, polynucleotide phosphorylase (PNPase), which catalyzes the polymerization of nucleotide diphosphate (32). Because poly I and C are synthesized with inositol diphosphate and cytidine diphosphate as substrates, poly I:C does not contain a 5′-triphosphate nucleotide, and dsRNA fragments generated by RNase III digestion contain a 5′-monophosphate end, showing absence of triphosphates in the prepared poly I:C solution. Furthermore, IFN induction by poly I:C treated with RNase III for 60 min was not altered by additional treatment with RNase A for the removal of ssRNA contamination (unpublished data), indicating that RIG-I recognizes dsRNA without triphosphates, in addition to 5′-triphosphate ssRNAs. Collectively, these results suggest that the length of poly I:C determines the differential recognition by RIG-I and MDA5.
Differential regulation of ATPase activity by long and short poly I:C
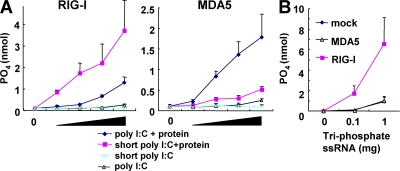
It has been shown that RIG-I has the ability to catalyze ATP in the presence of dsRNA. To determine if this ATPase activity of these helicases is differentially activated by dsRNAs in a length-dependent manner, we examined the ATPase activity of purified RIG-I and MDA5 proteins in the presence of poly I:C or poly I:C treated with RNase III for 60 min (designated as short poly I:C). The ATPase activity of RIG-I increased in the presence of short poly I:C in a dose-dependent manner, and the activation was higher than that with untreated poly I:C. In contrast, untreated poly I:C strongly activated the ATPase activity of MDA5 compared with short poly I:C (Fig. 2 A), indicating that the ATPase activities of RIG-I and MDA5 are selectively activated by the short poly I:C and untreated long poly I:C, respectively. As expected, 5′-triphosphate ssRNA induced the ATPase activity of RIG-I, but not of MDA5 (Fig. 2 B). Thus, the ATPase activity clearly correlated with the IFN responses triggered by these RNA helicases, indicating that RIG-I and MDA5 directly distinguish between short and untreated long poly I:C molecules, respectively.
Figure 2.
Long and short poly I:C preferentially activate ATPase activities of MDA5 and RIG-I. ATPase activity of RIG-I or MDA5 protein was measured in the presence of the indicated RNAs. The x axis shows the concentration of RNAs. (A) Long and short poly I:C. (B) 5′-tri-phosphate ssRNA. Several quantities (1, 0.2, 0.04, and 0.008 μg) of poly I:C were used. Error bars show SDs between triplicates.
Association of poly I:C with RIG-I and MDA5
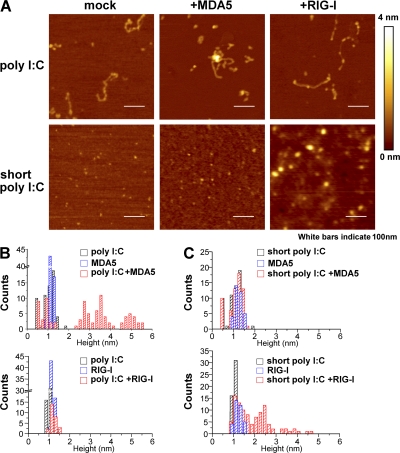
To investigate whether RIG-I and MDA5 directly distinguish between the lengths of poly I:C, we examined the binding of purified RIG-I and MDA5 proteins to poly I:C by atomic force microscope (AFM). As shown in Fig. 3 A, untreated poly I:C was detected as strings with lengths of 100–200 nm, and short poly I:C was visualized as dots (∼1 nm high). MDA5 mixed with untreated poly I:C formed a thick structure indicated by bright white dots, representing the binding between MDA5 and poly I:C (Fig. 3 A). On the other hand, the mixture of RIG-I and untreated poly I:C failed to form such complexes. Reciprocally, short poly I:C mixed with RIG-I, but not MDA5, formed the thick structures. Next, we statistically analyzed the thickness of molecules visualized by AFM. Whereas the heights of poly I:C, short poly I:C, MDA5, or RIG-I alone were 0.5–1.5 nm, 2.5–5.5 nm structures appeared when poly I:C was mixed with MDA5, but not with RIG-I, protein (Fig. 3 B). In contrast, as shown in Fig. 3 C, an association of short poly I:C with RIG-I, but not with MDA5, was detected. These data indicate that RIG-I and MDA5 distinguish the lengths of poly I:C and specifically bind to their cognate ligands.
Figure 3.
RIG-I and MDA5 specifically bind to short and long poly I:C. (A) Complex of indicated poly I:C and protein (MDA5 or RIG-I) was observed by AFM. Height is on a scale from 0 to 4 nm, with a low area depicted in dark brown and a higher area depicted in brighter color. Scale area, 500 nm. Bars, 100 nm. Those are representative images from several pictures. (B and C) Statistical height analyses of molecules corresponding to pictures in A.
