Abstract
Bioassay-guided fractionation of an EtOH extract obtained from the roots of the Madagascan plant Albizia gummifera led to the isolation of three new cytotoxic oleanane-type triterpenoid saponins, gummiferaosides A-C (1-3). The structures of these new compounds were elucidated using 1D and 2D NMR experiments and mass spectrometry. Compounds 1-3 showed cytotoxicity against the A2780 human ovarian cancer cell line with IC50 values of 0.8, 1.5, and 0.6 μg/mL, respectively.
In our continuing search for bioactive molecules from the Madagascar rainforests as part of an International Cooperative Biodiversity Group (ICBG) program,1 we obtained an extract of the roots of Albizia gummifera (J. F. Gmel.) C. A. Sm. var. gummifera (Fabaceae). This extract, designated MG 1012, showed reproducible cytotoxicity to the A2780 ovarian cancer cell line, with an IC50 value of 7.2 μg/mL. The extract was selected for bioassay-guided fractionation based on this activity.
The genus Albizia comprises about 150 species widely distributed in the tropics, with the greatest diversity in Africa and South America.2 Alkaloids,3 flavonoids,4 sterols,5 and triterpenoid saponins6-11 have been isolated from Albizia species, and Albizia gummifera in particular has been studied for its alkaloids3a,3c and triterpenoids.5,9 It has been reported that alkaloids from Albizia adinocephala inhibit plasmepsin II, an aspartyl proteinase crucial to the survival of the malaria parasite.3c Albiziasaponin B from Albizia myriophylla was found to show a potent sweetness intensity relative to sucrose.6b Some triterponoid saponins from Albizia species exhibited in vitro cytotoxicity against various cancer cell lines.2,6a,7,8
In this paper, we report the isolation, structure elucidation, and cytotoxicity of three new bioactive triterpenoid saponins (1-3) obtained from the roots of Albizia gummifera.
Results and Discussion
