Abstract
We developed a robust and reproducible methodology to amplify human sequences in parallel for use in downstream multiplexed sequence analyses. We call the methodology SMART (Spacer Multiplex Amplification Reaction), and it is based, in part, on padlock probe technology. As a proof of principle, we used SMART technology to simultaneously amplify 485 human exons ranging from 100 to 500 bp from human genomic DNA. In multiple repetitions, >90% of the targets were successfully amplified with a high degree of uniformity, with 70% of targets falling within a 10-fold range and all products falling within a 100-fold range of each other in abundance. We used long padlock probes (LPPs) >300 bases in length for the assay, and the increased length of these probes allowed for the capture of human sequences up to 500 bp in length, which is optimal for capturing most human exons. To engineer the LPPs, we developed a method that generates ssDNA molecules with precise ends, using an appropriately designed dsDNA template. The template has appropriate restriction sites engineered into it that can be digested to generate nucleotide overhangs that are suitable for lambda exonuclease digestion, producing a single-stranded probe from dsDNA. The SMART technology is flexible and can be easily adapted to multiplex tens of thousands of target sequences in a single reaction.
Keywords: human exons, multiplex PCR, padlock probe, single-strand DNA
One of the greatest challenges in the study of the human genome is our ability to extract information of interest in a timely and cost-effective manner. Advances in genomic technologies have made it possible for scientists to start to understand the contribution of genetic variations, within large sets of genes or even the entire genome, in an effort to better understand common human diseases (1–5). The value of those technologies shows promise in uncovering the contributions of particular genes not previously thought to be involved in the development of disease. Studies suggest there is a surprisingly large involvement of diverse sets of genes in many diseases, such as cancer and hypertension, and rare variants in many of these genes are likely to have a significant contribution to the diseased state (4, 6, 7). To gain better understanding of the genetic contribution to disease progression, we need to fully understand the genetic sequence variations in the structural and the regulatory sequences of the human genome. To address this need, several array-based (8–10) or sequence-by-synthesis technologies (11, 12) have been developed to determine the genotype or gene copy number or to sequence the human genome.
To take full advantage of these technologies, reducing the complexity of the human genome to smaller regions of interest is required, because the human genome is too large to sequence in a cost-effective manner using current technologies and too complex to be labeled directly for genome-wide genotyping or copy number assessment. The first successful nontargeted genome wide reduction of the complexity of the human genome was achieved by amplification of size-selected fragments of human DNA fractionated after a restriction endonuclease digestion. This enabled the development of a very successful genotyping and copy number methodology using allele-specific hybridization to oligonucleotide arrays (13–15). The Mismatch Repair Detection technology was successfully used for detection of polymorphisms in PCR-amplified amplicons (16). Genome reduction can be achieved by direct amplification of specific targets or amplification of targets selected by virtue of hybridization to sequences on an array. It has been shown that direct multiplex PCR amplifications can generate a high degree of nonspecific amplification products because of extensive mispriming by multiple primers (17, 18). Dahl et al. (19, 20) have shown that careful choice of primer pairs to avoid mispriming, combined with the selector technology that allows capture of regions of interest by digestion with restriction endonucleases, followed by circularization of genomic DNA can allow multiplex amplification of up to 170 amplicons. However, this methodology is well short of the thousands of simultaneous amplifications needed for resequencing the entire set of human exons.
A very successful targeted multiplex amplification strategy is the molecular inversion probe (MIP) technology, which has successfully adapted the padlock methodology to genotype >53,000 SNPs in a single assay (21–23). To identify the SNPs the MIP technology uses four steps: hybridization, polymerization, ligation, and amplification (24, 25). This technology has also been adapted to capture selected sequences by increasing the distance between the sequence-specific binding sites of the MIP probes (26). Recently, Porreca et al. (27) modified the MIP technology adapted to perform a multiplex amplification reaction of fragments ranging from 60 up to 200 bp in a high-throughput capture of ≈10,000 exons. They successfully captured only 20% of the probes as amplified products, and there was little reproducibility in replicates (27). Microarrays have also been used for selective capture of genomic targets and subsequent amplification of bound targets that are eluted from the array. These methods have an advantage, in that they do not require extensive synthesis of oligonucleotides; however, the success rate of target capture was limited (28–30).
It is a challenging task to evaluate whether the padlock probes and the MIP assay represent a suitable methodology to perform robust high-throughput multiplex amplification reactions using selected sequences of the human genomic DNA as templates. Several features of the human exonic sequence, like the variable length of exons and the varying GC composition, could interfere with the fidelity and efficiency of the enzymatic steps of the protocol and, consequently, diminish the performance and uniformity of the assay.
