Abstract
Ligases conduct the final stage of repair of DNA damage by sealing a single-stranded nick after excision of damaged nucleotides and reinsertion of correct nucleotides. Depending upon the circumstances and the success of the repair process, lesions may remain at the ligation site, either in the template or at the oligomer termini to be joined. Ligation experiments using bacteriophage T4 DNA ligase were carried out with purine lesions in four positions surrounding the nick site in a total of 96 different duplexes. The oxidized lesion 8-oxo-7,8-dihydroguanosine (OG) showed, as expected, that the enzyme is most sensitive to lesions on the 3′ end of the nick compared to the 5′ end and to lesions located in the intact template strand. In general, substrates containing the OG·A mismatch were more readily ligated than OG·C. Ligations of duplexes containing the OA·T base pair (OA=8-oxo-7,8- dihydroadenosine) that could adopt an anti-anti conformation proceeded in high efficiencies. An OI·Acontaining duplex (OI = 8-oxo-7,8-dihydroinosine) behaved similarly to OG·A. Due to its low reduction potential, OG is readily oxidized to secondary oxidation products, such as the guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) nucleosides; these lesions also contain an oxo group at the original C8 position of the purine. Ligation of oligomers containing Gh and Sp occurred when opposite A and G although the overall ligation efficiencies were much lower than most OG base pairs. Steady-state kinetic studies were carried out for representative examples of lesions in the template. Km increased by 90–100-fold for OG·C, OI·C, OI·A and OA·T containing duplexes compared to G·C. Substrates containing Gh·A, Gh·G, Sp·A and Sp·G base pairs showed Km values 20–70-fold higher than G·C while the Km value for OG·A was 5 times lower than G·C.
Reactive oxygen species (ROS)1 such as O2−•, HO• and H2O2 are continuously produced during normal metabolic processes, and their production is augmented by inflammation and exposure to certain agents (1, 2). DNA is sensitive to ROS, and in vivo oxidative damage results in DNA strand breaks, base modifications and DNA-protein cross-links (1). Unrepaired oxidative DNA damage can be mutagenic and is implicated in carcinogenesis, neurological disorders and aging (2–4). Oxidation of guanine, the most easily oxidized nucleobase (5), can lead to the commonly observed lesion 8-oxo-7,8-dihydro-2′-deoxyguanosine (OG) which is regarded as a biomarker of oxidative DNA damage in the cell (6). OG is mutagenic in the absence of repair leading to G→T transversions (7–10).
OG can be removed by the base excision repair pathway (BER) (11–14) involving glycosidic bond cleavage of the damaged base followed by excision of the remaining abasic (AP) site (15). A nucleotide is inserted by a DNA polymerase, and in a final stage, the nick is sealed by a DNA ligase (16–20). Although DNA repair of oxidized base lesions has been extensively investigated, only a few studies have appeared describing the effect of oxidized base lesions on DNA ligation, the last step of DNA repair (21, 22). Both human DNA ligases I (Ligl) and III are able to join OG·A more efficiently than OG·C when A or C are inserted at the 3′ end of the nick (23). Lig1 also joins Okazaki fragments and aids in long-patch base-excision repair (24–26). The crystal structure of Lig1 complexed with an adenylated DNA intermediate was recently reported and aids in the understanding of its interaction with the DNA substrate at the catalytic core (27).
The present study was designed to gain an understanding of how oxidized purines located at different positions surrounding the nick site in the DNA duplex influence the ligation efficiency of bacteriophage T4 DNA ligase (EC 6.5.1.1). T4 DNA ligase is widely used as a tool in gene manipulation, and its low fidelity makes it a good starting point to study DNA ligation. Ligases catalyze the formation of a phosphodiester bond between the 5′-phosphoryl and 3′-hydroxyl termini in order to close a nick in duplex DNA or RNA (28). T4 DNA ligase is purified from Escherichia coli and uses ATP as a cofactor in this process (28, 29). The ligation mechanism involves three steps: (a) activation of the enzyme by adenylation (30), (b) formation of the intermediate by transfer of the AMP residue from the enzyme to the 5′-phosphate of the nicked DNA (30), and (c) sealing of the DNA by a transesterification reaction (28, 31). The existence of clustered DNA damage (32–35) and the insertion of dOGMP during replication suggests the possibility that an oxidized lesion could appear near a nick site. Furthermore, the DNA repair enzyme MutY excises adenine opposite guanine or 8-oxoG during BER (11, 36, 37), and complete repair requires the further action of a polymerase for insertion of dCTP opposite OG, followed by ligation (38). In this case, ligase would be required to seal the nick with an OG:C base pair at the junction. T4 DNA ligase is a low fidelity enzyme, which allows it to seal various mismatches particularly the G·T mismatch (39, 40); however, mismatches at the duplex terminus are known to slow the ligation reaction (41). The efficiency of T4 DNA ligase to ligate two oligodeoxynucleotides greatly decreases when single base pair mismatches appear on either side of the ligation junction (42, 43). Previous studies reported that T4 DNA ligase is capable of ligating C:T and G:T mismatches on the 3′ end of the nick and can tolerate pyrimidine/pyrimidine mismatches better than purine-containing mismatches (44–46).
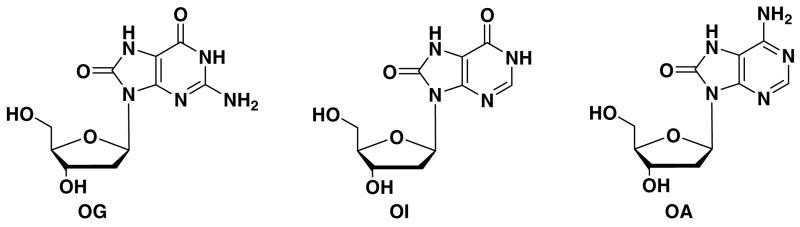
The purpose of this study was to investigate the ability of T4 DNA ligase to seal a nick that contains an oxidized base lesion at or near the ligation site. OG and other oxidized purines, such as 8-oxo-7,8- dihydroadenine (OA), occur frequently in mammalian cells (Figure 1) (47, 48). A previous study showed that OA may contribute to carcinogenesis by the observation that it appears in human tumor tissue at an increased level (49). OA:T and OG:A base pairs with the oxidized lesion positioned at the 3′ end of the nick were reported to have high ligation efficiency with T4 DNA ligase (50). 8-Oxo-7,8- inosine (OI) has a similar structure to OG but lacks the 2-amino group and serves as a structural probe (Figure 1). Both OG and OI have carbonyl groups at C6 and can base pair with A at a syn-anti conformation (51). OA, with an amino group at C6, preferentially pairs with T in an anti-anti conformation, although it can also mispair with G, leading to A→C transversions (52, 53). The OA·C mismatch was also observed in a syn-anti conformation that distorts the DNA duplex (54, 55). In this work, a comparison of ligation efficiency with different oxidized lesions provides information concerning the influence of purine modifications on base stacking and base pairing related to enzyme activity.
Figure 1.
Structures of 8-oxo-7,8-dihydro-2′-deoxyguanosine (OG), 8-oxo-7,8-dihydro-2′-deoxyinosine (OI) and 8-oxo-7,8-dihydro-2′ deoxyadenosine (OA).
