Abstract
The type I, 55-kDa tumor necrosis factor receptor (TNFR1) is released to the extracellular space by two mechanisms, the constitutive release of TNFR1 exosome-like vesicles and the inducible proteolytic cleavage of TNFR1 ectodomains. Both pathways appear to be regulated by an interaction between TNFR1 and ARTS-1 (aminopeptidase regulator of TNFR1 shedding). Here, we sought to identify ARTS-1-interacting proteins that modulate TNFR1 release. Co-immunoprecipitation identified an association between ARTS-1 and RBMX (RNA-binding motif gene, X chromosome), a 43-kDa heterogeneous nuclear ribonucleoprotein. RNA interference attenuated RBMX expression, which reduced both the constitutive release of TNFR1 exosome-like vesicles and the IL-1β-mediated inducible proteolytic cleavage of soluble TNFR1 ectodomains. Reciprocally, over-expression of RBMX increased TNFR1 exosome-like vesicle release and the IL-1β-mediated inducible shedding of TNFR1 ectodomains. This identifies RBMX as an ARTS-1-associated protein that regulates both the constitutive release of TNFR1 exosome-like vesicles and the inducible proteolytic cleavage of TNFR1 ectodomains.
Keywords: Cytokine Receptors, Inflammation, Endothelial cells, Cytokines
Introduction
Tumor necrosis factor (TNF, TNFSF2) activity can be regulated by extracellular TNF receptors that function as TNF-binding proteins. The 55-kDa, type I TNF receptor (TNFR1) can be released from cells by two pathways. The first involves proteolytic cleavage of TNFR1 ectodomains to generate 27–34 kDa soluble TNFR1 (sTNFR1)[1]. The second involves the constitutive release of full-length TNFR1 within the membranes of 20–50 nm exosome-like vesicles that are capable of binding TNF[2]. The translocation of intracytoplasmic TNFR1 vesicles appears to play an important role in both pathways. We previously identified ARTS-1 (Aminopeptidase Regulator of TNFR1 Shedding), also known as endoplasmic reticulum-associated aminopeptidase 1, as a type II integral membrane protein that binds full-length TNFR1 and regulates both the constitutive release of TNFR1 exosome-like vesicles and the IL-1β-mediated inducible proteolytic cleavage of soluble TNFR1 ectodomains[3, 4]. Nucleobindin 2 (NUCB2), a putative DNA- and calcium-binding protein, was subsequently identified as a calcium-dependent ARTS-1 binding partner that associates with cytoplasmic TNFR1 prior to its commitment to either release pathway[3]. Here, we sought to identify additional ARTS-1-associated proteins that regulate TNFR1 release. Using a mass spectroscopy approach, we show that RBMX (RNA binding motif protein, X-linked), a 43-kDa RNA-binding motif protein (heterogeneous nuclear ribonucleoprotein (hnRNP) G), associates with ARTS-1 and regulates both the constitutive release of TNFR1 exosome-like vesicles and the IL-1β-mediated inducible proteolytic cleavage of TNFR1 ectodomains. This identifies an unexpected role for a hnRNP in the regulation of TNFR1 release pathways.
Methods
Cells and Reagents
NCI-H292 cells (American Type Culture Collection, Manassas, VA) were grown in RPMI 1640 medium with 10% fetal bovine serum (Invitrogen Corporation, Carlsbad, CA). HUVEC were grown in EGM-2 medium (Cambrex, East Rutherford, NJ). Recombinant human IL-1β was from R & D Systems (Minneapolis, MN). Goat polyclonal anti-RBMX, murine monoclonal anti-TNFR1 (H5), and murine monoclonal anti-β-tubulin (D10) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA).
Immunoblotting
Immunoblotting was performed as previously described [3, 4]. For immunoblots of HUVEC-conditioned medium, cells were grown in exosome-depleted medium. For immunoblots of HUVEC-conditioned medium following IL-1β stimulation, cells were incubated in 1 ml of EGM-2 medium without fetal bovine serum or supplements for 2-h. Proteins present in 0.5 ml of conditioned medium were precipitated with 10% trichloroacetic acid. Densitometry was performed using NIH Image Software (version 1.63).