To address these challenges, we have developed a strategy that maximizes the ability of padlock probes to amplify multiple sequences in a single reaction. We have adapted the padlock probe technology to perform multiplex PCR using probes that are three times larger than in the conventional MIP assay and have developed a method to produce these probes from double-stranded molecules. These long padlock probes (LPPs) hybridize to the target DNA sequences where the 3′ end of the probe is up to 500 bases apart from the 5′ end. This gap-fill region is the sequence of interest, and the gap between the two ends of the probe is filled by polymerization from the 3′end using the target DNA sequence as the template. The efficiency of this step is crucial in making this a robust and reproducible assay; we have made several improvements in the current assay that now make it very suitable for high-throughput use. The specificity and abundance of each species of the expected product were evaluated independently by qPCR and by hybridizing the products to an Affymetrix resequencing array. We attempted to amplify 485 exons using 557 probes and were able to amplify >90% of the exons successfully in a single multiplex reaction.
Results
Padlock probes and the MIP technology have proven very successful in polymerizing short stretches of nucleic acids between the 3′ and 5′end of the probes that were hybridized to target DNA (21, 25). To test whether these methodologies are capable of successfully polymerizing long gap-fill stretches of nucleic acids between the target-specific sequences, we designed probes where the size of the gap between the 3′ and 5′ ends of the probe ranged from 161 to 383 bp. In a series of singleplex assays, padlock probes that were ≈120 bases in length were annealed to human DNA, and the gap-fill reaction was performed as described in Materials and Methods using the conditions for the Stoffel DNA polymerase. After completion of the gap-fill reaction with the formation of the circular DNA molecule by ligation, the desired DNA sequences were amplified using common primers flanking the 3′ and 5′ target sequences of the padlock probe (Fig. 1D). We measured the performance of the assay by running the amplified products on a 2% agarose gel to determine the length of the amplified products. We concluded that the assay was successful and quite reproducible when the length of gap-fill sequences was <200 bases, but we found that the consistency and the reproducibility of the assay diminished as the size of the gap-fill region increased [supporting information (SI) Tables S1 and S2]. We hypothesized that the size of the padlock probes (≈120 bases) was posing a physical constraint in allowing a robust gap-fill reaction, and this effect became more pronounced as the size of the target DNA sequence increased.
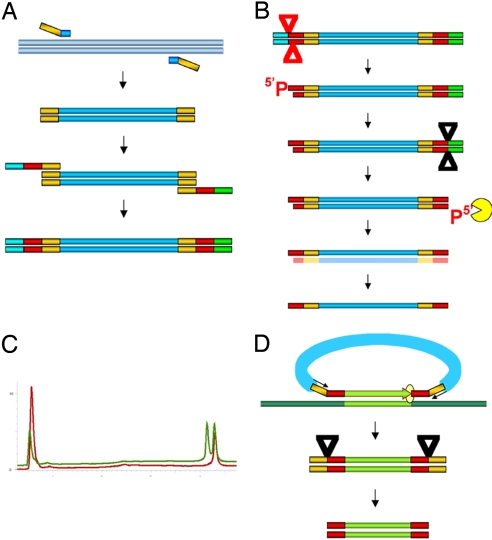
Fig. 1.
Construction of single-strand probes. (A) The bacteriophage lambda DNA (gray) was used as a template, and the primers containing the common amplification primers as adaptors on the 5′ end were used to PCR amplify the spacer that is common to all of the probes. This common spacer with the amplification sequences was the further template for building the double-stranded probe precursor. The left primer with the target sequence (red) had an adaptor with a BsaI site (blue) The right primer with the target sequences (red) has an adaptor (green) with the MlyI site. (B) The construct was digested with BsaI (red triangle) cutting 5 bases inwards from the recognition sequence. The digestion creates a phosphorylated 5′end and a recessed 3′ end. The phosphate group is removed by shrimp alkaline phosphatase digestion. A subsequent digestion with MlyI (black triangle) cuts 5 bases inwards from the right-hand recognition sequence to create a blunt end with a 5′ phosphate. The lambda exonuclease digests the lower strand from the 5′ phosphorylated end created by the MlyI digestion leaving the upper strand intact. (C) The dHPLC profile of the double-stranded template has two peaks (blue) at ≈4.5 min. After exonuclease digestion (red) there is only one peak at 4.5 min, and one large early peak that represent the digested products. (D) The LPP is hybridized to genomic DNA (gray) and gap-filled (orange) using the genomic DNA as template. Ligation occurs when the polymerase reaches the 5′ end of the probe (yellow circle) to form a circular molecule. The amplification sequences (yellow) in the circle are then used to amplify the targeted exons. Digestion with AscI and ClaI (sites are included in the amplification primers) separate the primers from the genomic target sequences.