Since the redox potential of OG (0.7 V) is lower than the unmodified bases T (1.7 V), C (1.6 V), A (1.4 V), and G (1.3 V), it is readily oxidized by one-electron oxidants, peroxynitrite, carbonate radicals and singlet oxygen (56, 57). Two oxidation products of OG, guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) have been identified arising from direct 4e− oxidation of G (e. g. by 1O2) or further oxidation of initially formed OG (58–60) (Figure 2). Sp was recently detected in repair-deficient bacteria exposed to chromate (61). The levels of Gh and Sp arising from OG during in vitro oxidation of calf thymus DNA with peroxynitrite (ONOO−) were 3.7% and 0.6% respectively (62). Both Gh and Sp can exist as two diastereomers (58), and recent computational studies have revealed that the R and S stereoisomers of Sp distort the duplex by perturbing the base stacking and neighboring hydrogen binding (63). OG, Gh, and Sp have been studied in primer extension experiments by the Klenow fragment (KF) of Escherichia coli Polymerase I (64). Unlike OG-containing templates for which dCMP and dAMP insertion is facile, the presence of Gh or Sp in a template is inhibitory to primer extension although small amounts of dAMP and dGMP are incorporated opposite these lesions (64). In vivo studies in E. coli showed that the lesions OG and Gh were easily bypassed, and OG showed low overall mutagenesis that was largely G→T (65). In contrast, Gh and Sp lesions were essentially 100% mutagenic (66, 67). Gh was highly specific for G→C mutations, while Sp led to both G→T and G→C mutations (65).
Figure 2.
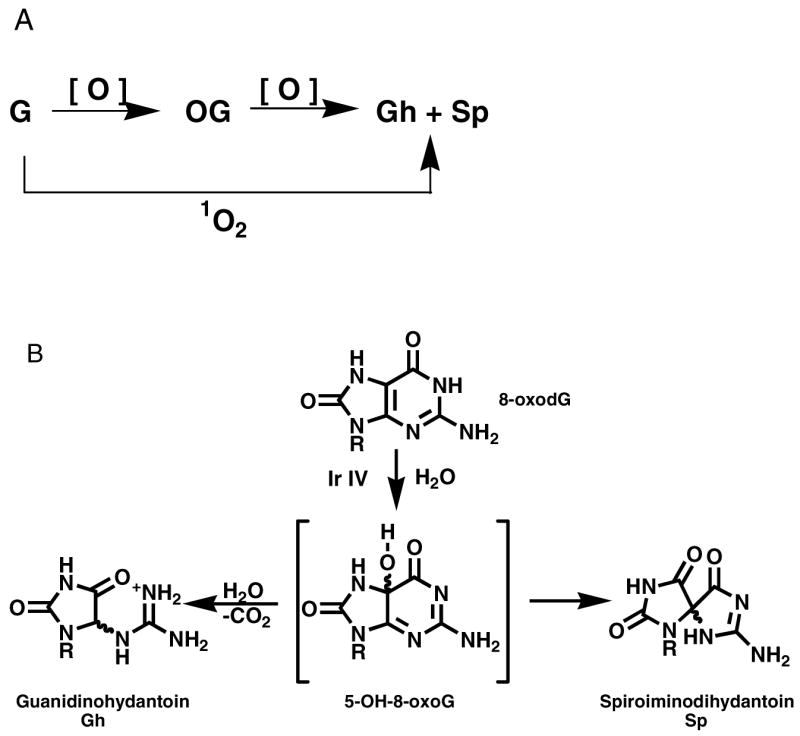
A: Oxidation from G to Gh and Sp can occur sequentially via OG or in one step using a 4-eoxidant such as singlet oxygen. B: Oxidation of OG with one-electron oxidants leads to Gh and Sp.
In this ligation study, both Gh and Sp were synthesized at four positions immediately surrounding the single-strand nick as comparisons to OG, OA and OI (Figure 3); all members of this oxidized purine family have a carbonyl group at the C8 position of the original purine. To further investigate the suppressed ligation efficiencies of lesion-containing duplexes, steady-state kinetic studies (68) and UV melting experiments (64) were performed with selected lesions in the template strand. Kinetic studies with Gh·G and T·G pairs at the 5′ end of primer1 were also studied to test our hypothesis that Gh mimics T in a T·G wobble pair. Based on these results, we discuss how base-stacking, hydrogen-bonding and protein-DNA interactions influence the ligation efficiency.
Figure 3.

Sequences used for Template/Primer1/Primer2 duplexes.
MATERIALS AND METHODS
Oligodeoxynucleotide Synthesis and Purification
Oligodeoxynucleotides containing A, G, T, C, OG, OI and OA were obtained either from the DNA/Peptide Core Facility (University of Utah) or synthesized on an Applied Biosystems Model 392 DNA/RNA synthesizer using solid phase DNA synthesis. Sequences of the oligodeoxynucleotides studied are presented in Figure 3. The dOG phosphoramidite (Glen Research) was used to synthesize OG containing oligodeoxynucleotides, which were cleaved off the column overnight in fresh 30% NH4OH containing 0.25% β-mercaptoethanol followed by deprotection for 17 h at 55°C. Oligodeoxynucleotides with a 3′-OG nucleotide were synthesized using a Universal Support II column (Glen Research) and cleaved using 3 mL 2 M ammonia in methanol for 30 min at room temperature followed by deprotection for 24 h at 55°C. The dOI phosphoramidite was synthesized by a modification of the method reported by Oka and Greenberg (69), the details of which will be reported elsewhere. The dOI and dOA (Glen Research) phosphoramidites were used to synthesize OI and OA-containing oligodeoxynucleotides, which were subsequently deprotected and cleaved by the same method as the OG-containing oligodeoxynucleotides. Purification of the oligodeoxynucleotides was accomplished using 15% or 20% denaturing polyacrylamide (19:1) gel electrophoresis (PAGE) with 6 M urea. ESI-MS analysis on a Micromass Quattro II mass spectrometer was used to verify the identity and purity of the oligodeoxynucleotide.
DNA Oxidation
OG-containing oligodeoxynucleotides were desalted twice using a G-25 column. The oligomer (12 μM) and Na2IrCl6 (100 μM Alfa Aesar) in a 500- μL reaction volume were used for the oxidation of OG to Gh or Sp. For the synthesis of Gh from OG, the oligodeoxynucleotides were oxidized by incubation with the oxidant for 30 min at room temperature in H2O, whereas Sp was generated from OG by incubation for 30 min at 65°C in 10 mM sodium phosphate (NaPi)/100mM NaCl buffer (pH 7.4). Oxidation reactions were quenched by addition of 4 μL 20 mM EDTA (pH 8.5). Oxidized products were dialyzed (3,500 MWCO) against water for 24–48 h to remove Na2IrCl6, NaPi, NaCl and EDTA. Oligomers containing OG, Gh, or Sp lesions were purified by HPLC (Figure S1, Supporting Information). All other lesion-containing sequences were also subjected to HPLC purification. The purity of the oligodeoxynucleotides used for ligation was >99% (Figure S2, Supporting Information). ESI-MS analysis confirmed the purity of the Gh and Sp oligodeoxynucleotides.
Mass Spectrometric Analysis and HPLC Analysis
Oligodeoxynucleotides (2 nmol) in 30 μL H2O were combined with 10 M NH4OAc (30 μL) for 2 h at RT. A 50:50 mixture of ethanol and isopropanol (180 μL) was added, and the sample was stored for more than 2 h in dry ice, followed by centrifugation for 30 min at 4°C. The supernatant was removed, and 1 mM NH4OAc was added to produce of solution with DNA concentration of 40 μM. Isopropanol was added to yield a final concentration of 20 μM. The resulting sample was then used for ESI-MS analysis.
End Labeling of Primers and Template/Primer Annealing
Primer 1 with a native base on the 5′ end was 5′ end-labeled with T4 polynucleotide kinase (New England Biolab) and [γ-32P] ATP (Amersham). Primer 1 with OG, OI, or OA on the 5′ end was first 5′ end-phosphorylated with T4 polynucleotide kinase and 1 mM non-radioactive ATP and then 3′ end-labeled with terminal transferase (New England Biolab) and [α-32P] dideoxyATP (Amersham). A microspin G-25 column (Amersham Pharmacia Biotech) was used to remove unreacted [γ-32P] ATP and [α-32P] dideoxyATP from the radioactive DNA. Complementary oligodeoxynucleotides shown in Table 1 were annealed in water with a template:primer 1:primer 2 ratio of 2:1:1 at 90°C for 5 min following by slow cooling to room temperature. The final concentration of the duplex was 0.25 μM.