Differential recognition of short and long dsRNAs by two helicases
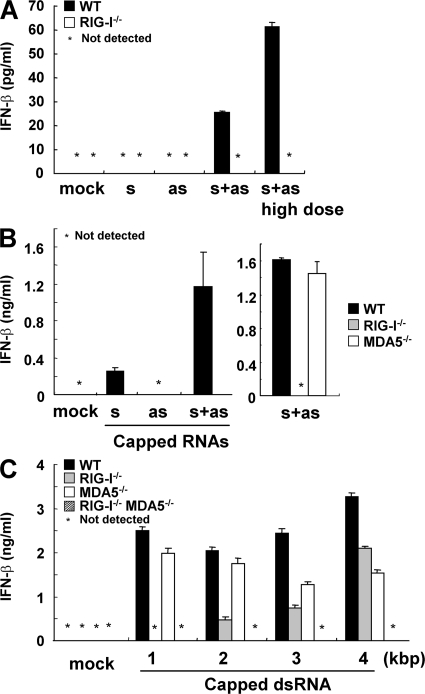
We examined if short dsRNAs other than poly I:C are also recognized by RIG-I. We chemically synthesized 70 base sense and antisense ssRNAs corresponding to the sequence of Lamin A/C harboring a 5′ hydroxyl end, annealed them, and examined their IFN-inducing ability. Whereas transfection of ssRNAs did not induce production of IFN-β, dsRNA generated by annealing sense and antisense ssRNAs (designated s and as in Fig. 4 A) induced IFN-β. Although the level of production was comparably low, it was clearly dependent on RIG-I, indicating that a 70-bp dsRNA without 5′ triphosphate is also a RIG-I ligand (Fig. 4 A). To further analyze longer dsRNA synthesized in vitro by T7 polymerase in the production of IFN-β, we generated a 400 base sense and antisense ssRNA corresponding to the sequence of Lamin A/C harboring a 5′ capping with 7mG (designated s and as in Fig. 4 B). Introduction of dsRNA, generated by annealing capped 400 base sense and antisense ssRNAs, induced greatly enhanced RIG-I–dependent IFN-β production compared with the transfection of cells with each ssRNA (Fig. 4 B). These data also supported the aforementioned observation that dsRNA can induce IFN responses in a RIG-I–dependent fashion. We further examined the IFN responses triggered by 1–4 kbp capped-dsRNA. Although 1 kbp dsRNA-induced IFN-β was dependent on RIG-I, but not on MDA5, 2 kbp capped-dsRNA induced IFN-β, even in Rig-I−/− cells. The production of IFN-β in response to 3 and 4 kbp capped-dsRNA was less dependent on RIG-I, and an impairment of IFN-β production was observed in Mda5−/− cells (Fig. 4 C). Of note, Rig-I−/−Mda5−/− cells did not produce any IFN-β in response to 1–4 kbp capped-dsRNA (Fig. 4 C). These data indicate that RIG-I and MDA5 preferentially recognize short and long dsRNAs synthesized by T7 polymerase, respectively.
Figure 4.
RIG-I and MDA5 selectively recognize dsRNA in a length-dependent manner. (A and B) Indicated genotype of MEFs were treated with 1 μg/ml of the indicated RNAs (10 μg/ml in the case of high dose) for 24 h. s, sense ssRNA; as, antisense ssRNA; s+as, dsRNA generated by annealing s with as. s and as are chemically synthesized 70-bp ssRNAs having a 5′ hydroxyl end in A. s and as are in vitro transcribed capped-ssRNAs in (B). The production of IFN-β in the supernatant was measured by ELISA. (C) WT, Rig-I−/−, Mda5−/−, and Rig-I−/−Mda5−/− MEFs were treated with 1 μg/ml of in vitro–transcribed capped dsRNAs for 16 h. The production of IFN-β in the supernatant was measured by ELISA. Error bars show SDs between triplicates.
