Abstract
The allele and genotype frequencies of the prion protein gene (PrP), known to have an impact on scrapie susceptibility, were determined by real-time PCR for 500 Quebec purebred rams. Molecular beacons were very efficient in discriminating the 5 alleles investigated. Polymorphisms at coding positions 136, 154, and 171 of the PrP gene were analyzed using 3 separate real-time PCR reactions and a total of 7 molecular beacons. A total of 4 different alleles (ARQ, ARR, AHR, and VRQ) were observed at different frequencies among the 7 breeds of sheep investigated. Results show that more than 50% of the rams in every breed carried at least one ARR allele, which is considered the most resistant to scrapie. The susceptibility ARQ allele was also present in every breed and together with the ARR allele, they were the most frequent alleles found in Quebec rams. The VRQ allele associated with the highest susceptibility to scrapie occurred in 5 of the 7 breeds, although at low frequencies. Overall, the results indicate that the frequencies of PrP alleles and genotypes in common breeds of sheep in Quebec make it feasible to reduce scrapie risk by selective breeding.
Résumé
Les fréquences alléliques et génotypiques du gène de la protéine prion (PrP), reconnu pour avoir un impact sur la susceptibilité envers la tremblante, ont été déterminées par PCR en temps réel pour 500 béliers pur-sang du Québec. Des balises moléculaires étaient très efficaces pour discriminer les 5 allèles étudiés. Le polymorphisme aux positions codantes 136, 154 et 171 du gène PrP a été analysé à l’aide de 3 analyses séparées de PCR en temps réel et un total de 7 balises moléculaires. Quatre allèles différents (ARQ, ARR, AHR et VRQ) ont été observés à des fréquences variables parmi les 7 races de moutons étudiées. Les résultats montrent que plus de 50 % des béliers dans chaque race sont porteurs d’au moins un allèle ARR, considéré comme étant le plus résistant à la tremblante. L’allèle de susceptibilité ARQ était également présent dans chaque race et avec l’allèle ARR ils étaient les allèles les plus fréquemment rencontrés chez les béliers du Québec. L’allèle VRQ associé avec la plus grande susceptibilité à la tremblante était rencontré chez 5 des 7 races, bien qu’à des fréquences peu élevées. De manière générale, les résultats indiquent que la fréquence des allèles PrP et des génotypes chez les races de mouton communes au Québec rendent possible une réduction du risque de tremblante en effectuant des accouplements sélectifs.
(Traduit par Docteur Serge Messier)
Introduction
Scrapie is a member of the transmissible spongiform encephal-opathies (TSEs) which include bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) in cervids, and Creutzfeldt-Jacob (CJD) disease in humans. While the exact etiology of TSEs is still debated, the disease is associated with the presence of an abnormal isoform of a host-encoded protein called a prion (1). In many species, including sheep, polymorphisms in the prion gene are associated with susceptibility to the disease. Considerable data now exist on the genetics of susceptibility to scrapie (2). It is generally accepted that susceptibility or resistance to scrapie is modulated by allelic variants of the prion protein (PrP) gene (3). Over 40 polymorphisms have been reported so far for the PrP gene, although only those at codons 136, 154, and 171 appear to be closely linked to scrapie susceptibility. These later polymorphisms give rise to 5 alleles that vary by 1 amino acid from what is considered to be the original or wild type allele (ARQ): ARR, AHQ, ARH, VRQ, and ARQ (4). Polymorphisms at codon 136 (alanine A or valine V) and at codon 171 (glutamine Q, histidine H, or arginine R) clearly influence susceptibility of developing scrapie (5–7). The association between the polymorphism at codon 154 (arginine R or histidine H) and susceptibility to scrapie is still unclear since some data suggest that a histidine at codon 154 offers some protection in certain breeds of sheep (7) but not in others (8). The genotype AA136RR154RR171 is associated with the highest resistance to scrapie, whereas the genotype VV136RR154QQ171 is associated with the highest susceptibility to infection (7). In breeds where the V136R154Q171 allele is absent, homozygous AA136RR154QQ171 animals are the most susceptible. Based on this linkage, a classification scheme has been established in the UK under the National Scrapie Plan (9), with NSP type 1 representing sheep that are genetically most resistant to scrapie and NSP type 5 representing those that are highly susceptible to scrapie and that should not be used for breeding. Several studies show that the distribution of scrapie genotypes varies widely according to breed or geographic origin.