Immunoprecipitations
Immunoprecipitations of NCI-H292 cellular proteins were performed as previously described[4]. Samples of membrane proteins (1 mg) were incubated overnight at 4 °C with 1 µl of rabbit anti-ARTS-1 antibody or non-immune serum, followed by addition of 200 µl of immobilized protein A/G beads (Pierce, Rockford, IL) for 2-h. After washing 6 times in lysis buffer, bound proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue R-250. Bands were excised and proteins were identified by MALDI-MS peptide mass mapping.
HUVEC were lysed in buffer containing 50 mM Tris, pH 7.5, 120 mM NaCl, and 0.1% Triton X-100, with aprotinin (2 ug/ml) and AEBSF (1 mg/ml). Protein A/G beads were blocked with 1% ovalbumin and incubated with 1 µl of rabbit anti-ARTS-1 antibody or pre-immune serum. Samples (300 ug) of HUVEC lysates were treated with or without SUPERase-In RNase inhibitor (1 unit/ul) or RNase cocktail (0.4 units/ul), which contains RNases A and T1 (Ambion, Austin, TX). Lysates were incubated with antibody-coated protein A/G beads, washed with ice-cold PBS, followed by Western blotting. Proteins remaining in supernatants were precipitated with 10% trichloroacetic acid prior to Western blotting.
RNA Interference and Quantitative Real-Time RT-PCR
The RBMX siRNA duplex, (GUGGAAGUCGAGACAGUUAUU and UAACUGUCUCGACUUCCACUU), was purchased from Dharmacon (Lafayette, CO). The control siRNA duplex that targets green fluorescent protein (GFP) was purchased from Qiagen-Xeragon (Germantown, MD). HUVEC were transfected with 25 nM siRNA using DharmaFECT #1 (Dharmacon, Chicago, IL) and assayed 2 days post-transfection.
RBMX Expression Plasmid
The full-length coding sequence of RBMX was generated by RT-PCR of HUVEC mRNA, cloned into pcDNA3.1/V5-His TOPO TA (Invitrogen), and sequence verified. HUVEC, grown in 6-well plates, were transfected with plasmids using FuGENE 6, (Roche, Indianapolis, IN), as previously described[5].
Quantification of Extracellular TNFR1 by ELISA
To assess constitutive TNFR1 release, HUVEC were transfected with siRNA targeting RBMX or GFP for 1 day and fresh, exosome-depleted medium was added for an additional 24-h. To assess IL-1β-induced TNFR1 release, HUVEC were transfected with siRNA targeting RBMX or GFP for 2 days and medium with IL-1β, but without FBS or supplements, was added for 2-h. Conditioned medium was collected and cleared of cells and debris prior to TNFR1 quantification using a Quantikine sandwich ELISA kit (R & D Systems).
Statistical Analysis
Data were analyzed by a paired Student’s t test. A P value ≤ 0.05 was considered significant.
Results
RBMX Co-immunoprecipitates with ARTS-1
RBMX co-immunoprecipitated with ARTS-1 from membrane proteins isolated from NCI-H292 human pulmonary epithelial cells and was identified by mass spectroscopy (Fig. 1A). Cytokeratin 18, which was pulled-down by both the ARTS-1 antibody and pre-immune serum, was considered to be bound non-specifically. The association between endogenous ARTS-1 and RBMX was confirmed by immunoprecipitation of HUVEC proteins (Fig. 1B).
Figure 1. Co-immunoprecipitation of Endogenous RBMX and ARTS-1.