Recognition of viral genomic dsRNA by RIG-I and MDA5
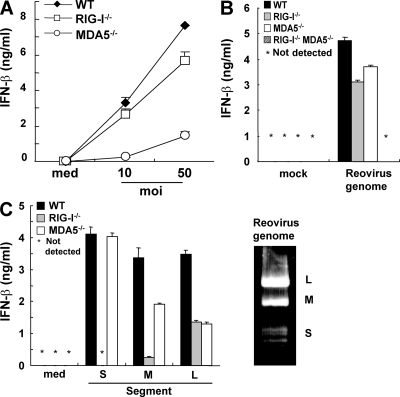
We then examined whether viral dsRNAs are also differentially recognized by RIG-I and MDA5. Reovirus, a dsRNA virus, was used for further analysis. The production of IFN-β in response to reovirus was severely impaired in Mda5−/− conventional DCs (cDCs; Fig. 5 A), although this was totally abolished in Rig-I−/−Mda5−/− cDCs (not depicted). This result indicates that both RIG-I and MDA5 are involved in the recognition of reovirus, which is consistent with a recent study (24). Next, we stimulated MEFs with whole genomic RNA prepared from reovirus. Reovirus genome RNA induced production of IFN-β in either Rig-I−/− cells or Mda5−/− MEFs, but not in Rig-I−/−Mda5−/− MEFs (Fig. 5 B). These findings suggest that the reovirus genome RNA contains both RIG-I and MDA5 ligands. Reovirus genome RNA consists of 10 segments in three distinct classes called L, M, and S corresponding to their size. Segment sizes are 3.9 kbp (L), 2.2–2.3 kbp (M), and 1.2–1.4 kbp (S; Fig. 5 C). These fragments were purified from the whole reovirus genome and each fragment was separately transfected into MEFs. As shown in Fig. 5 C, the production of IFN-β in response to S segments was dependent on RIG-I, but not on MDA5. The response to M fragments was not abrogated in Rig-I−/− MEFs, and was modestly impaired in Mda5−/− MEFs. In contrast, the production of IFN-β induced by L segments was reduced in both Rig-I−/− and Mda5−/− MEFs, suggesting that MDA5 contributed more to the recognition of longer segments of reovirus genomic dsRNA, whereas shorter segments were preferentially recognized by RIG-I.
Figure 5.
Reovirus genome dsRNA includes both RIG-I and MDA5 ligands. (A) WT, Rig-I−/−, and Mda5−/− GM-CSF-DCs were infected with the indicated multiplicity of infection of reovirus. The production of IFN-β in the supernatant was measured by ELISA. (B and C) The indicated genotypes of MEFs were treated with 1 μg/ml of reovirus genome RNA (B) or 0.1 μg/ml of dsRNA segments (C) for 16 h. The production of IFN-β in the supernatant was measured by ELISA. The reoviral genome is shown on the ethidium bromide-stained gel (C, right), and the S (1.2–1.4 kbp), M (2.2–2.3 kbp), and L (3.9 kbp) segments are indicated. Error bars show SDs between triplicates.
MDA5 and RIG-I recognize viral dsRNA generated during replication
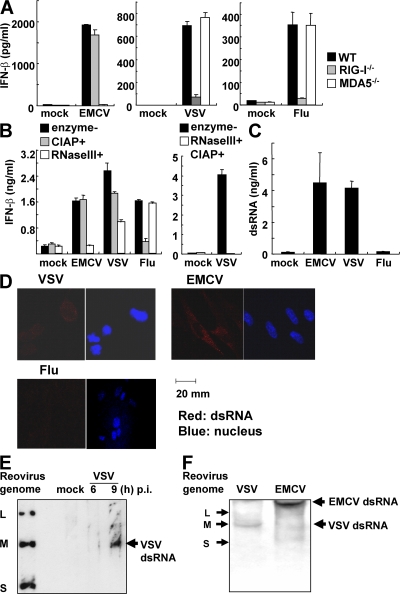
We have reported that EMCV is recognized by MDA5, whereas RIG-I detects VSV and influenza virus. In response to influenza virus infection, triphosphate ssRNA was considered to be the source of induction of type I IFNs, and no dsRNA was detectable during its replication (30, 33). To investigate whether triphosphate ssRNA is also the source of type I IFN induction in VSV infection, we harvested whole RNA from noninfected cells or virus-infected cells, and examined the IFN-β responses to the RNAs. RNA prepared from EMCV-infected cells induced IFN-β production in an MDA5-dependent manner (Fig. 6 A). Treatment of this RNA with calf intestine alkaline phosphatase (CIAP) did not reduce IFN-β production in MEFs (Fig. 6 B). In contrast, RNA from influenza virus-infected cells induced IFN-β production in a RIG-I–dependent manner, which was severely reduced after CIAP treatment (Fig. 6, A and B). IFN-β production in response to RNA from VSV-infected cells was dependent on RIG-I (Fig. 6 A) and this production was only partially reduced by treatment with CIAP (Fig. 6 B). These data suggest that 5′-triphosphate ssRNA is not the sole RIG-I ligand involved in VSV-induced IFN-β production. Furthermore, degradation of dsRNA from virus-infected cells by RNase III treatment abolished IFN-β–inducing activity in RNA from EMCV-infected cells (Fig. 6 B). Also the level of IFN production was impaired in RNA from VSV-infected cells by RNase III treatment, whereas the response to RNA from influenza virus-infected cells was not altered after treatment. Moreover, treatment