Liquid-liquid partitioning of a portion of an EtOH extract of the roots of Albizia gummifera into hexane, CH2Cl2 and aqueous MeOH fractions indicated that the aqueous MeOH fraction (1 g) was the most active fraction, with an IC50 value of < 6.25 μg/mL (the lowest concentration tested). Purification of the aqueous MeOH fraction using a C18 open column, followed by preparative HPLC on a phenyl bonded column, and final purification by HPLC on an analytical size C8 bonded column, led to the isolation of compounds 1-3.
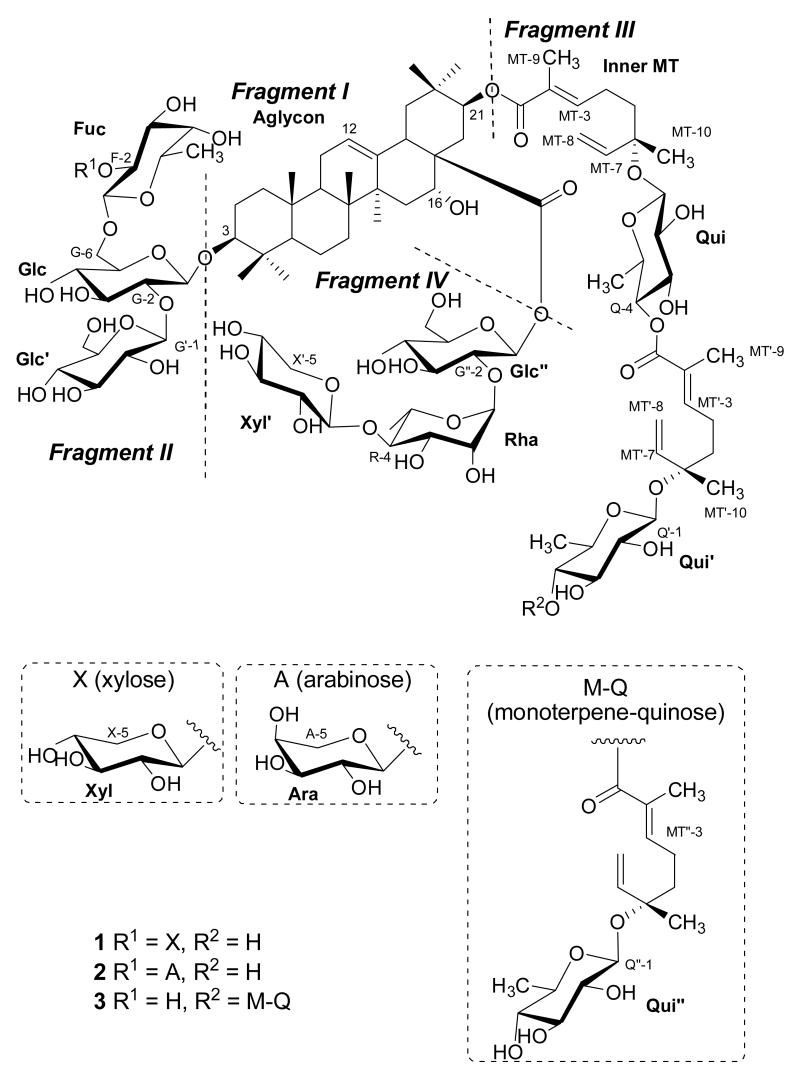
Compound 1 was obtained as a white solid. Its HRFABMS (positive-ion mode) exhibited a quasimolecular ion peak at m/z 2177.9998, consistent with a molecular composition of C102H162O48Na (calcd for C102H162O48Na+, 2178.0128).12 The aglycon of 1 (fragment I) was identified as acacic acid by analysis of 1H and 13C NMR spectra (Tables 1 and 2) and from observation of connectivities in the COSY, TOCSY, ROESY, HSQC, and HMBC NMR spectra. The NMR spectra indicated the presence of one trisubstituted double bond (12-position) and three oxygenated methines (3-, 16-, and 21-positions) in the oleanane-type aglycon of 1. Out of the seven methyl groups in the aglycon, only H3-27 (δH 1.42, s) exhibited a 3J HMBC correlation to C-13 (δC 143.7), which confirmed the location of the double bond. In the HMBC spectrum of 1, H3-23 (δH 1.09, s) and H3-24 (δH 0.86, s) showed correlations to C-3 (δC 90.3). The H2-22 (δH 1.67 and δH 2.07) signals correlated to both C-16 (δC 74.1) and C-21 (δC 78.6), while H2-22, H3-29 (δH 0.85, s), and H3-30 (δH 1.03, s) exhibited 2J, 3J, and 3J HMBC correlations to C-21, respectively. ROESY correlations between H3-23 and H-3 (δH 3.33), and H3-25 (δH 0.96, s) and H3-24 revealed that H-3 has an α-axial orientation. In turn, H-16 (δH 4.45, dd, J = 5.0 and 5.0 Hz) of 1 was assigned as a β-equatorial proton from its coupling constants. In corroboration of this assignment, H-16α of the 16β-oxygenated triterpenoid gymnemagenin is a doublet of doublets with coupling constants of 11.5 and 5.0 Hz, respectively. 13 The orientation of H-21 (δH 5.43, dd, J = 10.8 and 5.5 Hz) was determined as α-axial, which was confirmed by a ROESY correlation between H3-29 and H-21. Further, the NMR data of fragment I of 1 were in full agreement with those reported in the literature for acacic acid, supporting an acacic acid aglycon.10,14
Table 1.