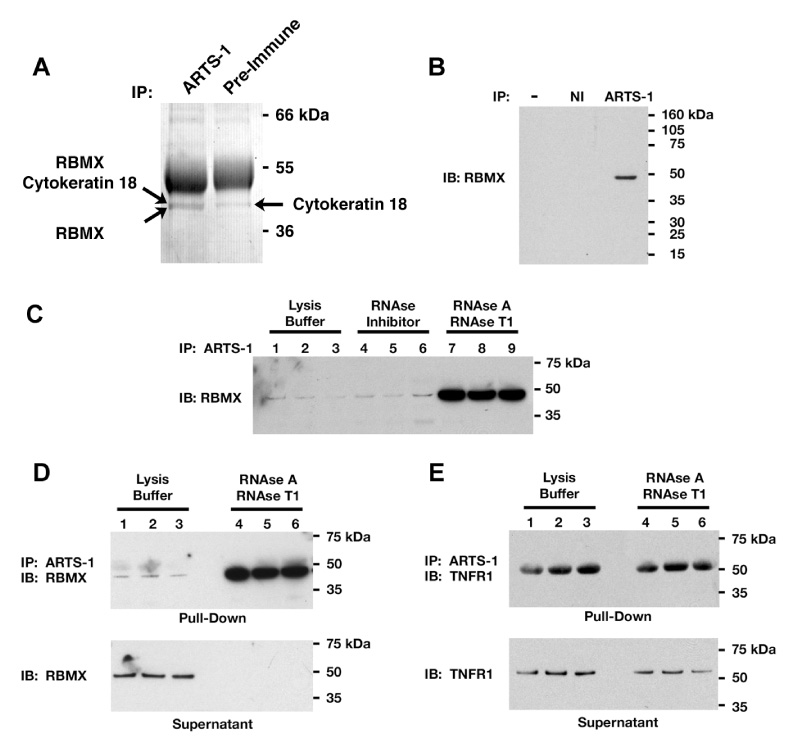
Panel A. NCI-H292 cell membrane proteins were immunoprecipitated (IP) with anti-ARTS-1 antibodies or pre-immune serum and stained with Coomassie blue. This gel is representative of two experiments. Panel B. HUVEC lysates were immunoprecipitated with the anti-ARTS-1 antibody or non-immune serum (NI) and RBMX was detected by Western blotting. Panel C. HUVEC lysates, in triplicate, were incubated with RNase inhibitor or a mixture of RNases A and T1 for 1-h prior to immunoprecipitation with the anti-ARTS-1 antibody. RBMX was detected by Western blotting. Panel D. Immunoprecipitation of proteins in HUVEC lysates were performed, in triplicate, as in Panel C. Proteins pulled-down by the ARTS-1 immunoprecipitation are shown in the top panel, while proteins remaining in lysates are shown in the bottom panel labeled supernatant. Panel E. Immunoprecipitation of proteins in HUVEC lysates, performed in triplicate, as described in Panels C and D. TNFR1 was detected by immunoblotting.
A Subset of RBMX that is not bound by RNA Associates with ARTS-1
Since RBMX is a RNA-binding protein, we hypothesized that its association with ARTS-1 was modulated by RNA binding. HUVEC lysates were incubated without or with RNase inhibitor or RNases, prior to ARTS-1 immunoprecipitation. Treatment with RNase inhibitors had no effect on the association between ARTS-1 and RBMX, whereas treatment with RNases significantly increased the amount of RBMX that was pulled-down by ARTS-1 (Fig. 1C). This result was confirmed in Fig. 1D, which showed that before exposure to RNases, only a subset of total cellular RBMX associated with ARTS-1, whereas following the RNase-mediated degradation of RNA, all the RBMX present in HUVEC lysates associated with ARTS-1. The RNase-mediated degradation of RNA had no effect on the ability of ARTS-1 to pull-down TNFR1 (Fig. 1E).
RBMX Mediates the Constitutive Release of TNFR1 Exosome-like Vesicles
Cells transfected with siRNA targeting RBMX showed a significant reduction in constitutive TNFR1 release as detected by ELISA (Fig. 2A–C). Similarly, Western blots of culture medium showed a significant reduction in the amount of full-length, 55-kDa TNFR1, which is consistent with decreased release of exosome-like vesicles. As previously reported, a minor approx. 40-kDa band was also detected, which was similarly decreased in medium from cells transfected with siRNA targeting RBMX[3].
Figure 2. RBMX Modulates the Constitutive Release of TNFR1 Exosome-like Vesicles.