of RNA from VSV-infected cells with both CIAP- and RNase III abolished IFN-β–inducing activity (Fig. 6 B). These results further suggest that the dsRNA generated in VSV-infected cells is recognized by RIG-I to induce IFN-β production. To determine if dsRNA is generated during VSV infection, we performed quantification of dsRNA in viral-infected cells by ELISA. Interestingly, infection with VSV and EMCV produced high amounts of dsRNA, despite no dsRNA being detected after infection with influenza virus (Fig. 6 C). As shown in Fig. 6 D, dsRNA (red dot) was observed in EMCV-infected cells, but not in influenza virus-infected cells, consistent with previous reports (30, 33). The presence of dsRNA was also detected in VSV-infected cells (Fig. 6 D). These results suggest that dsRNA is generated in cells infected with VSV, a virus recognized by RIG-I, and dsRNA, in addition to 5′-triphosphate ssRNA, might be a target for RIG-I–mediated recognition in vivo. On the other hand, long dsRNA could be generated in EMCV-infected cells and recognized by MDA5. Next, we examined the length of dsRNA generated after VSV infection. RNA harvested from cells infected with VSV for 6 or 9 h was electrophoresed, transferred to a nylon membrane, and blotted with anti-dsRNA antibody. As shown in Fig. 6 E, a dsRNA band was detected in the RNA sample (9-h infection of VSV), and the dsRNA migrated similar to M fragment of reovirus genomic RNA, suggesting that dsRNA generated in VSV infection is ∼2.2 kbp. We compared the lengths of dsRNAs generated in VSV- and EMCV-infected cells. As shown in Fig. 6 E, dsRNA found in EMCV-infected cells was much longer than that detected in VSV-infected cells. Although it is difficult to determine precise length of dsRNAs, the dsRNA from EMCV-infected cells migrated much slower than the L fragment of reovirus genomic RNA (3.9 kbp). These data suggest that the lengths of dsRNAs generated in RNA virus-infected cells varied depending on the virus species, and this difference could explain the differential recognition of viruses by RIG-I and MDA5.
Figure 6.
dsRNA generated during VSV replication induces IFNs in a RIG-I–dependent manner. (A) RNA samples harvested from uninfected (mock), EMCV-, VSV-, or influenza virus (flu)–infected cells were transfected into WT, Rig-I−/−, and Mda5−/− MEFs. The production of IFN-β in the culture supernatant 10 h after transfection was measured by ELISA. (B) RNA harvested from noninfected (mock) or EMCV-, VSV-, or influenza virus–infected cells with CIAP-, RNase III-, both CIAP-, and RNase III-treatments or nontreatment (enzyme-) was transfected into WT MEFs. The production of IFN-β in the supernatant 10 h after transfection was measured by ELISA. (C) dsRNA in uninfected (mock), EMCV-, VSV-, or influenza virus-infected cells was measured by ELISA. (D) Immunostaining for dsRNA in MEFs infected with EMCV, VSV, and influenza virus for 8 h. Red, dsRNA; blue, nucleus. Error bars show SDs between triplicates. (E) RNA harvested from noninfected (mock) or VSV-infected cells (indicated periods) was electrophoresed in 1.5% agarose gel, transferred to a nylon membrane, and blotted by anti-dsRNA antibody. Reovirus genome RNAs were indicated as the size control. The arrow shows VSV dsRNA. (F) dsRNA blotting of RNA harvested from EMCV- or VSV-infected cells. RNAs were electrophoresed in nondenaturing 10% polyacrylamide gel. Reovirus genome RNAs were indicated (left). Arrows (right) show EMCV and VSV dsRNA.
DISCUSSION
Our data indicate that RIG-I recognizes dsRNA in addition to 5′-triphosphate end ssRNA, and the length of dsRNA determines the utilization of RIG-I and MDA5 for the recognition. Genomic dsRNAs of Reovirus are also differentially recognized by RIG-I and MDA5 depending on the length of the dsRNA. The results also indicate that VSV, a rhabdovirus recognized by RIG-I, generates dsRNA in infected cells, whereas influenza virus does not generate dsRNA in the cells as reported previously (33).