Several techniques have been used by different laboratories to genotype the PrP gene. These include polymerase chain reaction restriction fragment length polymorphisms (PCR-RFLP), PCR followed by dot blot hybridization, sequencing and real-time PCR using Taqman technology. The novel method used in this study utilizes molecular beacons. This technique was selected because it offers very high specificity and high throughput capacity. Due to their hairpin structure, molecular beacons are uniquely suited for the detection of single nucleotide polymorphisms (SNPs) because they are much more specific in their target recognition than linear probes (10,11). Moreover, since molecular beacons can possess different fluorophores, multiple targets can be detected in the same reaction tube (multiplex).
Scrapie is widespread and is found in most parts of the world. Diagnosed in Canada for the first time in 1938, it was made a reportable disease in 1945, and a control program was put in place. Since 1984, over 140 infected and exposed flocks have been depopulated, mostly in Quebec. The control program has evolved since 1945 and genotyping is now part of the different strategies offered to the owner of an infected flock (12). There are over 180 000 reproductive ewes in Quebec flocks, about 10 000 to 12 000 of which are purebred; there are approximately 3500 purebred rams. Most of the flocks are commercial flocks in which mainly crossbred or hybrid females are used with registered or nonregistered purebred rams; lambs are sold to the meat market. Rams are either terminal (meat) or maternal/ prolific type breeds.
The aim of this study was to investigate prion gene polymorphisms at 3 codons of interest in common breeds of sheep in Quebec to provide information about their susceptibility to scrapie and about the genetic potential within each breed to reduce this susceptibility.
Materials and methods
Sheep
A total of 500 purebred rams from 49 flocks were selected for this study (Table I). Rams were chosen because they are thought to represent the PrP genotypes found within each of their respective breeds. The rams were from the most common paternal (Suffolk, Hampshire, and Canadian Arcott), maternal (Dorset and Polypay) and prolific breeds (Rideau Arcott and Romanov) in Quebec. All flocks had to be participating in the Quebec genetic Scheme (GenOvis). The sampling size for each breed reflected the different proportion of these breeds in GenOvis (2000–2002). In 2003, when the study took place, there were over 1100 rams and ram lambs available in 67 GenOvis participating flocks. Moreover, considering that maternal and prolific breeds have a larger incidence on the whole reproductive population (breeding ewes), it was decided to have these breeds overrepresented.
Table I.
Number of rams sampled from each breed
| Breed | Number of flocks | Number of rams |
|---|---|---|
| Canadian Arcott (CD) | 5 | 28 |
| Suffolk (SU) | 13 | 84 |
| Hampshire (HA) | 5 | 40 |
| Dorset (DP) | 22 | 142 |
| Polypay (PO) | 7 | 97 |
| Rideau Arcott (RI) | 9 | 52 |
| Romanov (RV) | 6 | 57 |
| Totals | 67 | 500 |
Note: There was more than one breed on some farms.
DNA samples
All analyses were performed at TransBIOtech in Lévis, Québec. For each sheep, 5 mL of blood was collected by jugular venipuncture into an EDTA-vacutainer and kept cold until deoxyribonucleic acid (DNA) extraction. Genomic DNA was extracted from white blood cells in 200 μL of whole blood using the DNA blood kit (Qiagen, Mississauga, Ontario) following the manufacturer’s instructions.