To test whether short padlock probes were limiting the gap-fill reaction, we needed to make longer probes. To this end, we developed a methodology for making probes that were >300 bases. We posited that the LPPs should (potentially) overcome the limitations of the smaller-size probes. In our current protocol, the ssDNA probe is produced from a specifically designed dsDNA template using a series of enzymatic reactions and selective removal of one of the strands using lambda exonuclease digestion. We first built the “core” construct of the template that is common to all probes. The core is composed of a common nucleic acid sequence, the spacer, derived from the bacteriophage lambda flanked by the amplification primers (Fig. 1A). That sequence from bacteriophage lambda was chosen to avoid cross-hybridization occurring between the spacer sequences and human genomic DNA. The target-specific ends that are unique to each probe were added to this template by PCR amplification. In addition, as part of the same PCR amplification, we placed Type IIS restriction endonuclease recognition sequences at the 5′ ends of the construct to generate precise ends after restriction digestion. The advantage of Type IIS restriction endonucleases is that, although they digest the DNA at a given sequence, the cleavage site is outside of their recognition sequence.
In the first step for making the LPP, we digested the double-stranded template with the restriction enzyme BsaI to generate a 5′ overhang 5 bases inward from the recognition site (Fig. 1B). It was important that this end of the molecule has a recessed 3′ end to ensure it remains resistant to lambda exonuclease digestion. To ensure production of ssDNA of high quality, the 5′ phosphate group on the overhang was removed by a phosphatase. Digestion with the second restriction enzyme MlyI generated the desired structure at other end of the molecule: a blunt end with a phosphorylated 5′ end. Digestion with lambda exonuclease selectively degraded the strand with the phosphorylated 5′ end, whereas the other strand remains intact. The quality of the digestion was monitored by denaturing HPLC (dHPLC) (Fig. 1C)
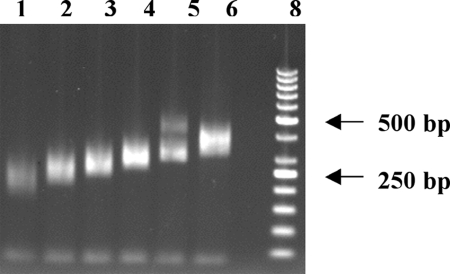
With this reliable methodology for making LPPs, we adapted the padlock technology to successfully polymerize, ligate, and subsequently amplify gap-filled sequences, which are >200 bp, using the LPPs that have a large lambda spacer between the 5′ and 3′ recognition sequences. We were successful in obtaining very reliable and robust amplifications with target sequences gapped as large as 500 bp (Tables S1 and S2). To test whether the LPPs can amplify a large set of sequences in a uniform manner, targets were chosen from sequences of exons present in a set of 30 kinase genes relevant to the study of cancer (31). In that study, mismatch repair detection was used to detect variations in the exons of these genes, and standard PCR was used to amplify each exon. These same sequences were used in our study to design the 5′ and 3′ targeting sequences of the LLPs. We synthesized the LPPs as described above and excluded those exons in which in the primer sequences contained either a BsaI or an MlyI restriction site. Initially, pools of LPPs were grouped together by expected product size and used in the assay, with the Stoffel polymerase for the gap-fill reaction as described in Materials and Methods. Using these LPPs, we were able to successfully polymerize, ligate, and subsequently amplify gap-filled sequences. The resulting PCR products were in the size ranges expected (Fig. 2). The specificity of the reaction is evident in lane 5, where two distinct sets of bands appear, corresponding to the sizes of products we expected to amplify. We were successful in obtaining very reliable and robust amplifications with target sequences gapped large as 500 bp. Because the larger spacer was instrumental in the success of the reaction, we named our technology spacer multiplex amplification reaction (SMART).
Fig. 2.
Multiplex amplification using the SMART assay of (557) human targets divided in six pools of ≈96 amplicons per pool. After completion of the assay, the PCR products from each pool were purified and digested with AscI and ClaI to remove the flanking amplification primers from the amplified genomic products, and run on a 2% agarose gel. Lane 1, 96 amplicons of size ranging from 150 to 190 bp; lane 2, 96 amplicons 190–220 bp; lane 3, 96 amplicons 220–240 bp; lane 4, 96 amplicons 240–280 bp; lane 5, 48 amplicons 280–300 bp, 29 amplicons 400–460 bp, lane 6, 96 amplicons 300–390 bp, lane 8; 50-bp ladder (Fermentas).