The 5′-triphosphate ssRNA has been considered to be the sole ligand for RIG-I because dsRNA has been generated in vitro by a T7 polymerase using nucleotide-triphosphates as substrates (34). However, we discovered that RNase III-treated short poly I:C is another RIG-I ligand (Fig. 1). Poly I:C, a synthetic MDA5 ligand, was converted to a RIG-I ligand, when the length of dsRNA was shortened. The 5′ ends of short poly I:C harbor either monophosphate or diphosphate, showing that dsRNA without triphosphate is also a RIG-I ligand. RNase III and Dicer are dsRNA specific endonucleases generating 15–25 nt short dsRNAs, which mediate RNAi in a cell. Indeed, completely digested poly I:C lost the activity to induce type I IFNs, indicating that a certain length of dsRNA is required for RIG-I–mediated recognition. It was reported that host RNA digested by RNase L could be recognized by RIG-I for inducing IFN-β (35). Although it is not clear if the RNase L-digested RNA could harbor 5′-triphosphate for activating immune cells, the length of RNA might be important for RIG-I–mediated recognition in RNA digested with RNase L. Furthermore, chemically synthesized RNAs which do not possess a phosphate at the 5′ end also stimulated cells in a RIG-I–dependent manner, clearly indicating that dsRNA elicits RIG-I–mediated IFN responses independent of 5′-triphosphate. The amount of IFN-β produced in response to synthetic 70 nt dsRNA was lower than that in response to 5′ triphosphate end ssRNA and short poly I:C. It may be possible that 70 nt dsRNA is not long enough to efficiently induce IFN-β in MEFs. Alternatively, the presence of mono- or diphosphate at the 5′ end of dsRNAs might enhance recognition by RIG-I.
We also clarified that MDA5 and RIG-I directly distinguish between long and short dsRNAs generated by T7 polymerase in the presence of 7mG. ∼1 kbp dsRNAs induced IFNs in a RIG-I–dependent manner. ∼2 kbp dsRNAs induced IFNs even in RIG-I–deficient cells, and the induction was MDA5 dependent, showing that MDA5 preferentially recognizes long dsRNAs and RIG-I is involved in the recognition of short dsRNAs. This explains why poly I:C is an MDA5 ligand and poly I:C is shifted from being an MDA5 ligand to being a RIG-I ligand, after shortening of the length of dsRNA by RNase III treatment. Also, our data indicate that viral short and long dsRNA are also differentially recognized by RIG-I and MDA5. Reovirus genome RNA contains several lengths of dsRNA fragments and long dsRNA fragments induced IFN production in an MDA5-dependent manner, whereas IFN induction by short dsRNAs was RIG-I dependent. These data indicate that MDA5 and RIG-I differentiate between the lengths of dsRNA and the mechanism of how these helicases distinguish the length is an interesting issue.
Long and short poly I:C distinctly activated the ATPase activity of MDA5 and RIG-I, respectively. The ATPase activity was found to correlate with biological IFN responses triggered by these RNA helicases, suggesting that the induction of ATPase activity is the key to revealing the mechanism of how RIG-I and MDA5 recognize viral RNA and transduce signaling to downstream molecules. In addition to dsRNAs, triphosphate ssRNA activates the ATPase activity of RIG-I, but not of MDA5, suggesting that this ATPase activity is necessary not just for unwinding dsRNAs. Thus, the ATPase activity might be essential for conformational changes of these helicases to signal downstream, such as opening the CARD position masked by their helicases and RD domains. Whether this ATPase activity correlates with conformational changes of RIG-I and MDA5 remains to be determined and structural analysis of these helicases is desirable. Furthermore, we observed, with AFM, the specific association of MDA5 with poly I:C and also of RIG-I with the short poly I:C. It is interesting that the binding of these helicases to dsRNA also correlates with the biological IFN responses triggered by these RNA helicases. The mechanism of how these two helicases distinguish the length of dsRNA, leading to the specific binding to dsRNAs is an exciting issue for future studies.
Using Mda5−/− and Rig-I−/− mice, it has been shown that poly I:C is preferentially detected by MDA5 in inducing type I IFNs, and the contribution of RIG-I in poly I:C recognition was low (26). However, several reports showed that RIG-I was also involved in poly I:C recognition in vitro. Overexpression of RIG-I in cell lines activates an IFN-β promoter and its activation level is clearly augmented by poly I:C stimulation. RIG-I, as well as MDA5, is shown to bind to poly I:C (23). We tested poly I:Cs purchased from several companies and found that the induction of IFNs by some poly I:Cs was impaired in Rig-I−/− cells when the size of these poly I:Cs was comparably small (unpublished data). These observations are also consistent with our conclusion that the length of poly I:C is important for the differential recognition by RIG-I and MDA5.