Genotyping
All primers and beacons were synthesized and purified by high performance liquid chromatography (HPLC) at Integrated DNA Technologies (IDT), Coralville, Iowa, USA. Three independent real-time PCR reactions were developed, 1 for each of the 3 codons of interest. Each reaction was optimized and validated with DNA from animals of known genotypes (ARQ/ARQ, ARQ/VRQ, VRQ/VRQ, AHQ/ARQ, AHQ/AHQ, ARR/ARQ, ARR/ARR, ARH/ARQ, ARH/ARR, ARH/ARH). For each reaction, the conditions were as follows: in a final volume of 25 μL, containing 100 μM of each nucleotide, 400 μM of each primer, 200 μM of each beacon, 3 mM MgCl2, 100 mM Tris-HCl (pH 8.3), 50 mM KCl and 0.5U Platinum Taq (Invitrogen, Burlington, Ontario), 50–100 ng of genomic DNA (determined at 260 nM in a spectrophotometer) was subjected to PCR amplification in an M × 3000p instrument (Stratagene, La Jolla, California). Each sample was tested in duplicate, and appropriate controls from animals whose genotypes were determined by automated sequencing were included in each run. Furthermore, 142 animals had their genotype determined in a blind fashion by TransBioTech Lévis, Quebec and a French accredited laboratory (Labogena, 78352 Jouy-en-Josas, France). All results were fully congruent.
Codon 136
The thermocycling protocol was: 2 min at 94°C followed by 40 cycles at 94°C for 15 s, 62°C for 45 s, and 72°C for 30 s. Fluorescence was measured at each cycle at the end of the annealing step. The nucleotide sequences of each beacon and primer are as follows: Beacon 136A: 5′ FAM - TCG GGC TGG GAA GTG CCA TGA GCA GGC GCC CGA - BHQ1 3′, Beacon 136V: 5′-HEX AGC GAT TGG GAA GTG TCA TGA GCA GGA TCG CT - BHQ2 3′, sens-primer: - 5′-ATG AAG CAT GTG GCA GGA GC - 3′, anti sens primer: - 5′ - ACG GTC CTC ATA GTC ATT GCC - 3′.
Codon 154
The thermocycling protocol was: 2 min at 94°C followed by 40 cycles at 94°C for 15 s, 60°C for 45 s, and 72°C for 30 s. Fluorescence was measured at each cycle at the end of the annealing step. The nucleotide sequences of each beacon and primer are as follows: beacon 154H: 5′ HEX - CGC GAG ACC GTT ACT ATC ATG AAA ACA TGT CGC G - BHQ2 3′, beacon 154R: 5′ FAM - CGC GAA CCG TTA CTA TCG TGA AAA CAT GTC GCG - BHQ 1 3′, sens primer: 5′ - ATG AAG CAT GTG GCA GGA GC - 3′, anti-sens primer: 5′ - CTG ATC CAC TGG TCT GTA GTA CAC - 3′.
Codon 171
The thermocycling protocol was: 2 min at 94°C followed by 40 cycles at 94°C for 15 s, 59°C for 45 s, and 72°C for 30 s. Fluorescence was measured at each cycle at the end of the annealing step. The nucleotide sequences of each beacon and primer are as follows: beacon 171Q: 5′ - FAM CGC GAT CCA GTG GAT CAG TAT AGT AAC CAT CGC G - BHQ1 3′, beacon 171R: 5′ - HEX CGC GAT CCA GTG GAT CCG TAT AGT AAC CAT CGC G - BHQ2 3′, beacon171H: 5′ - CY5 CGC GAT CCA GTG GAT CAT TAT AGT AAC CAT CGC G - BHQ2 3′, sens primer: 5′ - CGT GAA AAC ATG TAC CGT TAC CCC - 3′, and antisens primer: 5′ - GGT GAC TGT GTG TTG CTT GAC TG - 3′.
Results
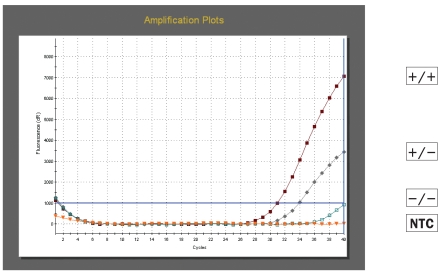
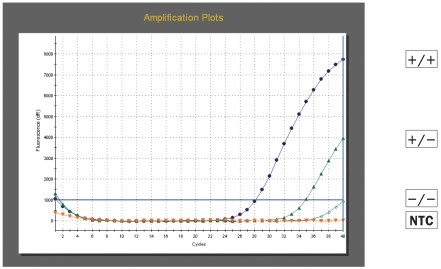
All molecular beacons were very efficient in discriminating the single base mismatches responsible for the allelic differences under study. An example of the results obtained with codon 136 molecular beacons is shown in Figures 1a and 1b. The beacons only fluoresced and gave a strong signal in the presence of their perfectly complementary target making allele scoring unambiguous. In the absence of their perfectly complementary target, the probes remained dark and no or very weak signals were detected (samples −/− Figures 1a and 1b).