Table 1.
Steady-State Kinetics Parameters of Ligation with Lesions on the 3′the Nick (Fig 5B)
| Km(μM) | kcat(min −1) | kcat/Km(min−1• μM−1) | Tm(°C) | |
|---|---|---|---|---|
| G·C | 0.064 ± 0.008 | 0.51 ± 0.02 | 7.99 | 58 ± 1 |
| OG·C | 5.8 ± 1.3 | 1.15 ± 0.21 | 0.20 | 58 ± 3 |
| OG·A | 0.36 ± 0.11 | 0.61 ± 0.06 | 1.69 | 57 ± 1 |
| OI·C | >10* | 58 ± 2 | ||
| OI·A | 5.9 ± 2.1 | 0.55 ± 0.02 | 0.09 | 59 ± 4 |
| OA·T | 6.5 ± 2.7 | 0.69 ± 0.23 | 0.11 | 57 ± 1 |
| Gh·A | 1.2 ± 0.4 | 0.45 ± 0.007 | 0.36 | 58 ± 1 |
| Gh·G | 2.8 ± 0.5 | 0.22 ± 0.02 | 0.08 | 60 ± 1 |
| Sp·A | 4.5 ± 1.3 | 0.52 ± 0.11 | 0.11 | 61 ± 1 |
| Sp·G | 2.0 ± 0.5 | 0.08 ± 0.01 | 0.04 | 59 ± 1 |
Estimated Km value. The Michaelis –Menten curves for some duplexes do not reach a plateau.
Ligation with T4 DNA Ligase
Ligation of the duplex was carried out with T4 DNA ligase (New England Biolabs) in 1 × T4 DNA ligase buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM DTT, 1mM ATP, 25 μg/ml BSA, pH 7.5, 25°C). The reaction mixture included 0.125 μM duplex and 0.625 units T4 DNA ligase. Reaction mixtures were incubated at 25°C for 20 min. All reactions were inactivated by incubating at 90°C for 10 min and then lyophilized to dryness. Loading buffer (8 μL, 5% Bromophenol Blue and 5% Xylene Cyanol FF in 6 M urea) was added to the sample and a 4 μL aliquot was then taken out and loaded onto 20% denaturing PAGE with 6 M urea. Gels were then analyzed by storage phosphor autoradiography on a Typhoon and quantified with ImageQuant software. Data presented are the averages of 3 or 4 independent experiments.
Determining the Concentration of Active T4 DNA ligase
Primer2 containing 3′ amino group on the 3′-terminal nucleotide was synthesized by transferring a 3′-amino-2′,3′-dideoxycytidine-5′-triphosphate (TriLink BioTechnologies) using terminal transferase. After purification by HPLC, primer2 was annealed with primer1 and template to form a nicked substrate containing a 5′-phosphate and a 3′-amino group with no lesion. A T4 DNA ligase burst assay was performed with 100-fold dilution of the enzyme (400 unit/μL) and 50 nM substrate. Data was fit in the burst equation to calculate the active enzyme concentration.
Steady-State Kinetics for Overall Ligation Assay
To investigate the substrate specificity of T4 DNA ligase, ligation reactions with various DNA substrates containing lesions in the template were performed under the same conditions used for qualitative studies. Steady-state kinetics experiments were performed in a saturating concentration of ATP (1 mM) with various amounts of DNA substrates (7.5 nM –2 μM) and different enzyme concentrations. The annealed duplex containing a normal G·C pair at the nick site required 6 units of T4 DNA ligase. Ligation with an OG·A base pair utilized 20 units while duplexes containing Gh and Sp required 40 units enzyme. Kinetics studies with OI and OA-containing duplexes were performed with 80 units of enzyme. Various amounts of enzyme (6–80 unit) and different reaction times (1–3 min) were used to ensure single turnover conditions (70). Reaction samples were quenched with loading buffer and applied to a 20% denaturing PAGE. Ligation products were visualized by Typhoon and quantified by ImageQuant. Km and kcat values were calculated by Grafit 5. Kinetics studies of Gh·G and T·G were compared using the same method.
UV Melting Studies
DNA duplexes with lesions in the template were used for UV melting studies. Tm values for each duplex were measured with a Beckman DU7400 UV-vis spectrophotometer. Methods were the same as described by Kornyushyna et al (64).
RESULTS
Oligodeoxynucleotides were synthesized with each of six lesions incorporated at one of four positions, either on the 3′ terminus or the 5′ terminus of the nick, or in the template opposite the two sites. For each position, a C, A, T, or G nucleotide was synthesized in the opposite strand totaling 96 different nicked duplexes that were studied. Oligodeoxynucleotides containing lesions on the 3′ end of the nick were synthesized with a Universal Support II column using an input sequence with one extra nucleotide on the 3′ end. OG, OA and OI-containing oligodeoxynucleotides were synthesized using the corresponding phosphoramidites. Gh and Sp-containing oligodeoxynucleotides were prepared from OG-containing oligodeoxynucleotides by oxidation. All synthetic oligodeoxynucleotides were purified by PAGE and their purity was checked by ESI-MS. After further purification by HPLC, the purities of oligodeoxynucleotides were greater than 99%. The diastereomers of Gh were used in the experiments as a mixture because they slowly interconvert and are not isolable (71). Sp diastereomers were also used as a mixture because they behaved similarly in an in vivo lesion bypass assay (72). Primer 1 was labeled using [β-32P] ATP and T4 polynucleotide kinase and then annealed with the template strand and Primer 2. Bacteriophage T4 DNA ligase was added, and the product was visualized and quantified by PAGE storage phosphor autoradiography.
Ligation with OG, OI and OA Lesions
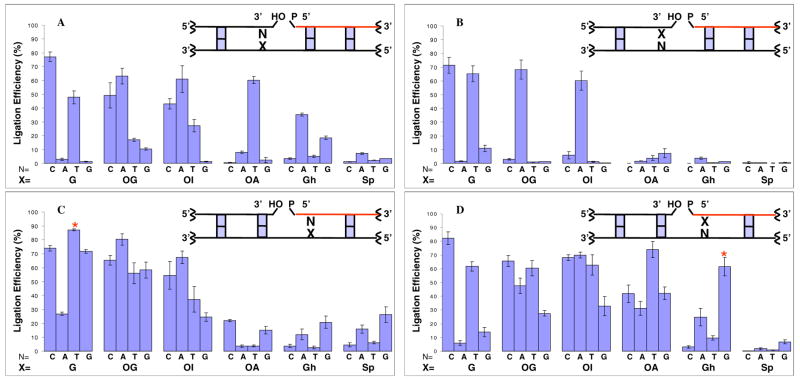
The extent of ligation by T4 DNA ligase of oligodeoxynucleotides containing mismatches and lesions was first investigated using template X with primer 1T and primer 2N (X = G, OG, N = C, A, T or G) (Figure 3). Template strands were annealed with the appropriate primers and ligated with T4 DNA ligase at 25°C for 20 min. The same reaction system without enzyme was used as a control. The Watson-Crick base pair G·C, in which C occupies the 3′ side of the nick, had the highest percent ligation at about 77% whereas 47% of the mismatched G·T base pair was ligated (Figures 4 and 5A). G·A and G·G mismatches were barely ligated with levels less than 3%. For base pairs containing OG, both OG·C and OG·A ligated to relatively large extents at about 49% and 63%, respectively. The positioning of OG opposite T or G resulted in low ligation efficiencies (Figures 4 and 5A).
Figure 4.