Neither dsRNA nor 5′ triphosphate end RNA are present in the cytoplasm of host resting cells, and these RNA structures are the target of recognition by innate immunity. In response to RNA virus infection, dsRNA is generated in the course of viral RNA replication. However, it was also shown that dsRNA is barely present in infected cells, and 5′ triphosphate RNA was shown to be the major target of recognition. Indeed, dsRNA was not detected in influenza virus infected cells as shown previously (33). Consistently, IFN-β production in response to RNA from influenza virus–infected cells depended on the presence of the 5′-triphosphate end as determined by CIAP treatment. On the other hand, EMCV and VSV generated dsRNA in the cells, which were recognized by MDA5 and RIG-I, respectively. RNase III treatment of RNAs from VSV-infected cells impaired the IFN-β–inducing activity, indicating that dsRNA, generated in the course of VSV infection, contributed to RIG-I–mediated recognition. As reported previously, dsRNA generated by EMCV infection appears to be responsible for MDA5-mediated recognition. The length of dsRNA generated by VSV appeared to be close to 2.0–2.5 kbp by the immunoblot analysis with anti-dsRNA antibody. Given that the length of VSV genomic RNA is 11 kb, the dsRNA was not the replication intermediate of VSV. It has been reported that defective interfering (DI) particles are generated in VSV-infected cells, and the size of reported DI particles is ∼2.2 kb (36). Thus, dsRNA generated in the course of VSV replication might be derived from DI particles, although further studies are needed to clarify what is the source of the dsRNA. On the other hand, dsRNA found in EMCV–infected cells was much longer than that detected in VSV-infected cells. This long dsRNA is assumed as the replication intermediates of EMCV genomic RNA (∼8 kb); however, the characteristic of dsRNA generated by EMCV remains to be determined. These data suggest that MDA5 and RIG-I distinctly recognize long and short dsRNAs generated in a cell after RNA virus infection. RNA viruses recognized by RIG-I could possibly be subclassified based on the contribution of 5′-triphosphate end ssRNA and short dsRNA. In the future, it would be interesting to explore if the lengths of dsRNA differ between several RNA viruses recognized by RIG-I and MDA5.
In summary, we characterized the RNA molecules recognized by RIG-I and MDA5, and showed that dsRNAs differentially induce RIG-I– and MDA5-mediated IFN responses depending on length. Viral RNAs were also differentially recognized by RIG-I and MDA5, suggesting that the two helicases evolved for covering a broad spectrum of RNA viruses. The identification of the nature of dsRNA recognition will lead to the discovery of small molecules efficiently activating RIG-I and/or MDA5, and will lead to the development of novel vaccines and therapies against viral infection.
MATERIALS AND METHODS
Cells and viruses.
Mda5−/− and Rig-I−/− MEFs and GM-CSF DCs were generated as previously described (26, 37). EMCV, influenza virus, and VSV were obtained as previously described (26). Reovirus was as previously described (38).
Purification of recombinant RIG-I and MDA5.
RIG-I protein was purified as previously described (24). For the synthesis of MDA5, 2xFlag-MDA52-1025 (MDA5) was expressed as a GST-fusion protein using a BaculoGold GST-Expression System (BD Biosciences). GST-MDA5 bound to Glutathione–Sepharose 4B (GE Healthcare) was eluted by digestion with AcTEV protease (Invitrogen) and passed through Ni-NTA Agarose (QIAGEN). MDA5 was further purified by Q-Sepharose (GE Healthcare) chromatography.
ATPase assays.
ATPase assays were performed in 25 μl of ATPase reaction buffer (20 mM Tris-HCl, pH 8.0, 1.5 mM MgCl2, and 1.5 mM DTT) including 1 μg of purified RIG-I or MDA5 protein and the indicated amounts of RNAs (tri-p-ssRNA, etc.). After a 15-min incubation at 37°C, 50 nmol of ATP was added and the mixture was further incubated at 37°C for 1 h. BIOMOL GREEN Reagent (BIOMOL) was added for 5 min, and the absorbance at 620 nm was determined.
Immunofluorescence analysis.
MEFs were infected with the indicated virus for 8 h and fixed with 4% paraformaldehyde. Cells were then permeabilized with 0.5% Triton X-100 dissolved in PBS. For the detection of dsRNA, the mouse monoclonal antibody J2 and Alexa Fluor 594 anti–mouse secondary antibody (Invitrogen) were used.
Sample solution for AFM imaging.
RNAs and proteins were diluted with sterile purified water to the final concentration of 25 ng/μl and 2.5 ng/μl, respectively. Each solution was incubated for 30 min at room temperature.
AFM imaging.
5 μl of each sample solution was dropped on freshly cleaved mica (Nilaco). After 1 min, the mica surface was rinsed with 100 μl of sterile purified water and dried in air. AFM observation was performed using a commercial microscope operating in the dynamic force mode (model SPI3700-SPA300; Seiko) with an Si micro-cantilever (model SI-DF20; Seiko; spring constant = 15 N/m, resonance frequency = 135 kHz) (39). AFM images were analyzed using the SPIP Metrology software package (Image Metrology).
Preparation of RNAs.