| position | 1 | 2 | 3 | position | 1 | 2 | 3 |
|---|---|---|---|---|---|---|---|
| 1 | 1.62 m | 1.62 m | 1.62 m | MT′-4 | 2.30 m | 2.30 m | 2.30 m |
| 2 | 1.85 m | 1.85 m | 1.85 m | -5 | 1.70 m | 1.70 m | 1.70 m |
| 1.70 m | 1.70 m | 1.70 m | |||||
| 3 | 3.33 m | 3.33 m | 3.33 m | -7 | 5.93 dd (17.7, 11.0) | 5.92 dd (17.7, 11.0) | 5.92 dd (17.7, 11.0) |
| 5 | 0.78 br d (10.0) | 0.78 m | 0.78 m | -8 | 5.27 dd (17.7, 1.2) | 5.27 dd (17.7, 1.2) | 5.27 dd (17.7, 1.2) |
| 5.20 dd (11.0, 1.2) | 5.20 dd (11.0, 1.2) | 5.21 m | |||||
| 6 | 1.50 m | 1.50 m | 1.52 m | -9 | 1.89 s | 1.82 s | 1.82 s |
| 1.28 m | 1.28 m | 1.38 m | |||||
| 7 | 1.36 m | 1.38 m | 1.38 m | -10 | 1.38 s | 1.38 s | 1.38 s |
| 9 | 1.68 m | 1.66 m | 1.66 m | MT″-3 | 6.73 t (7.3) | ||
| 11 | 1.92 m | 1.90 m | 1.90 m | -4 | 2.30 m | ||
| 12 | 5.34 br s | 5.34 br s | 5.35 br s | -5 | 1.70 m | ||
| 15 | 1.50 m | 1.45; 1.55 | 1.50 m | -7 | 5.73 dd (17.7, 11.0) | ||
| 16 | 4.45 dd (5.0, 5.0) | 4.48 br s | 4.48 br s | -8 | 5.21 m | ||
| 18 | 2.96 dd (10.5, 5.5) | 2.97 m | 2.97 m | -9 | 1.79 s | ||
| 19 | 2.50 dd (12.0, 10.5) | 2.50 dd (14.0, 14.0) | 2.50 dd (14.0, 14.0) | -10 | 1.33 s | ||
| 1.18 dd (12, 5.5) | 1.17 dd (13.1, .5.0) | 1.17 dd (13.1, 5.0) | |||||
| 21 | 5.43 dd (10.8, 5.5) | 5.44 dd (11, 5.5) | 5.43 dd (11, 5.5) | Q-1 | 4.42 d (7.6) | 4.41 d (7.8) | 4.42 d (8.0) |
| 22 | 2.07 dd (12.0, 5.5) | 2.10 dd (13.8, 5.5) | 2.10 dd (13.8, 5.5) | -6 | 1.10 d (6.1) | 1.09 d (6.0) | 1.09 d (6.2) |
| 1.67 m | 1.67 m | 1.67 m | |||||
| 23 | 1.09 s | 1.09 s | 1.09 s | Q′-1 | 4.35 d (7.6) | 4.34 d (7.8) | 4.54 d (8.0) |
| 24 | 0.86 s | 0.85 s | 0.85 s | -6 | 1.23 d (6.1) | 1.22 d (6.2) | 1.23 d (6.2) |
| 25 | 0.96 s | 0.96 s | 0.96 s | Q″-1 | 4.35 d (7.8) | ||
| 26 | 0.75s | 0.76s | 0.76s | -6 | 1.26 d (6.2) | ||
| 27 | 1.42 s | 1.41 s | 1.40 s | G-1 | 4.47 m | 4.43 d (7.6) | 4.43 d (7.6) |
| 29 | 0.85 s | 0.85 s | 0.85 s | G′-1 | 4.66 d (7.8) | 4.66 d (7.7) | 4.66 d (7.8) |
| 30 | 1.03 s | 1.03 s | 1.03 s | F-1 | 4.47 m | 4.46 d (7.1) | 4.32 d (7.1) |
| MT-3 | 6.75 t (7.3) | 6.75 t (7.3) | 6.75 t (7.3) | -6 | 1.26 d (6.4) | 1.26 d (6.2) | 1.26 d (6.2) |
| -4 | 2.30 m | 2.30 m | 2.30 m | X-1 | 4.47 m | ||
| -5 | 1.70 m | 1.70 m | 1.70 m | A-1 | 4.55 d (6.4) | ||
| -7 | 5.95 dd (17.7, 11.0) | 5.93 dd (17.7, 11.0) | 5.93 dd (17.7, 11.0) | G″-1 | 5.32 d (7.8) | 5.32 d (7.8) | 5.32 d (7.8) |
| -8 | 5.29 dd (17.7, 0.8) | 5.29 dd (17.7, 1.2) | 5.29 dd (17.7, 1.2) | R-1 | 5.41 d (1.4) | 5.40 d (1.5) | 5.40 d (1.5) |
| 5.21 dd (11.0, 0.8) | 5.21 dd (11.0, 1.2) | 5.21 m | |||||
| -9 | 1.83 s | 1.82 s | 1.84 s | -6 | 1.31 d (6.4) | 1.30 d (6.2) | 1.30 d (6.2) |
| -10 | 1.36 s | 1.36 s | 1.37 s | X′-1 | 4.47 m | 4.50 d (7.7) | 4.50 d (7.6) |
| MT′-3 | 6.80 t (7.3) | 6.80 t (7.3) | 6.83 t (7.3) |
δ (ppm) 500 MHz; multiplicities; J values (Hz) in parentheses.