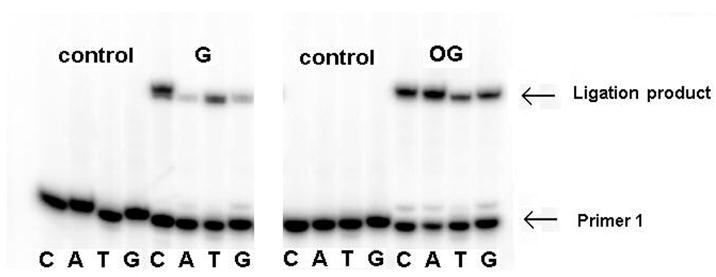
Ligation with lesions in the template opposite the 3′ end of the nick. G, OG, Gh and Spcontaining oligomers were each base paired with C, A, T, and G in the opposite strand. Lanes without ligation products were control studies with no enzyme added. Sequences used are indicated in Figure 3a.
Figure 5.
Ligation efficiencies of oligodeoxynucleotides containing lesions X=OG, OI, OA, Gh or Sp around the nick; X = lesion; N = C, A, T, G. A: lesions in the template opposite the 3′ end of the nick; B: lesions on the 3′ end of the nick; C: lesions in the template opposite the 5′ end of the nick; D: lesions on the 5′ end of the nick. DNA sequences used in Figure 5A, 5B, 5C and 5D are indicated in Figure 3a, 3b, 3c and 3d respectively. Primer1 shown in red was 5′-end labeled for PAGE analysis. Asterisks (panels C and D) indicate a comparison of 5′-T:G and 5′-Gh:G, both of which are ligated with high efficiency.
To gain a greater understanding of the structural features influencing helical integrity and enzyme activity, we examined the ligation with OI and OA in place of OG. Figure 5A shows the extent of ligation of substrates containing OI and OA in the template opposite the 3′ end of the nick, with OG used as a comparison. OI led to a high level of ligation when base paired with C and A at about 43% and 61%, which is similar to OG·C and OG·A substrates (49% and 63%, respectively). OI·T was moderately ligated at about 27% while the ligation of OI·G was less than 2%. As expected, OA behaved very differently from OG. Its base pair with a T-containing oligomer showed a relatively high degree of ligation (about 60%), since it is analogous to a Watson-Crick A·T base pair. Other bases opposite OA led to very low levels of ligation (≤8%).
Quantitative analysis of lesions located on the 3′ end of the nick is presented in Figure 5B. G·C, G·T, and OG·A base pairs were well ligated to an extent of approximately 70%. Other base mismatches containing the OG lesion were very poorly ligated (≤ 3%). When OI and OA are on the 3′ end of a nick, the difference between the two nucleotide analogs becomes obvious. The only base pair that ligated well was OI·A (about 60%), which is similar to OG·A. In the case of OA, none of the base pairs were tolerated by T4 DNA ligase (≤8%). Lesions, except for OG·A and OI·A, on the 3′ end essentially shut down the ligation activity of T4 DNA ligase, and apparently none of the 8-oxopurines is tolerated in a standard anti conformation.
Ligation of oligomers containing lesions at the position opposite the 5′ end of the nick in the template strand are revealed in Figure 5C. T4 DNA ligase catalyzed ligation of nearly all of the mismatches and base pairs containing the OG lesion with relatively high levels varying from 55% to 87%. For the ligation with OI, all the base pairs containing OI are well ligated (25% to 67%), with OI·A and OI·C higher than 50%. Surprisingly, when OA was paired with C and G, the ligation efficiency was about 22% and 15% respectively. OA·A and OA·T were both lower than 4%. Overall, OA-containing oligomers do not form good substrates at this position although OG and OI generally do so irrespective of the base opposite.
Figure 5D shows the extent of ligation when the lesion is on the 5′ end of the nick. Similar to other positions, G·C base pairs had the highest ligation efficiency of about 82%. The G·T mismatch, as for other positions, was ligated best (about 62%) among all the other non-canonical base pairs. All the nucleotides base paired with OG on the 5′ end are ligated readily from 28% to 65%. Base pairs containing OI showed ligation patterns similar to OG base pairs. The ligation efficiencies were high, from 60% to 70%, except for OI·G (about 32%). OA base pairs were also ligated well and OA·T had the highest efficiency among them (about 74%). Other OA base pairs displayed ligation efficiency to the range of 30% to 40%.
Ligation with Gh and Sp Lesions
Established procedures were used to generate the Gh and Sp lesions as diastereomeric mixtures. Gh diastereomers rapidly interconvert and cannot be purified separately (71, 73); Sp diastereomers are stable, but their activities with other enzymes have often been similar (64, 72). For hydantoin lesions in the template opposite the 3′ end of the nick, Gh·A and Gh·G were moderately ligated at about 35% and 19%, while only about 7% of Sp·A was ligated by T4 DNA ligase. Other duplexes with Gh and Sp in the template had extremely low ligation efficiencies (≤ 5%) (Figure 5A).
Quantitative analysis of lesions on the 3′ end of the nick is presented in Figure 5B. All the base pairs containing Gh, and Sp lesion had very low ligation efficiency (≤ 3%). Gh and Sp are very poor substrates in this position, and T4 DNA ligase is very sensitive to the proper base pairing on the 3′ end of the nick.
Ligation efficiencies of lesions at the position opposite the 5′ end of the nick in the template strand are revealed in Figure 5C. Duplex DNA containing Gh and Sp base pairs had relatively lower efficiencies than OG base pairs. Gh·G and Sp·G were the most effectively ligated at about 20% and 26% respectively. Gh·A and Sp·A were also ligated to some extent (12% and 16%). The duplexes containing base pairs of Gh and Sp with C or T were not good substrates for ligation (≤ 6%).
Figure 5D shows the extent of ligation when the hydantoin lesion is on the 5′ end of the nick. Here the difference between Gh and Sp is highlighted because certain Gh-containing substrates ligated well, but none of the Sp duplexes were good substrates. The Gh·G base pair had a high ligation efficiency of about 62% and Gh·A base pair was moderately ligated (about 25%). Duplexes containing Gh·C, Gh·T and Sp lesions were barely ligated (≤10%); although the Gh substrates were competent for being adenylated (see Supporting Information, Fig. S3C), their adenylated intermediates were not able to complete the ligation process. In a related experiment, the Sp·G base pair was only slightly ligated (about 7%), and little adenylation was observed for any Sp·N base pair. Since Gh and Sp lesions, when positioned on the 5′ end of the oligodeoxynucleotides, enhance the exonuclease activity of T4 polynucleotide kinase, degradation was observed for oligodeoxynucleotides with Gh and Sp at that position. In order to verify if oligodeoxynucleotides containing 5′-phosphorylated Gh and Sp lesions are efficiently ligated, experiments were carried out with 3′ end labeled primer 1, and the results yielded similar ligation abilities (see Supporting Information).
Concentration of Active T4 DNA Ligase
In order to pursue kinetics studies and obtain accurate kcat/Km data, we performed experiments to determine the concentration of active T4 DNA ligase following a protocol developed by Tsai and coworkers (44). The enzyme was precharged to form an enzyme-AMP intermediate by incubating a nicked DNA substrate containing a 5′-phosphate and a 3′-amino group at the nick site. The adenylation of the 5′-phosphate resulted in a rapid single turnover followed by a slow steady-state phase. Data was fit to a burst equation in which the burst amplitude corresponds to the concentration of active enzyme, determined to be 3.11 ±0.06 μM.