Figure 1a.
Amplification plots obtained with dual labelled molecular beacon A136 (FAM) for PrP codon 136 genotyping. Results are shown for an AA 136 homozygote (+/+), a VA136 heterozygote (+/−), a VV136 homozygote (−/−) and a no template control (NTC).
Figure 1b.
Amplification plots obtained with dual labelled molecular beacon VA136 (HEX) for PrP codon 136 genotyping. Results are shown for a VV136 homozygote (+/+), a V136 heterozygote (+/−), an AA136 homozygote (−/−) and a no template control (NTC).
The PrP allelic frequencies of the 500 sampled rams are summarized for each breed in Table II. Four of the 5 PrP alleles associated with susceptibility or resistance to scrapie were found: ARR, ARQ, AHQ, and VRQ — no ARH. Codon 171 was polymorphic (R/Q) in every breed but no histidine (H) was found meaning that every breed sampled has access to the ARR allele associated with the greatest resistance to scrapie. Codon 154 was also found to be polymorphic (R/H) in 4 of the breeds (Rideau Arcott, Polypay, Dorset, and Canadian Arcott) but with a very low proportion of the AHQ allele (< 2%). The remaining 3 breeds had only arginine (R) and no H at position 154. At codon 136, all breeds but 2 (Rideau Arcott and Polypay, which had no VRQ allele) were polymorphic (V/A) although the frequency of the VRQ allele (associated with the highest susceptibility to scrapie) was always very low.
Table II.
Percentage frequencies of prion protein alleles (PrP) in 7 breeds of sheep in Quebec (n = 1000 alleles − 500 rams)
| Breed | CD | DP | HA | PO | RI | RV | SU |
|---|---|---|---|---|---|---|---|
| ARR | 60.7 | 52.5 | 36.25 | 52.6 | 45.2 | 28.9 | 49.4 |
| AHQ | 1.8 | 0.7 | 0 | 0.5 | 1.9 | 0 | 0 |
| ARQ | 30.4 | 40.8 | 62.5 | 46.5 | 52.9 | 66.7 | 49.4 |
| VRQ | 7.1 | 6.0 | 1.25 | 0 | 0 | 4.4 | 1.2 |
CD — Canadian Arcott; DP — Dorset; HA — Hampshire; PO — Polypay; RI — Rideau Arcott; RV — Romanov; SU — Suffolk.
In all breeds, the dominant alleles were ARQ and ARR and the less frequent were AHQ and VRQ, but there was considerable variation among breeds. In all breeds, the frequency of the ARQ allele exceeded 30% and it reached 66.7% in Romanov. For the most resistant ARR allele, all breeds showed a relative frequency of > 25% and it reached or exceeded 50% for Canadian Arcott, Dorset, Suffolk, and Polypay. The VRQ allele, which is associated with the highest susceptibility to scrapie, was found at low frequencies (between 1.2% and 7.1%) in Suffolk, Hampshire, Romanov, Dorset, and Canadian Arcott. The AHQ allele was found in very low proportion (< 2%) in Canadian Arcott, Dorset, Polypay, and Rideau Arcott.