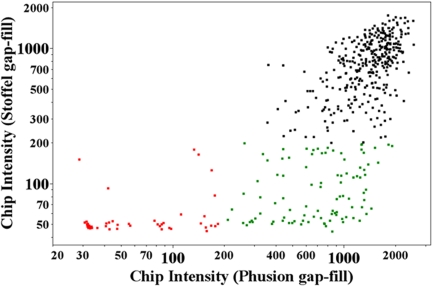
One of the applications of multiplex PCR amplification is for targeted resequencing. An Affymetrix resequencing chip was used to determine whether these products were of sufficient quality to provide a high-quality resequencing for all of the targets. The resequencing array has 25-mer oligonucleotides tiled across all of the amplicons used in this study. The median intensity value for each exon was used as a measure of the success of multiplex amplification. The background for the chip was between 30 and 40, and therefore a signal threshold of ≥200 indicated successful amplification. All probes were combined in a single pool, gap-filled using Stoffel polymerase, and amplified in a multiplex reaction. The products were labeled and hybridized to the resequencing chip (Fig. 3). Seventy-five percent of the amplicons successfully amplified, based on the intensity values on the chip (Fig. 3A). To further improve the performance and fidelity of the SMART assay, a variety of other theromostable polymerases that have the ability to overcome the constraints of sequences with high GC content and a higher proofreading activity than either the Stoffel or the Amplitaq Gold polymerases were tested. We tried Pfu DNA polymerase, Pfu Ultra II fusion HS DNA polymerase, DyNAzyme EXT DNA polymerase, and Phusion DNA polymerase in the gap-fill reaction using the Ampligase buffer. In each trial, the reaction buffer and reaction conditions in the multiplex amplification step were changed according to the optimum conditions of each polymerase (data not shown). Using Phusion polymerase in both the gap-fill and amplification reaction, the success of the SMART assay was enhanced to 90% based on our previous thresholds. The amplification reaction was performed using the Phusion GC buffer that improves the uniformity of PCR amplification in templates with high GC content. The Phusion amplification outperformed the extension with Stoffel polymerase: 60% of the amplicons that failed with Stoffel were successful with Phusion (Fig. 3). All subsequent experiments were performed with Phusion polymerase and buffer.
Fig. 3.
The median CEL intensities for each amplicon obtained by using Stoffel DNA polymerase and Phusion DNA polymerase in the gap-fill reaction are plotted against each other. The CEL intensities that were <200 in both the Stoffel and Phusion are indicated in red, and failures with Stoffel polymerase that were successful with Phusion polymerase are marked with green.
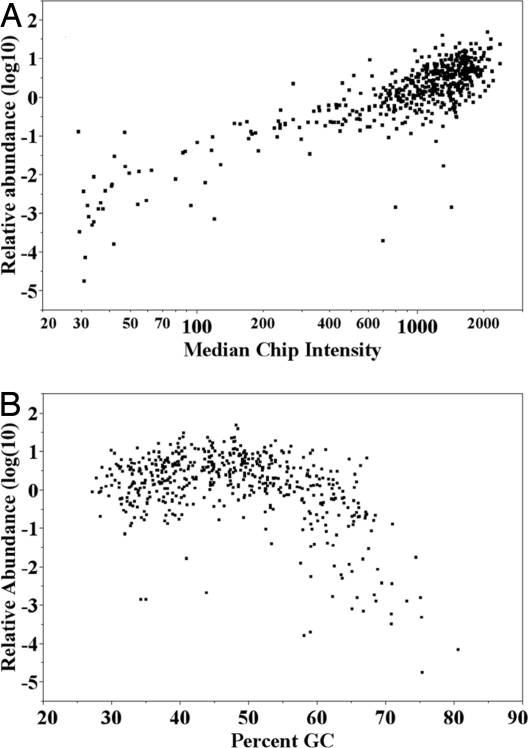
Several resequencing technologies require that the PCR products be evenly represented when amplified from a mixture of probes. The PCR products from the SMART reaction were diluted and used as the template for qPCR using primers that were specific to the expected product (SI Text and Dataset S1). We calculated the fold differences from the mean using the lowest threshold cycle values (CT)) values from all of the individual qPCRs (Fig. 4). To verify the quality of the primers and to avoid any systemic biases, they were first tested against genomic DNA templates, although no quantitative adjustments were made for sequence-dependent differences in amplification. The qPCR results indicate that 83% of the amplicons varied in a range of two logarithms in abundance, and only 9.6% of the amplicons were represented <10-fold difference from the mean. The majority of these dropouts were amplicons that had a higher percentage of GC (Fig. 4). The failures were pooled together and amplified in a separate reaction using the same probe concentration as before to determine whether they would be successful in a subset of the larger pool. Successful amplification resulted in 60% of the failures in the larger pool (Fig. S1). We were not able to get any significant improvement in the performance of these probes by spiking them at a higher concentration in the larger pool (data not shown).