Steady-State Kinetics and UV Melting Experiments
Steady-state kinetics studies using T4 DNA ligase to seal a nick were carried out with duplexes bearing lesions in the template opposite the 3′ end of the nick. An overall ligation assay was performed under a fixed and saturating concentration of ATP (1 mM) while the concentration of another substrate, the DNA duplex, was varied (7.5 nM –2 μM). The values of Km, kcat, and kcat/Km were calculated for each lesion (Table 1). The relative catalytic efficiency of G·C was 4-fold higher than OG·A. The ligation of OG·C demonstrated a similar efficiency to the OIand OA-containing duplexes, which is around a 70-fold difference compared to that of the analogous G·C substrate. The substrates bearing further oxidized OG lesions yielded 66–180-fold lower efficiencies than G·C except Gh·A, which is only about 20-fold lower. G·C had the lowest Km value at 0.064 μM; the Km of OG·A is 5-fold higher than G·C, while still fairly low when compared with other lesions. Duplexes containing Gh and Sp lesions yielded much higher Km values than G·C by about 18– 70-fold. The Michaelis-Menten curves for OI- and OA-containing duplexes did not reach plateaus and so the Km values were estimated at >5 μM for these DNA substrates. kcat values were similar for all the duplexes.
UV melting experiments were conducted with all the duplexes used in the kinetic studies. 40-mer templates containing lesions were annealed with a 20-mer primer and a 17-mer primer. Most of duplexes had Tm values around 58° (Table 1). Within experimental error, the Tm data showed little difference among various DNA duplexes.
Gh·G and T·G comparison
Steady-state kinetics were studied with T or Gh on the 5′ end of primer 1 with G in the template opposite (* in Fig 5, panels C and D). The ligation efficiencies for Gh·G and T·G base pairs were similar with kcat/Km of 0.31 and 0.28 min-1•μM-1respectively (Table 2), although the individual kcat and Km values were quite different with Gh·G displaying weaker binding but a compensatingly higher kcat. Among the 32 different orientations of hydantoin lesion base pairs (Gh·N or Sp·N), Gh·G with the Gh lesion at the 5′ end of primer 1 (Fig. 5D*) displayed the highest ligation efficiency.
Table 2.
Steady-State Kinetics Parameters of Ligation with Gh•G and T•G on the 5′end of the Nick
| Km(μM) | kcat(min−1) | kcat/Km(min −1 • μM−1) | |
|---|---|---|---|
| Gh·G | 1.5 ± 0.1 | 0.45 ± 0.02 | 0.31 |
| T·G | 0.055 ± 11 | 0.015 ± 0.001 | 0.28 |
DISCUSSION
In the present study, we investigated the influence of the guanine oxidation product 8-oxo-7,8- dihydroguanine (OG) and the related oxidized purines, 8-oxo-7,8-dihydroadenine (OA) and 8-oxo-7,8- dihydrohypoxanthine (OI), on the ligation reaction catalyzed by bacteriophage T4 DNA ligase. Since guanosine has a lower redox potential than A, C, or T, it is the most subject to oxidation by reactive oxygen species, leading to formation of OG in, for example, the template strand of DNA. Numerous studies have established that dCTP and dATP are inserted opposite OG during replication and repair. Although misinserted dATP opposite template OG can be excised by the action of MutY in E. coli or hMYH in humans, a preliminary level of proofreading by inhibition of ligation of OG:A mispairs would also aid in maintaining fidelity. In contrast, previous studies have shown that duplexes containing a 3′-terminal A at the nick site opposite OG in the template are easily ligated. For example, human ligases I and III are able to join OG·A more efficiently than OG·C when OG is in the template opposite the 3′ end of the nick (Figure 5A) (23). The opposite base pair orientation is also biologically relevant if the dOGTP lesion is present in the nucleotide pool and is inserted opposite C or A during DNA replication (74, 75). This would also require ligase to seal the nick next to a lesion, in this case at the 3′ side of OG as in Figure 5B. It is also worthwhile to study the other two positions around the nick in which lesions occupy the site at the 5′ end of the nick or the position in the template opposite the 5′ end of the nick to give a complete understanding of how the lesions in different positions could influence the ligation reaction. Although lesions at these positions near a nick (Figures 5C and D) are less likely to occur, they could be relevant to repair of tandem lesions. We therefore designed DNA duplexes that contained lesion base pairs or mismatches at four positions surrounding the nick leading to the sequences shown in Figure 3.
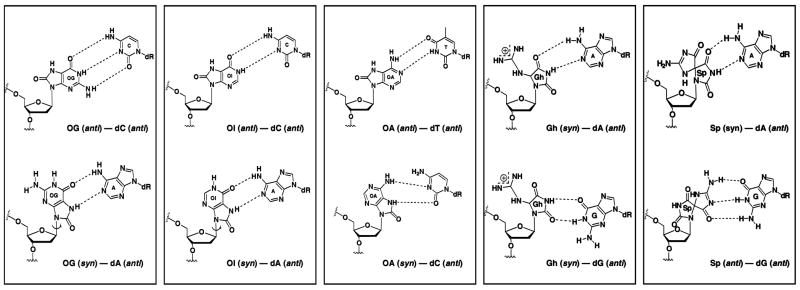
Possible base pair orientations (Figure 6) for OG include the Watson-Crick base pair with C in an anti-anti conformation, although the oxygen at C8 of the purine could have detrimental interactions with the sugar-phosphate backbone. A conformational change of OG to a syn orientation results in a Hoogsteen base pair with anti A. OI lacks the amino group at position 2, but retains the same ability to pair with C or A (69). Although OI is not a biologically significant lesion, it allows us to probe further the steric requirements of syn and anti 8-oxopurines in positions around the nick site. In contrast, OA has been reported to induce mutations both in vitro or in vivo (53). Although thymine is preferentially incorporated opposite 8-oxoA by KF and Taq pol, pol → and → can also incorporate dGMP as well as dCMP opposite 8-oxoA (76, 77). Figure 6 shows the structure in which OA is paired with T in an antianti conformation. Additionally, OA can also base pair with C in a syn-anti conformation by adopting a Hoogsteen wobble structure (55). Thus, many possible base mismatches could be accommodated in duplex DNA if DNA ligase is able to seal nicks surrounding these lesions.
Figure 6.
Base pairing of OG, OI, OA, Gh and Sp. OG can base pair with C at an anti-anti conformation and with A at a syn-anti conformation. OI can form a similar base pair with the same conformation as OG. OA can only form an anti-anti base pair with T. Gh is proposed to base pair with A and G in a syn-anti conformation. Sp is proposed to base pair with A or G in a syn-anti conformation or potentially with G in an anti-anti conformation (shown).
The results obtained in the present study lead to several important conclusions. Consistent with other studies, the ligation reactions showed that T4 DNA ligase is more affected by a 3′-terminal lesion or mismatch at the nick site (Fig. 5B), and to a lesser extent, to lesions in the template opposite the 3′ end of the nick (Fig. 5A). The data shown in the OG panel of Fig. 5A are similar to the results of Moriya and coworkers who studied human DNA ligases I and III and found more efficient ligation with A at the 3′ terminus opposite OG (23). In the present study, substrates containing 3′-terminal A were preferred to those containing 3′-terminal C, although the differences were not large. However, a dramatic difference was seen when the positions were reversed, i.e. when the 8-oxopurine was present at the 3′ terminus next to the nick (Figure 5B). These substrates were either very efficiently ligated (>65%) or not at all (<5%). For example, the OG•A base-paired substrate was very efficiently ligated, while the OG•C substrate was not. Similarly the OI•A substrate was efficiently ligated while the OI•C base pair was not a substrate. These results suggest that the active site of the enzyme cannot accommodate the oxo group protruding at C8 of the purine and must flip to a syn conformation. Additional evidence is seen with the fact that OA at the 3′ end of the nick is not a substrate no matter what base is opposite in the template (Figure 5B), although it is efficiently ligated when it resides in the template and has T at the 3′ nick site (Figure 5A). Thus, data for the 8-oxopurines show that only the syn orientation is allowed when the purine is at the 3′ end of the nick, but additionally, a reasonably stable syn-anti base pair is also a requirement for ligation since OA was not well ligated with any base opposite. In contrast, 8-oxopurines can adopt either the syn or anti conformation when they reside in the template opposite the 3′ end of the nick.