The PrP genotypes frequencies for the 7 breeds are shown in Table III. Eight different genotypes were identified in the sampled rams: ARR/ARR, ARR/ARQ, ARR/AHQ, ARQ/ARQ, ARQ/AHQ, ARR/VRQ, ARQ/VRQ, and VRQ/VRQ. Canadian Arcott, Dorset, and Polypay breeds had the highest percentage of the NSP type 1 genotype (ARR/ARR) (39%, 29%, and 27%, respectively), whereas the Romanov breed had the lowest percentage (7%) of the most resistant rams. Of note is that for all breeds, > 50% of the rams tested were of NSP type 1 (ARR/ARR) or 2 (ARR/ARQ and ARR/AHQ) which are all considered to confer resistance to scrapie. In most breeds, the cumulative frequencies of these 3 genotypes were over 70%. The most susceptible genotypes (VRQ/VRQ, VRQ/ARQ), which correspond to NSP type 5, were found in small proportions in Romanov (7.0%), Dorset (4.9%), Canadian Arcott (3.6%), and Suffolk (1.2%).
Table III.
Percentage frequencies of prion protein (PrP) genotypes in 7 Quebec sheep breeds (n = 500 rams)
| GENOTYPES | All breeds | CD | SU | HA | DP | PO | RI | RV |
|---|---|---|---|---|---|---|---|---|
| ARR/ARR | 21.8 | 39.3 | 19.1 | 12.5 | 28.9 | 26.8 | 11.5 | 7.0 |
| ARR/AHQ | 0.2 | 0 | 0 | 0 | 0 | 0 | 1.9 | 0 |
| ARR/ARQ | 48.8 | 32.1 | 59.5 | 45.0 | 40.9 | 51.6 | 65.4 | 43.9 |
| ARR/VRQ | 2.8 | 10.7 | 1.2 | 2.5 | 6.3 | 0 | 0 | 0 |
| AHQ/ARQ | 1.0 | 3.6 | 0 | 0 | 1.4 | 1.0 | 1.9 | 0 |
| ARQ/ARQ | 22.8 | 10.7 | 19.1 | 40.0 | 17.6 | 20.6 | 19.2 | 42.1 |
| ARQ/VRQ | 2.2 | 3.6 | 1.2 | 0 | 4.2 | 0 | 0 | 5.3 |
| VRQ/VRQ | 0.4 | 0 | 0 | 0 | 0.7 | 0 | 0 | 1.8 |
CD — Canadian Arcott; DP — Dorset; HA — Hampshire; PO — Polypay; RI — Rideau Arcott; RV — Romanov; SU — Suffolk.
Discussion
Although their design is challenging, molecular beacons can be very efficient in distinguishing between single base pair mismatch allelic variants (Figures 1a and 1b). The fact that 142 samples from this study gave identical results in our laboratory and the French laboratory Labogena, supports this finding. The assays are simple, rapid, and amenable to multiplexing and automation which make them ideally suited for routine diagnosis. Moreover, since the amplification, hybridization, and analysis are all performed simultaneously in a closed tube, the risk of cross-contamination is greatly reduced.
The first objective of this study was to estimate the frequencies of PrP alleles and genotypes among the major sheep breeds in Quebec. One other goal was to give breeders an indication of the genetic potential within each breed or flock in breeding animals with low genetic susceptibility to scrapie. This study represents a starting point for breeders interested in a breeding process that includes the selection of ARR/ARR animals and the elimination of VRQ-carrying animals.
The ARR allele, which plays an important role in scrapie resistance, was found in each breed and more than 50% of the rams in every studied breed carried at least 1 copy of this allele (NSP type 1 and 2). The breeds with the highest proportion of ARR/ARR rams were the Canadian Arcott with 39.3%, Dorset with 28.9%, and Polypay with 26.8%. The high percentage of resistant rams in some breeds probably reflects the fact that some breeders in Quebec have been selecting for scrapie resistant rams for several years. Another explanation for this high percentage of ARR/ARR rams observed in some breeds might be the founder effect. This might be the case for the Canadian Arcott where, to our knowledge, no selection based on scrapie genotypes had been done prior to our study. In this particular breed, the relatively small size of the sample might also cause a bias (few source flocks and less genetic variability).
Such high frequency of the ARR allele is common in many countries and in different breeds (12–15). The ARH allele was not found in any of the animals tested in this study, which is not unexpected since the ARH allele is usually found in a small proportion of animals of very few breeds like the Texel and Suffolk (16–18).