Fig. 4.
Quantitative distribution of exons in the multiplex PCR. (A) The relative abundance of each amplicon in the multiplex PCR products was estimated by quantitative PCR by using PCR primers specific to each amplicon. The log10 distribution of the amplicons was plotted against the median chip intensities for each amplicon. (B) The log10 distribution of the relative abundance with respect to the mean was calculated and plotted for each amplicon sorted by increasing percentage GC content.
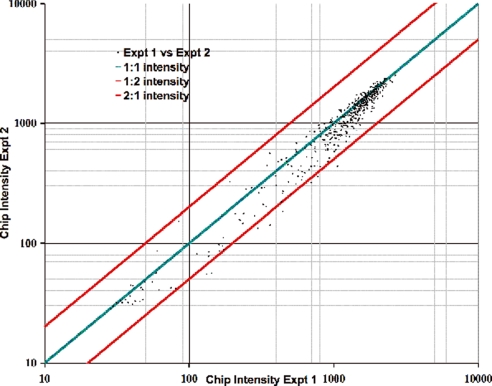
To determine the reproducibility of the chip hybridization, we performed multiple repetitions of the assay, and the chip intensities from two representative experiments are compared (Fig. 5). The multiplex PCRs were extremely reproducible (r2 = 0.96), indicating that there was no systemic bias in the amplification between repetitions. A comparison of the qPCR data with the chip hybridization data shows a very good correlation between the chip intensity values and qPCR abundance of the amplicons (r2 = 0.93). Products within a 10-fold difference from the mean provide adequate amounts of products to be analyzed in a resequencing chip (Fig. 4A). Four probes that worked well in the chip did not work in the qPCR assay because of primer design in the qPCR. In general, the amplicons that were distributed within 10-fold of the mean gave higher signal intensity on the chip than the products that were less abundant in the PCR mix.
Fig. 5.
The multiplex PCR products from 557 probes were labeled and hybridized to an Affymetrix resequencing chip. The median intensity values for each amplicon were used as the intensity value for each amplicon. The intensity values from two separate experiments were plotted against each other to determine their reproducibility on the chip. The 1:1 correlation line is shown in green, and the 1:2 and 2:1 correlation lines are shown in red.
Discussion
We have developed a method called SMART technology, based on padlock and MIP assays, for simultaneous multiplex amplification of DNA sequences in a single tube for targets that can range from <100 to 500 bp. As a proof of principle, we applied the SMART technology to amplified simultaneously 575 target sequences representing human exons present in 30 kinase genes. This assay is highly specific for the selected target sequences and is robust in nature. More than 90% of the targets were consistently amplified, whereas 10% that failed to amplify show a remarkable reproducibility in the failures, indicating these represent sequence-specific complexities. A significant advantage of our technology is the high degree of uniformity of the amplified targets, where 70% of targets are distributed within 10-fold, and all of the products are within 100-fold, of each other in abundance (Fig. 3). The high level of reproducibility and uniformity in the amplification of the target sequences makes SMART technology eminently suitable for use in several resequencing platforms. This technology should easily be scalable to include all of the exons in the genome as the amounts of each probe required for successful amplification is low. The multiplicity can expand to tens of thousands of targets, reaching similar levels of multiplex success as seen for the MIPs technology where >53,000 parallel reactions can be performed in one tube (23). The success of the SMART technology relies on the improvement of at least two essential elements of the padlock assay: (i) by applying a methodology to generate LPPs, where we have significantly increased the size of the spacer connecting the 3′ and 5′ prime target recognition sequences of the unimolecular padlock probe; and (ii) by optimization of the reaction conditions of the assay by selection of appropriate thermostable DNA polymerases for the gap-fill reactions and amplification reactions from the circularized probes. Our hypothesis is that the larger length of the nucleotide sequence (spacer) that connects the 3′ and the 5′ of the padlock probe provides more flexibility and stability during the polymerization step of the gap-fill reaction. We reason that the gap-fill polymerization creates a progressively more rigid nucleic acid structure that decreases the efficiency of polymerization as the length of the gap is increased.