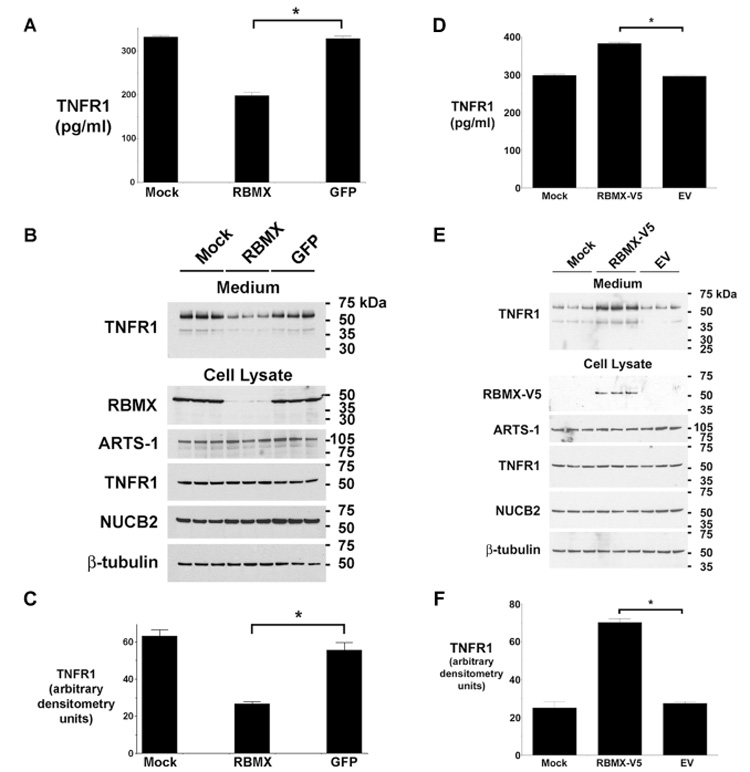
Panels A and B. TNFR1 in medium from HUVEC transfected with siRNA targeting RBMX or GFP was quantified by ELISA (Panel A) or Western blotting (shown in triplicate in Panel B). Mock denotes cells treated with transfection reagent alone. The asterisk denotes decreased TNFR1 release from cells transfected with siRNA targeting RBMX (P < 0.0001, n = 6). Western blots of HUVEC lysates are also shown. Panel C. Quantification of Western blots from Panel B. The asterisk indicates a significant decrease in the quantity of the 55-kDa TNFR1 present in conditioned medium from cells transfected with siRNA targeting RBMX (P < 0.003, n = 3). Panel D. The quantity of TNFR1 in conditioned medium from HUVEC transfected with plasmids encoding a V5-tagged RBMX (RBMX-V5), empty vector (EV), or the transfection reagent alone (Mock) was detected by ELISA. The asterisk denotes a significant increase in the quantity of TNFR1 present in medium from cells expressing RBMX-V5 (P < 10−9, n = 6). Panel E. Western blot showing, in triplicate, TNFR1 in HUVEC conditioned medium (top panel) and RBMX-V5 (detected with an anti-V5 antibody), ARTS-1, TNFR1, NUCB2, and β-tubulin in cell lysates (bottom panel). Panel F. Quantification of Western blots from Panel E by densitometry. The asterisk denotes significant increase in the quantity of 55-kDa TNFR1 present in medium from cells transfected with the RBMX-V5 plasmid (P < 10−4, n = 3).
To confirm the role of RBMX in the constitutive release of TNFR1 exosome-like vesicles, HUVEC were transiently transfected with a plasmid expressing human RBMX containing an amino-terminal V5 epitope tag. As shown in Fig. 2E, the RBMX-V5 protein was detected by the anti-V5 antibody only in cells transfected with the RBMX-V5 plasmid, but not in cells transfected with empty vector or following mock transfection. Medium from RBMX-V5-transfected cells contained significantly greater quantities of TNFR1 exosome-like vesicles, as measured by ELISA (Fig. 2D) and Western blotting (Fig. 2 E and F), as compared to cells transfected with the empty vector.
RBMX Regulates the IL-1β-mediated Inducible Proteolytic Cleavage of Soluble TNFR1 Ectodomains
The quantity of TNFR1 released from HUVEC in response to IL-1β stimulation for 2h, as detected by ELISA, was significantly reduced from cells transfected with siRNA targeting RBMX (Fig. 3A). This result was confirmed by Western blots that showed a reduction in the release of a 34-kDa TNFR1, which is consistent with a proteolytically cleaved sTNFR1 (Fig. 3B and 3C). To confirm the role of RBMX in the inducible proteolytic cleavage of TNFR1 ectodomains, HUVEC were transiently transfected with the RBMX-V5 plasmid and stimulated with IL-1β (20 ng/ml) for 2-h. Medium from cells transfected with the RBMX-V5 plasmid contained significantly greater quantities of the 34-kDa sTNFR1, as quantified by ELISA (Fig. 3D) and Western blotting (Fig. 3E and F), as compared to cells transfected with the empty plasmid.
Figure 3. Down-regulation of RBMX Expression by RNA Interference Attenuates the IL-1β-mediated Proteolytic Cleavage of TNFR1 Ectodomains.