These preferences in ligation are consistent with nucleotide insertion and primer extension studies in which dAMP and dCMP were found to be incorporated opposite OG and OI (69), and dTMP was incorporated opposite OA (64, 77). Lesions in the template are relatively dangerous to cells since they may not be readily removed by short-patch base excision repair (BER) (23). Also, the 3′ →5′ exonuclease activity of human apurinic endonuclease 1 (APE1) does not discriminate OG·A from OG·C (41). Furthermore, human DNA ligase I and III ligated OG·A more efficiently than OG·C when OG was located in the template (analogous to Figure 5A) (23).
A previous study showed that the 3′ terminal OG:C pair inhibits ligation by DNA ligase I or DNA ligase IIIα (78); our results suggest that the 3′ end OG:A is much more efficiently ligated. This is also consistent with ligation data reported by Wallace and coworkers, although we did not observe significant ligation with substrates containing an OA:T base pair (50). OA could also form an OA·C base pair with a syn-anti conformation; however, it is believed to be a Hoogsteen wobble base pair that distorts the DNA duplex and is unsuitable for normal base pairing in A or B forms of DNA (55).
One of the major differences between an anti and a syn conformation of OG (or OI) is that the oxo group at C8 is oriented in a different groove. This hydrogen bond acceptor is extended in the major groove in an anti conformation while flipped to the minor groove in a syn conformation. An explanation for their difference in reactivity is that the ligase may stabilize the 3′ terminal hydrogen bond with an acceptor in the minor groove that would position it for “in line” attack at the 5′-phosphate. This hydrogen bond acceptor would normally be O2 of a pyrimidine in a T•A (or C•G) base pair, but the 8- oxo group of OG or OI could play this role in the OG•A or OI•A mispairs. An alternative explanation is that both anti conformations of OG and OI have a carbonyl group in the major groove that may clash with the protein. It has been found that DNA ligase fidelity is ensured by interrogating minor groove contacts (79). A pyrimidine shape analog, difluorotoluene (DFT), was used to replace thymine, and although DFT paired with adenine approximates Watson-Crick geometry, a minor groove hydrogen bond acceptor is lost, and the ligation was inhibited (79). This result may help explain why T4 DNA ligase favors OG and OI in the syn conformation, although the possibility of steric interaction with the major groove is not ruled out. Overall, the 3′ end of the nick is the most important position in the ligation process, since it increases the fidelity of DNA ligase by its strict requirement for “in-line” attack to the 5′-phosphate (28, 41).
Lesions occurring on the 5′ terminus and in the template opposite the 5′ end of the nick were also studied, although their existence in vivo is considered minor. T4 DNA ligase does not differentiate mismatches at these positions and efficiently ligates all the mismatches containing OG, OI and OA (Fig. 5D). In the position opposite the 5′ end of the nick, OA·C and OA·G mismatches were weakly ligated while ligation of OA·A and OA·T was virtually nonexistent (Fig. 5C). This differs from OG and OI, which also suggests that OA behaves differently with an amino group at position 6 and its poor ability to flip over to form a syn conformation. Overall, the 5′ terminus of the nick contributes less to the fidelity of ligase in comparison with 3′ end of the nick.
Although a crystal structure of T4 DNA ligase has not been published, it is reasonable to assume that the AMP covalently bound to the 5′-phosphate is stabilized by a number of amino acids in the adenylation domain of ligase in comparison of active site amino acids of T4 DNA ligase with human DNA ligase I (27). It is stabilized and ready to be attacked by the 3′-OH despite the poor base pairing resulting from the presence of OG, OI and OA. The 3′ end of the nick may have less contact with the enzyme when a lesion is present, making it sensitive to the base pair stability and increasing the enzyme’s fidelity. The published crystal structure of human DNA ligase I in association with an adenylated DNA substrate demonstrated that the 5′-phosphorylated end is positioned in the active site, and the 5′ AMP interacts extensively with Glu621, Arg573 and Lys568 (27). These interactions lower the energetic requirement of the base pairing at the 5′ end of the nick while the 3′-OH has few interactions with the enzyme, and therefore is more dependent on the proper base pair chemistry to help the OH group attack the 5′ phosphate group (27).
Once OG forms in the DNA duplex, it has the potential to be oxidized to the secondary oxidation products Gh and Sp; the latter has been detected in repair-compromised cells exposed to chromate (80). In primer extension experiments, dATP and dGTP are inserted opposite Gh and Sp, which suggests a reason that these oxidative lesions are highly mutagenic to E. coli (64, 72, 81, 82). Ligation with Gh and Sp in the four positions around the nick was also studied (Figure 5). Except for the 3′ end of the nick, Gh and Sp in other positions were able to be ligated when placed opposite A and G, which conforms to the preference observed in primer extension experiments. Gh and Sp on the 3′ end of the nick totally blocked the ligation, likely because of poor base pairing with nucleotides and disruption of the helical structure. Although Gh and Sp might pair with A and G, the ligation efficiencies of Gh- and Spcontaining base pairs were found to be very low (mostly <30%).
To further investigate the ligation inhibition of lesion-containing duplex DNA, we measured the kinetic parameters kcat and Km for a representative series of base pair combinations, and examined thermal stability via UV melting experiments. For the DNA duplexes used in our ligation study, including Watson-Crick and Hoogsten base pairs and base mismatches, UV melting experiments showed very similar Tm data providing evidence that one unmatched base pair at the nick site did not destabilize the overall 37-mer DNA duplex to a large extent although local fraying could be occurring.
Steady-state kinetics studies were carried out with TemplateX/Primer1T/Primer2N (Figure 3). We found that kcat values for all the duplexes were similar, which suggested T4 DNA ligase converts the same amount of substrates to product per second despite the presence of lesions. The main factor influencing ligation efficiency is Km. The OG·A-containing nicked duplex had a Km value 5-fold higher than its G·C base counterpart. OG·C, OI·C, OI·A and OA·T had much higher Km values(>90-fold than G·C) while Gh·A, Gh·G, and Sp·G had Km values inbetween (20–40-fold higher than G·C). Km values measure the binding ability between the enzyme and substrate, and since the kcat values were similar for each lesion-containing duplex, we suggest that oxidized purine lesions inhibit enzyme binding to these DNA substrates, lowering the ligation efficiencies. The altered hydrogen bonding of an unmatched base pair distorts the duplex, which influences the formation of enzyme-substrate complex. The hydrogen bonding of base pairs has a minor influence on the chemical reaction to form products. Consistent with the structures of lesion base pairs (Figure 6), T4 DNA ligase may favor a hydrogen acceptor in the minor groove or potentially a hydrogen donor in major groove. OG·A, Gh·A, Gh·G, and Sp·G are all proposed to have syn-anti conformations in the duplex, which all have an oxo group in the minor groove and an amino group in the major groove. Their relatively low Km data suggested that they are good substrates for T4 DNA ligase. OI·A also adopts a syn-anti conformation.
Gh and Sp make weak base pairs with normal nucleotides due to the presence of an sp3-hybridized carbon that prevents normal base stacking and must lead to helix distortion. However, Gh is apparently able to pair somewhat with A and G compared to other bases. We propose that Gh adopts the syn orientation when paired with A and G, in which the hydantoin ring is positioned to mimic a T in hydrogen bonding. This hypothesis easily explains the high ligation efficiency of the substrates containing the Gh·A mismatch as well as the Gh·G mismatch, which may mimic the T·G wobble pair. For a T·G (or U·G in RNA) wobble pair, the pyrimidine is shifted toward the major groove to provide an appropriate hydrogen bonding partner with G. The same orientation in a Gh·G pair would shift the nonplanar Gh residue toward the major groove creating more space for the sp3 carbon at the same time as providing hydrogen bond complementarity with G. The observation of similarly high ligation efficiencies with Gh·G and T·G pairs at the 5′ terminus is an interesting result that may support this hypothesis (compare * in Figures 5C and 5D).