VRQ-carrying animals are considered to be the most susceptible to scrapie. In the present study, the VRQ allele was found in 5 of the 7 breeds but at low frequency similar to what has been found in other sheep breeds around the world (12–15). With a proportion of 14.3% and 7.0% of VRQ-carrying rams respectively, the Canadian Arcott and the Romanov breeds had the highest frequency of highly susceptible animals. It is interesting to note that although the Canadian Arcott had the highest proportion (14.3%) of very susceptible VRQ-carrying animals, it also had the highest proportion (39.3%) of the most resistant rams (ARR/ARR).
In non-VRQ breeds, animals with the ARQ/ARQ genotype are considered to be the most susceptible to scrapie. In this regard, Romanov and Hampshire rams must be considered at risk with a proportion of 42% and 40% of ARQ/ARQ animals, respectively. The proportion of ARQ homozygous rams in the other breeds was between 10% and 20%.
Among the rams studied here, the Romanov and the Hampshire appeared to be the most susceptible to natural scrapie with 49.1% and 42.5% of rams being of susceptible genotypes respectively (NSPtype 4 and 5). The Canadian Arcott rams also appeared to be at great risk with 14.3% of VRQ-carrying rams, but the high proportion of ARR-carrying rams in this breed (71.4%) makes it possible to rapidly diminish or eliminate the VRQ allele by selective breeding.
The results reported here can serve as a starting point for Quebec breeders in developing breeding strategies to increase overall resistance to the common strains of scrapie. Breeding towards scrapie resistance is clearly possible for the 7 breeds included in this study. However, it will certainly take more time in some breeds where less homozygous ARR rams are available. In these breeds, ARR heterozygous rams will have to be used for the first few generations. To ensure that no production trait is lost as the frequency of ARR allele is increased in the population, it is important to keep in mind that improving scrapie resistance should not be the sole, neither the first, selection criterion. So far, little or no association has been found between scrapie genotypes and important reproduction or production traits such as weight gain, ovulation rate, or litter size (19–21).
When selecting toward genetic resistance to natural scrapie, one must also consider the occurrence of atypical scrapie, such as the Nor98 isolate, in usually resistant sheep (22,23). Even though these rare isolates have not yet been found in Canada, they confirm the fact that ARR/ARR resistance is relative and not absolute and that the selection process must continuously adjust to new scientific findings.
Acknowledgments
This project was financially supported by a grant from Le Conseil pour le développement de l’agriculture du Québec (CDAQ). The authors wish to thank all the sheep producers that participated in the study. Special thanks are also given to Rebecca Guy for reviewing the manuscript.
References
- 1.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Baylis M, Goldmann W. The genetics of scrapie in sheep and goats. Curr Mol Med. 2004;4:385–396. doi: 10.2174/1566524043360672. [DOI] [PubMed] [Google Scholar]
- 3.Hunter N, Goldmann W, Marshall E, O’Neill G. Sheep and goats: Natural and experimental TSEs and factors influencing incidence of disease. Arch Virol Suppl. 2000;16:181–188. doi: 10.1007/978-3-7091-6308-5_17. [DOI] [PubMed] [Google Scholar]
- 4.Hunter N, Foster JD, Goldmann W, Stear MJ, Hope J, Bostock C. Natural scrapie in a closed flock of Cheviot sheep occurs only in specific PrP genotypes. Arch Virol. 1996;141:809–824. doi: 10.1007/BF01718157. [DOI] [PubMed] [Google Scholar]
- 5.Belt PB, Muileman IH, Schreuder BE, Bos-de Ruijter J, Gielkens AL, Smits MA. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol. 1995;76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 6.Clouscard C, Beaudry P, Elsen JM, et al. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. J Gen Virol. 1995;76:2097–2101. doi: 10.1099/0022-1317-76-8-2097. [DOI] [PubMed] [Google Scholar]
- 7.Elsen JM, Amigues Y, Schelcher F, et al. Genetic susceptibility and transmission factors in scrapie: Detailed analysis of an epidemic in a closed flock of Romanov. Arch Virol. 1999;144:431–445. doi: 10.1007/s007050050516. [DOI] [PubMed] [Google Scholar]
- 8.Billinis C, Psychas V, Leontides L, et al. Prion protein gene polymorphisms in healthy and scrapie-affected sheep in Greece. J Gen Virol. 2004;85:547–554. doi: 10.1099/vir.0.19520-0. [DOI] [PubMed] [Google Scholar]
- 9.Anonymous [Last accessed April 28, 2008];National Scrapie Plan for Great Britain. 2006 (NSP, DEFRA, 2006) [homepage on the Internet]. Available from http://www.defra.gov.uk/animalh/svj/vol1401/svj-vol1401.pdf.