Uncapped in vitro–transcribed dsRNA, capped RNAs, and 5′-triphosphate ssRNA were synthesized using a T7 RiboMAX Express RNAi System (Promega), the T7 Megascript Ultra kit (Ambion; 7mGpppG/GTP ratio of 8:1), and Silencer siRNA Construction kit (Ambion), respectively, following the manufacturers' instructions. Depending on the case, poly I:C was generated in vitro using PNPase with IDP or CDP. CIAP (TaKaRa) treatment was performed as previously described (29). Processing of poly I:C (GE Healthcare) was performed in 10 μl reaction buffer containing 2 μl of 5x buffer (siRNaseIII; TaKaRa), 2.5 mM MgCl2, 10 μg of poly I:C, and 1 U of RNaseIII, for the indicated periods, and the reaction was stopped by 2 μl of 120 mM EDTA. RNase A and Bal31 (TaKaRa) treatment was performed according to the manufacturer's instructions. Chemically synthesized ssRNAs (70 base) with a 5′ hydroxyl were purchased from Gene Design, Inc.
Preparation of viral RNAs.
Vero cells plated on 20 × 15-cm dishes were infected with multiplicity of infection 0.01 of reovirus. At 1 h after infection, medium was removed and replaced with DME containing 10% FCS and the cells were incubated for 2 d at 37°C. Then the supernatants were collected and centrifuged at 3,000 rpm for 15 min to remove cells for avoiding cellar RNA contamination. The supernatants were harvested and centrifuged at 25,000 rpm for 90 min in an SW28 rotor at 4°C. The viral pellet was suspended in TRIzol reagent (Invitrogen) and RNA was extracted. 30 μg of reovirus RNA was obtained.
ELISA.
The amount of dsRNA in viral-infected MEFs was determined by sandwich ELISA (30) using the mAb K1 as the capture antibody and biotinylated mAb J2 for detection, followed by streptavidin alkaline phosphatase. A 400-bp in vitro–transcribed dsRNA was used as a standard to calculate dsRNA concentrations. ELISA for IFN-β was performed as previously described (26).
dsRNA blot.
RNA harvested from noninfected, VSV-, or EMCV-infected cells (20 μg) was electrophoresed in 1.5% agarose gel or 10% nondenaturing polyacrylamide gel, transferred to a nylon membrane (Hybond N+), blotted with mouse anti-dsRNA antibody (J2; English and Scientific Consulting) and visualized with an enhanced chemiluminescence system (PerkinElmer). Reovirus genome RNAs were electrophoresed in the same gel and indicated as the size control.
Online supplemental material.
Fig. S1 shows IFN-β production from GM-CSF–induced WT, RIG-I−/−, and MDA5−/− cDCs in the stimulation with poly I:C or short poly I:C (1 μg/ml). Fig. S2 shows IFN-β production in sera of mice 6 h after intravenous injection of 20 μg poly I:C or short poly I:C. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20080091/DC1.
Supplementary Material
Acknowledgments
The authors wish to thank Y. Fujiwara, M. Shiokawa, and N. Kitagaki for technical assistance and M. Hashimoto for secretarial assistance.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology in Japan; from the Ministry of Health, Labour and Welfare in Japan; from the 21st Century Center of Excellence Program of Japan; and from the National Institutes of Health (AI070167).
The authors have no financial conflict of interests.
Abbreviations used in this paper: AFM, atomic force microscope; CARD, caspase-recruitment domain; cDC, conventional DC; CIAP, calf intestine alkaline phosphatase; DI, defective interfering; dsRNA, double-stranded RNA; EMCV, encephalomyocarditis virus; IPS-1, IFN-β stimulator-1; MEF, mouse embryonic fibroblast; MDA5, melanoma differentiation–associated gene 5; pDC, plasmacytoid DC; PNPase, polynucleotide phosphorylase; poly I:C, polyinosine-polycytidylic acid; PRR, pattern recognition receptor; RIG-I, retinoic acid-inducible gene-I; RLH, RIG-I–like helicase; ssRNA, single-stranded RNA; TIR, Toll/IL-1 receptor homology; TLR, Toll-like receptor; VSV, vesicular stomatitis virus.