Our current data suggests that, except for the OG·A mismatch, OG, OA, OI, Gh and Sp are poor substrates for ligation especially when they are located at the 3′ end of the oligomer to be ligated. Discrimination against ligation of a 3′ terminal lesion provides a means of proofreading of single nucleotide misincorporation during BER. Suppression of DNA ligation at oxidative sites is one possible mechanism for reducing spontaneous mutation. However, oxidative lesions occurring in the template are still potential threats to the cell. Whenever C and A are inserted opposite OG, or A and G inserted opposite Gh, these mismatches will be efficiently ligated potentially resulting in cellular mutation.
Supplementary Material
SUPPORTING INFORMATION AVAILABLE
HPLC analysis of crude and purified OG, Sp and Gh-containing oligomers, PAGE analysis of OI and OA-containing oligomers, and comparison of ligation of oligomers with lesions on the 5′ terminus of primer 1. This material is available free of charge via the Internet at http://pubs.acs.org.
Footnotes
This work was supported by the National Cancer Institute (CA090689).
Abbreviations: ROS, reactive oxygen species; OG, 8-oxo-7,8-dihydroguanine; Gh, 5-guanidinohydantoin; Sp, spiroiminodihydantoin; OI, 8-oxo-7,8-hypoxanthine; OA, 8-oxo-7,8-dihydroadenine; BER, base excision repair; PAGE, polyacrylamide gel electrophoresis; ESI-MS, electrospray ionization mass spectrometry; Ligl, human DNA ligase I; APE1, human apurinic endonuclease 1.
References
- 1.Shigenaga MK, Gimeno CJ, Ames BN. Urinary 8-hydroxy-2′-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc Natl Acad Sci USA. 1989;86:9697– 9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 3.Feig DI, Loeb LA. Oxygen radical induced mutagenesis is DNA polymerase specific. J Mol Biol. 1994;235:33–41. doi: 10.1016/s0022-2836(05)80009-9. [DOI] [PubMed] [Google Scholar]
- 4.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: Role in inflammatory disease and progression to cancer. Biochem J. 1996;313:17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steenken S, Jovanovic SJ. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 6.Shigenaga MK, Aboujaoude EN, Chen Q, Ames BN. Assays of oxidative DNA damage biomarkers 8-oxo-2′-deoxyguanosine and 8-oxo-guanine in nuclear DNA and biological fluids by high-performance liquid chromatography with electrochemical detection. Meth Enzymol. 1994;234:16–33. doi: 10.1016/0076-6879(94)34073-0. [DOI] [PubMed] [Google Scholar]
- 7.Cadet J, Bellon S, Berger M, Bourdat AG, Douki T, Duarte V, Frelon S, Gasparutto D, Muller E, Ravanat JL, Sauvaigo S. Recent aspects of oxidative DNA damage: Guanine lesions, measurement and substrate specificity of DNA repair glycosylases. Biol Chem. 2002;383:933–943. doi: 10.1515/BC.2002.100. [DOI] [PubMed] [Google Scholar]
- 8.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 10.Greenblatt MS, Bennett SE, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 11.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg E, Walker GC, Seide W. DNA Repair and Mutagenesis. ASM Press; Washington, DC: 1995. [Google Scholar]
- 13.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 14.Lomax ME, Cunniffe S, O’Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017– 11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 15.David-Cordonnier MH, Boiteux S, O’Neill P. Excision of 8-oxoguanine within clustered damage by the yeast OGG1 protein. Nucleic Acids Res. 2001;29:1107–1113. doi: 10.1093/nar/29.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Showalter AK, Lamarche BJ, Bakhtina M, Su MI, Tang KH, Tsai MD. Mechanistic comparison of high-fidelity and error-prone DNA polymerases and ligases Involved in DNA repair. Chem Rev. 2006;106:340–360. doi: 10.1021/cr040487k. [DOI] [PubMed] [Google Scholar]
- 17.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: Structure, reaction mechanism, and function. Chem Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 18.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 19.Braves RJ, Felzenszwalb I, Laval J, O’Connor T. Excision of 5′-terminal deoxyribose phosphate from damaged DNA is catalyzed by the Fpg protein of Escherichia coli. J Biol Chem. 1992;267:14429–14435. [PubMed] [Google Scholar]
- 20.Bernards AS, Miller JK, Bao KK, Wong I. Flipping duplex DNA inside out-- A double base flipping reaction mechanism by Escherichia coli MutY adenine glycosylase. J Biol Chem. 2002;277:20960–20964. doi: 10.1074/jbc.C200181200. [DOI] [PubMed] [Google Scholar]
- 21.Fortini P, Pascucci B, Parlanti E, D’Errico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053, 1–71. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto K, Tominaga Y, Nakabeppu Y, Moriya M. Futile short-patch DNA base excision repair of adenine: 8-oxoguanine mispair. Nucleic Acids Res. 2004;32:5928–5934. doi: 10.1093/nar/gkh909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase 1 is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 25.Levin DS, Bai W, Yao N, O’Donnell M, Tomkinson AE. An interaction between DNA ligase! and proliferating cell nuclear antigen: Implications for Okazaki fragment synthesis and joining. Proc Natl Acad Sci USA. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomkinson AE, Lasko DD, Daly G, Lindahl T. Mammalian DNA ligases. J Biol Chem. 1990;265:12611–12617. [PubMed] [Google Scholar]
- 27.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 28.Lehman IR. DNA ligase: Structure, mechanism, function. Science. 1974;186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 29.Deugau KV, Sande JH. T4 polynucleotide ligase catalyzed joining of short synthetic DNA. Biochemistry. 1978;17:723–729. doi: 10.1021/bi00597a026. [DOI] [PubMed] [Google Scholar]
- 30.Shuman S, Schwer B. In vitro mutagenesis and functional expression in Escherichia coli of a cDNA encoding the catalytic domain of human DNA ligase I. Nucleic Acids Res. 1995;19:6093–6099. doi: 10.1093/nar/19.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohle DS, Hansert B, Paulson SC, Smith BD. Biomimetic synthesis of putative cytotoxin peroxynitrite, ONOO−, and its characterization as a tetramethylammonium salt. J Am Chem Soc. 1994;116:7423–7424. [Google Scholar]
- 32.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 33.Lomax ME, Cunniffe S, O’Neill P. 8-oxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Bourdat AG, Douki T, Frelon S, Gasparutto D, Cadet J. Tandem base lesions are generated by hydroxyl radical within isolated DNA in aerated aqueous solution. J Am Chem Soc. 2000;121:4549–4556. [Google Scholar]
- 35.Lomax ME, Salje H, Cunniffe S, O’Neill P. 8-Oxo A inhibits the incision of an AP site by the DNA glycosylases Fpg, Nth and the AP endonuclease HAP1. Radiat Res. 2005;163:79–84. doi: 10.1667/rr3284. [DOI] [PubMed] [Google Scholar]
- 36.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo- 7,8-dihydrodeoxyguanosine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 38.Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 39.Harada K, Orgel L. Unexpected substrate specificity of T4 DNA ligase revealed by in vitro selection. Nucleic Acids Res. 1993;21:2287–2291. doi: 10.1093/nar/21.10.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landergren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- 41.Pritchard CE, Southern EM. Effects of base mismatches on joining of short oligodeoxynucleotides by DNA ligases. Nucleic Acids Res. 1997;25:3403–3407. doi: 10.1093/nar/25.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherepanov A, Yildirim E, Vries S. Joining of short DNA oligonucleotides with base pair mismatches by T4 DNA ligase. 2001 doi: 10.1093/oxfordjournals.jbchem.a002837. [DOI] [PubMed] [Google Scholar]
- 43.Wu DY, Wallace RB. Specificity of the nick-closing activity of bacteriophage T4 DNA ligase. Gene. 1989;76:245–254. doi: 10.1016/0378-1119(89)90165-0. [DOI] [PubMed] [Google Scholar]