- 10.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 11.Piatek AS, Tyagi S, Pol AC, et al. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 12.Canadian Food Inspection Agency. [Last accessed April 26, 2008]; [homepage on the Internet]. Available from http://www.inspection.gc.ca/english/anima/heasan/man/scrtre/scrtree.shtml.
- 13.Eglin RD, Warner R, Gubbins S, Sivam SK, Dawson M. Frequencies of PrP genotypes in 38 breeds of sheep sampled in the National Scrapie Plan for Great Britain. Vet Rec. 2005;156:433–437. doi: 10.1136/vr.156.14.433. [DOI] [PubMed] [Google Scholar]
- 14.Holko I, Novackova A, Holkova T, Kmet V. PrP genotyping of sheep breeds in Slovakia. Vet Rec. 2005;157:628–630. doi: 10.1136/vr.157.20.628. [DOI] [PubMed] [Google Scholar]
- 15.DeSilva U, Guo X, Kupfer DM, et al. Allelic variants of ovine prion protein gene (PRNP) in Oklahoma sheep. Cytogenet Genome Res. 2003;102:89–94. doi: 10.1159/000075731. [DOI] [PubMed] [Google Scholar]
- 16.Sild E, Volskiene R, Viinalass H, et al. Detection of prion protein gene polymorphisms in Baltic breeds of sheep. Vet Rec. 2006;159:247–250. doi: 10.1136/vr.159.8.247. [DOI] [PubMed] [Google Scholar]
- 17.O’Doherty E, Aherne M, Ennis S, Weavers E, Roche JF, Sweeney T. Prion protein gene polymorphisms in pedigree sheep in Ireland. Res Vet Sci. 2001;70:51–56. doi: 10.1053/rvsc.2000.0441. [DOI] [PubMed] [Google Scholar]
- 18.Drögemüller C, Leeb T, Distl O. PrP genotype frequencies in German breeding sheep and the potential to breed for resistance to scrapie. Vet Rec. 2001;149:349–352. doi: 10.1136/vr.149.12.349. [DOI] [PubMed] [Google Scholar]
- 19.de Vries F, Borchers N, Hamann H, et al. Associations between the prion protein genotype and performance traits of meat breeds of sheep. Vet Rec. 2004;155:140–143. doi: 10.1136/vr.155.5.140. [DOI] [PubMed] [Google Scholar]
- 20.Vitezica ZG, Moreno CR, Bodin L, et al. No associations between PrP genotypes and reproduction traits in INRA 401 sheep. J Anim Sci. 2006;84:1317–1322. doi: 10.2527/2006.8461317x. [DOI] [PubMed] [Google Scholar]
- 21.Sweeney T, Hanrahan JP, O’Doherty E. Is there a relationship between prion protein genotype and ovulation rate and litter size in sheep? Anim Reprod Sci. 2007;101:153–157. doi: 10.1016/j.anireprosci.2006.12.004. Epub 2006, Dec. 15. [DOI] [PubMed] [Google Scholar]
- 22.Benestad SL, Sarradin P, Thu B, Schönheit J, Tranulis MA, Bratberg B. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet Rec. 2003;153:202–208. doi: 10.1136/vr.153.7.202. [DOI] [PubMed] [Google Scholar]
- 23.Le Dur A, Béringue V, Andréoletti O, et al. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc Natl Acad Sci U S A. 2005;102:16031–16036. doi: 10.1073/pnas.0502296102. Epub 2005, Oct 20. [DOI] [PMC free article] [PubMed] [Google Scholar]