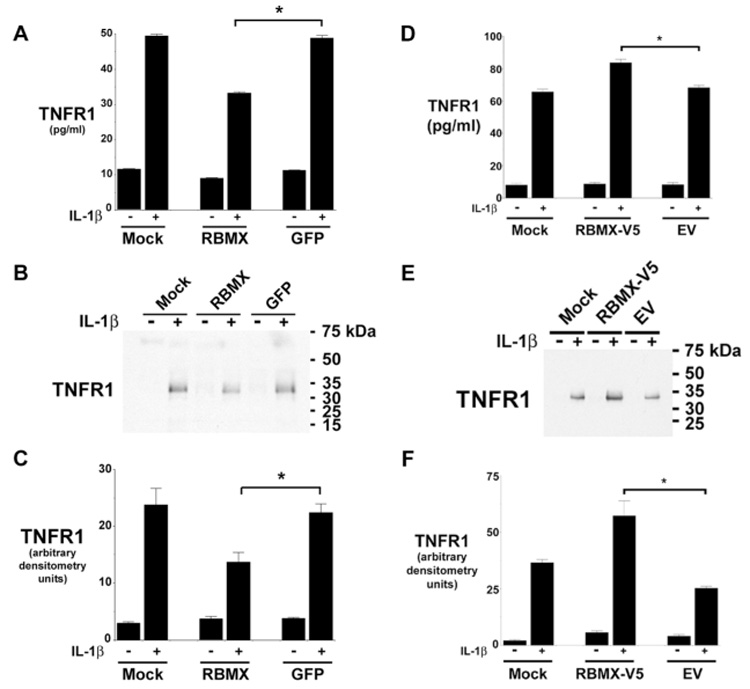
Panels A and B. HUVEC transfected with siRNA targeting RBMX or GFP were treated with 20 ng/ml of IL-1β for 2-h and the concentration of TNFR1 in conditioned medium was determined by ELISA (Panel A) or Western blotting (Panel B). Mock denotes treatment with transfection reagent alone. The asterisk indicates that the quantity of TNFR1 in conditioned medium after IL-1β stimulation from cells transfected with siRNA targeting RBMX was significantly reduced (P < 0.0001, n = 6). The blot in Panel B is representative of three individual experiments. Panel C. Quantification of Western blots from Panel B by densitometry. The asterisk indicates a significant decrease in the quantity of the 34-kDa sTNFR1 in conditioned medium after IL-1β stimulation from cells transfected with siRNA targeting RBMX (P < 0.02, n = 3). Panels D – E. HUVEC were transfected with plasmids encoding V5-tagged RBMX (RBMX-V5), the empty vector (EV), or the transfection reagent alone (Mock) for 2 days prior to the addition of fresh medium, without or with 20 ng of IL-1β for 2-h. The quantity of TNFR1 present in conditioned medium was detected by ELISA (Panel D) and Western blotting (Panel E). The asterisk denotes a significant increase in the quantity of TNFR1 present in medium after IL-1β stimulation from cells transfected with the RBMX-V5 plasmid (P < 0.00015, n = 6). This blot in Panel E is representative of three individual experiments. Panel F. Quantification of Western blots from Panel E by densitometry. The asterisk denotes a significant increase in the quantity of 34-kDa TNFR1 present following IL-1β stimulation from cells transfected with the RBMX-V5 plasmid (P < 0.008, n = 3).
Discussion
Here, we sought to identify ARTS-1-associated proteins that participate in cytoplasmic TNFR1 trafficking pathways that mediate the release of TNFR1 to the extracellular compartment. We identified that RBMX co-immunoprecipitated with ARTS-1 from both human airway epithelial and vascular endothelial cells. Using RNA interference, we showed that knock-down of RBMX expression reduced both the constitutive release of TNFR1 exosome-like vesicles and the inducible proteolytic cleavage of TNFR1 ectodomains. Reciprocally, over-expression of RBMX was associated with an increase in both the constitutive release of TNFR1 exosome-like vesicles and the inducible proteolytic cleavage of TNFR1 ectodomains. Taken together, these data are consistent with the conclusion that the association between endogenous RBMX and ARTS-1 regulates both TNFR1 release pathways to the extracellular compartment.
The ability of RBMX to associate with ARTS-1 and participate in the regulation of TNFR1 release identifies an unexpected function for a hnRNP. The human RBMX gene is located on the X chromosome and is the paralogue of the RBMY gene, which encodes a candidate spermatogenesis factor[6]. RBMX binds nascent RNA transcripts and may be associated with all hnRNP transcriptional complexes. Consistent with this, RBMX has been identified as a component of the spliceosome [7]. RBMX has been shown to interact with Tra2β, a SR-rich splicing activator, as well as the SR-like protein Htra2-β1[8, 9]. In contrast, RBMX and Tra2β have been shown to have opposite effects on exon splicing, with both being capable of acting as activators or repressors[10].