To ensure that the SMART technology is scalable, enabling it to achieve the ultimate goal of the successful amplification of all human exons, we investigated various approaches to amplify those targets that failed in the initial run. Because the only common denominator of all failed target sequences was their >60% CG content and not the length of the amplicon, we pooled together all these failed targets and repeated the assay using the same conditions. This resulted in the successful amplification of 50% of the failed targets. Combining the number of targets that amplified in the two reactions increases the success rate of amplification to 95%. Based on this finding, it is reasonable to assume that an optimized strategy to achieve successful amplification of all human exons would be to create pools where all of the exons will be distributed based on their GC content. The failures from these pools can then be assigned to smaller pools to achieve uniform amplification from all of the exons. Recently, a method based on the principles of the padlock and MIP assay was published by Porreca et al. (27), who performed a high-throughput multiplex amplification using ≈50,000 exons as target sequences in a single assay. The number of targets that successfully amplified was close to 20%. This indicates that the implementation of the previously known strategies and conditions are not sufficient to address such a complex assay. The limitations of the traditional padlock and MIP assays as tools for multiplex amplification of exons can be evaluated more precisely in the initial pilot experiment the authors performed using 480 probes. The amplicons generated were all of the same length, but no stratification of probes was made according to the CG content. Despite the relative small length of the target sequences, the overall uniformity of the assay was spread over a range of 4–5 logarithms, and <50% of the target sequences were within 2 logarithms. It is unlikely that the success rate and poor uniformity of the assay using the smaller probes could potentially be improved by increasing the concentration of targeting probes in the reaction. It is worth mentioning that the concentration of probes used in the SMART assay is 10-fold less than the one used in the 480-plex experiment, yet the multiplex amplification was greatly improved on the former relative to the latter.
We have tested the suitability of the SMART products in resequencing applications by hybridizing the products to a resequencing chip that was designed for these exons. The quality of sequences obtained from the multiplex PCR is comparable with that obtained by pooling PCR products from individual reactions. The sequence call rate is a function of the chip design and does not depend on the multiplex PCR. Several independent repetitions of the multiplex PCR and chip hybridization demonstrated that the procedure is highly reproducible (Fig. 5). The tight range of the quantitative distribution of exons suggests that this method should be very suitable in other resequencing platforms, such as the in the Genome Sequencer FLX System (Roche Applied Science, the SOLiD gene sequencer (Applied Biosystems), and Genome Analyzer (Illumina). In these platforms, it is important that the products have a narrow distribution because of the iterative nature of sequence determination.
A significant factor in the success of the SMART assay is the utilization of the LPPs. These single-stranded probes are much longer in length than any DNA molecule that can be chemically synthesized with the currently available technologies. To overcome this obstacle, we have developed a general method to produce the ssDNA from a double-stranded molecule. The double-stranded molecule is tailored to its desired configuration from custom made oligonucleotides in such a way that the final design can be stored and propagated indefinitely using various amplification methods. Using a series of enzymatic steps, we have been successful in generating high-quality single-stranded molecules as long as 500 bases (data not shown). In our study, we used the combination of BsaI and MlyI restriction endonucleases; however, there are other enzymes available that could be used to generate the desired template for lambda exonuclease digestion to produce ssDNA probes in a similar manner. We have tested our methodology with a variety other enzymes such as BsmBI and BciVI and the quality and accuracy of the ssDNA molecules generated after the lambda exonuclease treatment was similar to those LPPs used in this study (data not shown). It is likely that this method can be used to produce very large single-stranded molecules, because lambda exonuclease is a very processive enzyme (32, 33). Our method of producing single-stranded molecules has a number of applications where variable sizes or large quantities of long ssDNA molecules are needed. In addition, the method can be used to amplify minute amounts of large pools of appropriately designed oligonucleotides synthesized on arrays, and then the amplified products can be converted to single-stranded probes. Further advances in oligonucleotide synthesis technologies in the near future may dramatically decrease the cost of oligonucleotides required for the production of the designed double-stranded templates. The reduction in cost that results from using these methods of producing large quantities of single-stranded molecules, together with the robust SMART technology described here may make resequencing individual genomes feasible.
Materials and Methods
Preparation of LPPs.