References
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. [DOI] [PubMed] [Google Scholar]
- 2.Beutler, B., C. Eidenschenk, K. Crozat, J.L. Imler, O. Takeuchi, J.A. Hoffmann, and S. Akira. 2007. Genetic analysis of resistance to viral infection. Nat. Rev. Immunol. 7:753–766. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov, R. 2007. Recognition of microorganisms and activation of the immune response. Nature. 449:819–826. [DOI] [PubMed] [Google Scholar]
- 4.Fujita, T., K. Onoguchi, K. Onomoto, R. Hirai, and M. Yoneyama. 2007. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie. 89:754–760. [DOI] [PubMed] [Google Scholar]
- 5.Alexopoulou, L., A.C. Holt, R. Medzhitov, and R.A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738. [DOI] [PubMed] [Google Scholar]
- 6.Diebold, S.S., T. Kaisho, H. Hemmi, S. Akira, and C. Reis e Sousa. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 303:1529–1531. [DOI] [PubMed] [Google Scholar]
- 7.Heil, F., H. Hemmi, H. Hochrein, F. Ampenberger, C. Kirschning, S. Akira, G. Lipford, H. Wagner, and S. Bauer. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 303:1526–1529. [DOI] [PubMed] [Google Scholar]
- 8.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature. 408:740–745. [DOI] [PubMed] [Google Scholar]
- 9.Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 9:143–150. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643. [DOI] [PubMed] [Google Scholar]
- 11.Liu, Y.J. 2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23:275–306. [DOI] [PubMed] [Google Scholar]
- 12.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737. [DOI] [PubMed] [Google Scholar]
- 13.Andrejeva, J., K.S. Childs, D.F. Young, T.S. Carlos, N. Stock, S. Goodbourn, and R.E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA. 101:17264–17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenfusser, S., N. Goutagny, G. Diperna, M. Gong, B.G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K.A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260–5268. [DOI] [PubMed] [Google Scholar]
- 15.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K.J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, H., T. Kawai, H. Kato, S. Sato, K. Takahashi, C. Coban, M. Yamamoto, S. Uematsu, K.J. Ishii, O. Takeuchi, and S. Akira. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seth, R.B., L. Sun, C.K. Ea, and Z.J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 122:669–682. [DOI] [PubMed] [Google Scholar]
- 18.Sun, Q., L. Sun, H.H. Liu, X. Chen, R.B. Seth, J. Forman, and Z.J. Chen. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 24:633–642. [DOI] [PubMed] [Google Scholar]
- 19.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 437:1167–1172. [DOI] [PubMed] [Google Scholar]
- 20.Xu, L.G., Y.Y. Wang, K.J. Han, L.Y. Li, Z. Zhai, and H.B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 19:727–740. [DOI] [PubMed] [Google Scholar]
- 21.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 13:539–548. [DOI] [PubMed] [Google Scholar]
- 22.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777. [DOI] [PubMed] [Google Scholar]
- 23.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y.M. Loo, M. Gale Jr., S. Akira, et al. 2005. Shared and unique functions of the DExD/H-Box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858. [DOI] [PubMed] [Google Scholar]
- 24.Saito, T., R. Hirai, Y.M. Loo, D. Owen, C.L. Johnson, S.C. Sinha, S. Akira, T. Fujita, and M. Gale Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA. 104:582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkataraman, T., M. Valdes, R. Elsby, S. Kakuta, G. Caceres, S. Saijo, Y. Iwakura, and G.N. Barber. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444–6455. [DOI] [PubMed] [Google Scholar]
- 26.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K.J. Ishii, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105. [DOI] [PubMed] [Google Scholar]
- 27.Loo, Y.M., J. Fornek, N. Crochet, G. Bajwa, O. Perwitasari, L. Martinez-Sobrido, S. Akira, M.A. Gill, A. Garcia-Sastre, M.G. Katze, and M. Gale Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R.A. Flavell, M.S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA. 103:8459–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K.K. Conzelmann, M. Schlee, et al. 2006. 5′-triphosphate RNA is the ligand for RIG-I. Science. 314:994–997. [DOI] [PubMed] [Google Scholar]
- 30.Pichlmair, A., O. Schulz, C.P. Tan, T.I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 314:997–1001. [DOI] [PubMed] [Google Scholar]
- 31.Marques, J.T., T. Devosse, D. Wang, M. Zamanian-Daryoush, P. Serbinowski, R. Hartmann, T. Fujita, M.A. Behlke, and B.R. Williams. 2006. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 24:559–565. [DOI] [PubMed] [Google Scholar]
- 32.Grunberg-Manago, M., P.J. Oritz, and S. Ochoa. 1955. Enzymatic synthesis of nucleic acidlike polynucleotides. Science. 122:907–910. [DOI] [PubMed] [Google Scholar]
- 33.Weber, F., V. Wagner, S.B. Rasmussen, R. Hartmann, and S.R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, D.H., M. Longo, Y. Han, P. Lundberg, E. Cantin, and J.J. Rossi. 2004. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 22:321–325. [DOI] [PubMed] [Google Scholar]
- 35.Malathi, K., B. Dong, M. Gale Jr., and R.H. Silverman. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 448:816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pattnaik, A.K., L.A. Ball, A. LeGrone, and G.W. Wertz. 1995. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 206:760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 23:19–28. [DOI] [PubMed] [Google Scholar]
- 38.Connolly, J.L., and T.S. Dermody. 2002. Virion disassembly is required for apoptosis induced by reovirus. J. Virol. 76:1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikamo, E., C. Tanaka, T. Kanno, H. Akiyama, G. Jung, H. Tanaka, and T. Kawai. 2005. Native polysomes of Saccharomyces cerevisiae in liquid solution observed by atomic force microscopy. J. Struct. Biol. 151:106–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.