RBMX, which contains a RNA-binding domain, binds RNA non-specifically[8, 10]. We showed that RNase treatment markedly increased the association between endogenous RBMX and ARTS-1, whereas RNase inhibitors had no effect. This finding is consistent with the conclusion that RBMX primarily binds RNA and is thereby sequestered from associating with ARTS-1. This suggests that only the subset of RBMX that is not bound by RNA is available to interact with ARTS-1 and modulate TNFR1 release to the extracellular compartment. Furthermore, the RNAi-mediated decreases in RBMX did not affect the expression of ARTS-1, TNFR1, or NUCB2. This suggests that the mechanism by which RBMX participates in the regulation of TNFR1 release is mediated by its association with ARTS-1, rather than by regulating the expression or splicing of mRNAs encoding proteins known to participate in TNFR1 release pathways. It cannot be excluded, however, that the RNAi-mediated decreases in RBMX protein levels altered the expression or splicing of mRNAs encoding yet to be identified proteins that also are involved in TNFR1 release pathways.
In summary, we have shown that RBMX associates with ARTS-1 in human airway epithelial and vascular endothelial cells and thereby regulates both the constitutive release of TNFR1 exosome-like vesicles and the IL-1β-mediated inducible proteolytic cleavage of TNFR1 ectodomains. This defines a new and unexpected function for a hnRNP.
Acknowledgements
The authors thank Drs. Martha Vaughan, Joel Moss, Christian A. Combs, and Daniella Malide for their helpful discussions and critical review of the manuscript. This research was funded by the Division of Intramural Research, NHLBI, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schall TJ, Lewis M, Koller KJ, Lee A, Rice GC, Wong GHW, Gatanaga T, Granger GA, Lentz R, Raab H, Kohr WJ, Goeddel DV. Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell. 1990;61:361–370. doi: 10.1016/0092-8674(90)90816-w. [DOI] [PubMed] [Google Scholar]
- 2.Hawari FI, Rouhani FN, Cui X, Yu ZX, Buckley C, Kaler M, Levine SJ. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc Natl Acad Sci U S A. 2004;101:1297–1302. doi: 10.1073/pnas.0307981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islam A, Adamik B, Hawari FI, Ma G, Rouhani FN, Zhang J, Levine SJ. Extracellular TNFR1 release requires the calcium-dependent formation of a nucleobindin 2-ARTS-1 complex. J Biol Chem. 2006;281:6860–6873. doi: 10.1074/jbc.M509397200. [DOI] [PubMed] [Google Scholar]
- 4.Cui X, Hawari F, Alsaaty S, Lawrence M, Combs CA, Geng W, Rouhani FN, Miskinis D, Levine SJ. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J Clin Invest. 2002;110:515–526. doi: 10.1172/JCI13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islam A, Shen X, Hiroi T, Moss J, Vaughan M, Levine SJ. The brefeldin A-inhibited guanine nucleotide-exchange protein, BIG2, regulates the constitutive release of TNFR1 exosome-like vesicles. J Biol Chem. 2007;282:9591–9599. doi: 10.1074/jbc.M607122200. [DOI] [PubMed] [Google Scholar]
- 6.Delbridge ML, Lingenfelter PA, Disteche CM, Graves JA. The candidate spermatogenesis gene RBMY has a homologue on the human X chromosome. Nat Genet. 1999;22:223–224. doi: 10.1038/10279. [DOI] [PubMed] [Google Scholar]
- 7.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- 9.Venables JP, Elliott DJ, Makarova OV, Makarov EM, Cooke HJ, Eperon IC. RBMY, a probable human spermatogenesis factor, and other hnRNP G proteins interact with Tra2beta and affect splicing. Hum Mol Genet. 2000;9:685–694. doi: 10.1093/hmg/9.5.685. [DOI] [PubMed] [Google Scholar]
- 10.Nasim MT, Chernova TK, Chowdhury HM, Yue BG, Eperon IC. HnRNP G and Tra2beta: opposite effects on splicing matched by antagonism in RNA binding. Hum Mol Genet. 2003;12:1337–1348. doi: 10.1093/hmg/ddg136. [DOI] [PubMed] [Google Scholar]