The common spacer for all probes had a sequence devoid of MlyI and BsaI sites derived from bacteriophage lambda with two amplification primers that are used in the multiplex PCR. This spacer was used as the template for PCR amplification using a primer that had a BsaI site and one target-specific sequence, and a second primer that had an MlyI site and the other target-specific sequence. The purified PCR product was digested with BsaI (New England Biolabs) in buffer 3 (100 mM NaCl; 50 mM Tris·HCl, pH 7.9; 10 mM MgCl2; 1 mM DTT) at 50°C, followed by digestion with five units of shrimp alkaline phosphatase (USB Corporation) at 37°C. The products were subsequently digested with MlyI (New England Biolabs) to generate the other target-specific end of the molecule. The digested PCR product was treated with 0.1 units lambda exonuclease (New England Biolabs) at 37°C for 15 min in the same restriction enzyme buffer. The probes were phosphorylated with 5 units of T4 polynucleotide kinase (NEB) in 50 mM Tris·HCl, pH7.9; 10 mM MgCl2; 1 mM ATP; and 1 mM DTT. A list of all of the primer sequences is included in Dataset S1 and a detailed example is included in SI Text.
dHPLC analysis was used to monitor the efficiency of lambda exonuclease digestion. The digested PCR product was loaded on a DNASep column (Transgenomic). PCR products were eluted from the column using an acetonitrile gradient in a 0.1 M triethylamineacetate buffer (TEAA), pH 7, at a constant flow rate of 0.9 ml/min. The buffers used were eluent A, 0.1 M TEAA, 0.1 mM Na4EDTA; and eluent B, 25% acetonitrile in 0.1 M TEAA that were mixed for the gradient.
Gap Fill with Stoffel Fragment DNA Polymerase.
Fifty attomoles of the LPP were mixed with 350 ng of human genomic DNA (Promega) in 20 mM Tris·HCl (pH 8.3), 25 mM KCl, 10 mM MgCl2, 0.5 mM NAD, and 0.01% Triton X-100. The mixture was heated to 95°C for 2 min, followed by a gradual decrease in temperature of 1°C per min to 58°C and held for 16 h in a thermocycler. The extension reaction was performed with 0.15 mM dNTP, 0.05 units Stoffel polymerase (Applied Biosystems) and 5.0 units Ampligase (Epicentre Biotechnologies) for 15 min at 58°C. This was followed by digestion with 0.2 units Exonuclease I (Epicentre Biotechnologies) and 20 units of Exonuclease III (Epicentre Biotechnologies) at 37°C for 15 min. The PCR amplification was performed in 50 mM KCL, 10 mM Tris·HCl (pH 8.3), 2.5 mM MgCl2, 0.125 mM dNTP (New England Biolabs), 15 pmols each of the two amplification primers CGTCACATTATTTAGGTGACACTATAG and GCGTACTATTAACCCTCACTAAAGG, and 0.5 units Amplitaq Gold (Applied Biosystems). After 10 min of heat inactivation at 95°C, the reaction was cycled at 94°C for 15 sec, 63°C for 30 sec for 72°C for 30 sec for 40 cycles.
SMART Assay with Phusion.
Fifty attomoles of probe was mixed wit 360 ng of human genomic DNA (Promega) in 1× Ampligase buffer (Epicenter Biotechnologies) and held at 98°C and gradually decreased at the rate of 1°C min to 60°C and held overnight at that temperature. The extension was performed by addition of 0.4 units of Phusion High-Fidelity DNA Polymerase (New England Biolabs), 3 μl 1.0 mM dNTP, 5 units Ampligase (Epicenter Biotechnologies) in a 15-μl volume at 60°C for 15 min followed by 72°C for 15 min. The exonuclease digestions were performed as described above. The PCR was performed by the addition of 10 μl 5× GC buffer supplied wit the Phusion Polymerase Enzyme, 8 μl 1 mM dNTP, 25 pmol of each amplification primer, and 0.5 units Phusion polymerase (NEB). The reaction was held at 98°C for 2 min followed by cycles of 98°C, 15 sec, 63°C, 15 units, 72°C, and 90 sec for 40 cycles.
Real-Time PCR.
After the multiplex PCR, the reaction products were diluted 1:10,000 and used as the template in a reaction containing 1× SYBR green PCR Mastermix (Applied Biosystems) and 2.5 pmols of each pair of amplification primers that were specific to the expected product. The reactions were cycled in an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The complete list of primers is provided in the Dataset S1.
Resequencing.
The resequencing array has 25-mer oligonucleotides tiled across the all of the amplicons used. Every position on each strand of an amplicon is in the middle of one of these 25 mers. In addition, three additional oligos on each strand are laid on the arrays corresponding to the three possible mutations of the middle base. Therefore, for each position eight oligonucleotides (four on each strand) are tiled. The distance between two neighboring features (oligonucleotides) is 8 μM.