- 44.Lamarche BJ, Showalter AK, Tsai MD. An error-prone viral DNA ligase. Biochemistry. 2005;44:8408–8417. doi: 10.1021/bi047706g. [DOI] [PubMed] [Google Scholar]
- 45.Husain I, Tomkinson AE, Burkhart WA, Moyer MB, Ramos W, Mackey ZB, Besterman JM, Chen J. Purification and characterization of DNA ligase III from bovine testes. J Biol Chem. 1995;270:9683–9690. doi: 10.1074/jbc.270.16.9683. [DOI] [PubMed] [Google Scholar]
- 46.Tomkinson AE, Tappe NJ, Friedberg EC. DNA ligase I from Saccharomyces cerevisiae: Physical and biochemical characterization of the CDC9 gene product. Biochemistry. 1992;31:11762–11771. doi: 10.1021/bi00162a013. [DOI] [PubMed] [Google Scholar]
- 47.Jaruga P, Dizdaroglu M. Repair of products of oxidative DNA base damage in human cells. Nucleic Acids Res. 1996;24:1389–1394. doi: 10.1093/nar/24.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Olinski R, Zastawny T, Budzbon J, Skokowski J, Zegarski W, Dizdaroglu M. DNA base modifications in chromatin of human cancerous tissues. FEBS Lett. 1992;309:193–198. doi: 10.1016/0014-5793(92)81093-2. [DOI] [PubMed] [Google Scholar]
- 50.Hatahet Z, Purmal AA, Wallace SS. A novel method for site specific introduction of single model oxidative DNA lesions into oligodeoxyribonucleotides. Nucleic Acids Res. 1993;21:1563–1568. doi: 10.1093/nar/21.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng X, Kelso C, Hornak V, Santos C, Grollman AP, Simmerling C. Dynamic behavior of DNA base pairs containing 8-oxoguanine. J Am Chem Soc. 2005;127:13906– 13918. doi: 10.1021/ja052542s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shibutani S, Bodepudi V, Johnson F, Grollman AP. Translesional synthesis on DNA templates containing 8-oxo-7,8-dihydrodeoxyadenosine. Biochemistry. 1993;32:4615–4621. doi: 10.1021/bi00068a019. [DOI] [PubMed] [Google Scholar]
- 53.Kamiya H, Miura H, Murata-Kamiya N, Ishikawa H, Sakaguchi T, Inoue H, Sasaki T, Masutani C, Hanaoka F, Nishimura S, Ohtsuka E. 8-Hydroxyadenine (7,8- dihydro-8-oxoadenine) induces misincorporation in in vitro DNA synthesis and mutations in NIH 3T3 cells. Nucleic Acids Res. 1995;23:2893–2899. doi: 10.1093/nar/23.15.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barone F, Cellai L, Giordano C, Sala GL, Mazzei F. Influence of an 8- oxoadenine lesion on the structural and dynamic features of a 30-mer DNA fragment with and without a mismatch. Int J Radiat Biol. 2002;78:9–16. doi: 10.1080/09553000110087353. [DOI] [PubMed] [Google Scholar]
- 55.Cho BP, Evans FE. Structure of oxidatively damaged nucleic acid adducts. 3 Tautomerism, ionization and protonation of 8-hydroxyadenosine studied by 15N NMR spectroscopy. Nucleic Acids Res. 1991;19:1041–1047. doi: 10.1093/nar/19.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yanagawa H, Ogawa Y, Ueno M. Redox ribonucleosides. J Biol Chem. 1992;267:13320–13326. [PubMed] [Google Scholar]
- 57.Steenken S, Jovanovic SV, Bietti M, Bernhard K. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2′-deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J Am Chem Soc. 2000;122:2372–2374. [Google Scholar]
- 58.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo- 7,8-dihydroguanosine. Org Lett. 2000;2:613–617. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 59.Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin is the major product of 8-oxo-7,8-dihydroguanosine reaction with peroxynitrite in the presence of thiols, and guanosine photooxidation by methylene blue. Org Lett. 2001;3:763–766. [PubMed] [Google Scholar]
- 60.Adam W, Arnold MA, Nau WM, Pischel U, Saha-Moeller CR. J Am Chem Soc. 2002;124:3893–3904. doi: 10.1021/ja017600y. [DOI] [PubMed] [Google Scholar]
- 61.Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem Res Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu H, Venkatarangan L, Wishnok JS, Tannenbaum SR. Quantitation of four guanine oxidation products from reaction of DNA with varying doses of peroxynitrite. Chem Res Toxicol. 2005;18:1849–1857. doi: 10.1021/tx050146h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;40:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 64.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 65.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8- oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 66.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-Oxoguanine in DNA induces targeted GC->TA transversion in simian kidney cells. Proc Natl Acad Sci USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Page F, Margot A, Grollman AP, Sarasin A, Gentile A. Mutagenicity of a unique 8-oxoguanine in a human Ha-ras sequence in mammalian cells. Carcinogenesis. 1995;16:2779– 2784. doi: 10.1093/carcin/16.11.2779. [DOI] [PubMed] [Google Scholar]
- 68.Cherepanov A, Vries S. Kinetics and thermodynamics of nick sealing by T4 DNA ligase. Eur J Biochem. 2003;270:4315–4325. doi: 10.1046/j.1432-1033.2003.03824.x. [DOI] [PubMed] [Google Scholar]
- 69.Oka N, Greenberg MM. The effect of the 2-amino group of 7,8-dihydro-8-oxo-2′-deoxyguanosine on translesion synthesis and duplex stability. Nucleic Acids Res. 2005;33:1637–1643. doi: 10.1093/nar/gki305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 71.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. Formation of 13C-, 15N- and 18O-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J Am Chem Soc. 2003;125:13926– 13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 72.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8- oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 73.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo- 7,8-dihydroguanosine in a nucleoside model. Chem Res Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 74.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 75.Chmiel NH, Golinelli MP, Francis AW, David SS. Efficient recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain. Nucleic Acids Res. 2001;29:553–564. doi: 10.1093/nar/29.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wood ML, Esteve A, Morningstar ML, Kuziembo GM, Essigman JM. Genetic effects of oxidative DNA damage: Comparative mutagenesis of 7,8-dihydro-8- oxoguanine and 7,8 -dihydro-8-oxoadenine in Escherichia coli. Nucleic Acids Res. 1992;20:6023– 6032. doi: 10.1093/nar/20.22.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guschlbauer W, Duplaa AM, Guy A, Teoule R, Fazakerley GV. Structure and in vitro replication of DNA templates containing 7,8-dihydro-8-oxoadenine. Nucleic Acids Res. 1991;19:1753–1758. doi: 10.1093/nar/19.8.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parsons JL, Dianova II, Dianov GL. APE1-dependent repair of DNA singlestrand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res. 2005;33:2204–2209. doi: 10.1093/nar/gki518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu P, Burdzy A, Sowers LC. DNA ligases ensure fidelity by interrogating minor groove contacts. Nucleic Acids Res. 2004;32:4503–4511. doi: 10.1093/nar/gkh781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and ac cumulate the oxidized guanine lesion spiroiminodihydantoin. Chem Res Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kornyushyna O, Burrows CJ. Effect of the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin on proofreading by Escherichia coli DNA polymerase I (Klenow fragment) in different sequence contexts. Biochemistry. 2003;42:13008–13018. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION AVAILABLE
HPLC analysis of crude and purified OG, Sp and Gh-containing oligomers, PAGE analysis of OI and OA-containing oligomers, and comparison of ligation of oligomers with lesions on the 5′ terminus of primer 1. This material is available free of charge via the Internet at http://pubs.acs.org.