Purified PCR products were digested with DNase I to an average size of ≈60 bp and end-labeled with Bio-16-ddUTP (Enzo Life Sciences) using Terminal Transferase (New England Biolabs). The labeled products were hybridized according to the protocol provided by Affymetrix (GeneChip CustomSeq Resequencing Array Protocol). The array was washed and stained using the Affymetrix GeneChip Fluidics Station 450 and scanned using GeneChip Scanner 3000 according to the protocol supplied by Affymetrix with the exception that five washes with wash B were used instead of 24. The scanned probe array was analyzed to obtain a signal value for each amplicon. The median signal among all of the perfect match oligonucleotides corresponding to an amplicon was used as the signal of that amplicon. The background on the array in these experiments was ≈30–40 (based on the signal of unhybridized array features). The median intensity of hybridized features was ≈1,330 (log10 value = 3.123), following a relatively normal distribution in log-space with a standard deviation in log10 units of 0.413 (data not shown). Two standard deviations below the median log10 value correspond to an intensity of 102.30 = 200; therefore, a value of 200 was used as a threshold of signal falling well above background.
Supplementary Material
Acknowledgments.
We thank Ron Sapolsky, Evan Horowitz, and Kara Juneau for critical reading of the manuscript and valuable comments. This work was supported by National Institutes of Health Grants P01HG000205 and U54GM62119.
Footnotes
Conflict of interest statement: S.K., M.M., and R.D. are named on a patent application for work described in this paper.
Data deposition: The sequences reported in this paper have been deposited into ArrayExpress database (accession no. E-MEXP-1638).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803240105/DCSupplemental.
References
- 1.Spirin V, et al. Common single-nucleotide polymorphisms act in concert to affect plasma levels of high-density lipoprotein cholesterol. Am J Hum Genet. 2007;81:1298–1303. doi: 10.1086/522497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanna S, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romeo S, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen JC, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci USA. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 7.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007;296:50–57. [PubMed] [Google Scholar]
- 8.Mockler TC, et al. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85:1–15. doi: 10.1016/j.ygeno.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Weedon MN, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunderson KL, et al. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 11.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Matsuzaki H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 14.Matsuzaki H, et al. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 2004;14:414–425. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ZC, Buraimoh A, Iglehart JD, Richardson AL. Genome-wide analysis for loss of heterozygosity in primary and recurrent phyllodes tumor and fibroadenoma of breast using single nucleotide polymorphism arrays. Breast Cancer Res Treat. 2006;97:301–309. doi: 10.1007/s10549-005-9124-5. [DOI] [PubMed] [Google Scholar]
- 16.Peters BA, et al. Highly efficient somatic-mutation identification using Escherichia coli mismatch-repair detection. Nat Methods. 2007;4:713–715. doi: 10.1038/nmeth1081. [DOI] [PubMed] [Google Scholar]
- 17.Wang DG, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 18.Cho RJ, et al. Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat Genet. 1999;23:203–207. doi: 10.1038/13833. [DOI] [PubMed] [Google Scholar]
- 19.Dahl F, et al. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci USA. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahl F, et al. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardenbol P, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 22.Hardenbol P, et al. Highly multiplexed molecular inversion probe genotyping: Over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Performance of molecular inversion probes (MIP) in allele copy number determination. Genome Biol. 2007;8:R246. doi: 10.1186/gb-2007-8-11-r246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsson M, et al. Real-time monitoring of rolling-circle amplification using a modified molecular beacon design. Nucleic Acids Res. 2002;30:e66. doi: 10.1093/nar/gnf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson M, et al. Padlock probes: Circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 26.Akhras MS, et al. Connector inversion probe technology: a powerful one-primer multiplex DNA amplification system for numerous scientific applications. PLoS ONE. 2007;2:1298–1303. doi: 10.1371/journal.pone.0000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 28.Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 29.Okou DT, et al. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 30.Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 31.Bentivegna S, et al. Rapid identification of somatic mutations in colorectal and breast cancer tissues using mismatch repair detection (MRD) Hum Mutat. 2008;29:441–450. doi: 10.1002/humu.20672. [DOI] [PubMed] [Google Scholar]
- 32.Mitsis PG, Kwagh JG. Characterization of the interaction of lambda exonuclease with the ends of DNA. Nucleic Acids Res. 1999;27:3057–3063. doi: 10.1093/nar/27.15.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian K, Rutvisuttinunt W, Scott W, Myers RS. The enzymatic basis of processivity in lambda exonuclease. Nucleic Acids Res. 2003;31:1585–1596. doi: 10.1093/nar/gkg266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.