In CD3OD.
Table 2.
| carbon | 1c | 1d | 2c | 3c | carbon | 1c | 1d | 2c | 3c |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 39.9 | 39.9 | 40.2 | 40.1 | Q-1 | 99.3 | 99.6 | 99.5 | 99.4 |
| 2 | 27.4 | 27.1 | 27.6 | 27.4 | -2 | 76.4 | 75.8 | 76.6 | 76.5 |
| 3 | 90.3 | 89.0 | 91.2 | 91.6 | -3 | 76.4 | 75.8 | 76.4 | 76.7 |
| 4 | 40.6 | 40.3 | 40.7 | 40.6 | -4 | 77.9 | 77.5 | 78.1 | 78.0 |
| 5 | 57.1 | 56.4 | 57.2 | 57.3 | -5 | 70.9 | 70.5 | 71.1 | 71.3 |
| 6 | 18.3 | 18.7 | 18.5 | 18.5 | -6 | 18.3 | 19.1 | 18.4 | 18.4 |
| 7 | 34.3 | 33.7 | 34.5 | 34.5 | Q′-1 | 99.3 | 99.6 | 99.5 | 97.6 |
| 8 | 40.8 | 40.8 | 41.0 | 41.0 | -2 | 75.5 | 75.7 | 75.8 | 76.1 |
| 9 | 48.0 | 47.4 | 48.3 | 48.0 | -3 | 78.3 | 78.5 | 78.5 | 76.3 |
| 10 | 37.9 | 37.3 | 38.1 | 38.1 | -4 | 77.3 | 77.0 | 77 | 78.1 |
| 11 | 24.6 | 24.0 | 24.8 | 24.7 | -5 | 72.6 | 73.3 | 73.3 | 73.3 |
| 12 | 124.0 | 124.0 | 124.3 | 124.3 | -6 | 18.2 | 19.1 | 18.4 | 18.4 |
| 13 | 143.7 | 144.3 | 143.9 | 143.3 | Q″-1 | 99.4 | |||
| 14 | 42.6 | 42.3 | 42.8 | 42.8 | -2 | 75.8 | |||
| 15 | 35.9 | 36.2 | 36.1 | 36.1 | -3 | 78.5 | |||
| 16 | 74.1 | 73.8 | 74.3 | 74.3 | -4 | 77.5 | |||
| 17 | 52.3 | 52.0 | 52.5 | 52.5 | -5 | 73.0 | |||
| 18 | 41.5 | 41.3 | 41.8 | 41.9 | -6 | 18.4 | |||
| 19 | 48.0 | 48.0 | 48.3 | 48.0 | G-1 | 104.6 | 105.3 | 105.5 | 105.5 |
| 20 | 36.3 | 35.6 | 36.6 | 36.5 | -2 | 81.2 | 83.2 | 81.2 | 81.3 |
| 21 | 78.6 | 77.4 | 78.8 | 78.7 | -3 | 78.2 | 78.5 | 78.7 | 78.6 |
| 22 | 36.1 | 36.7 | 36.4 | 36.3 | -4 | 71.8 | 71.8 | 72.0 | 72.0 |
| 23 | 28.4 | 28.5 | 28.6 | 28.7 | -5 | 77.5 | 77.0 | 78.1 | 78.1 |
| 24 | 16.9 | 16.1 | 17.2 | 17.1 | -6 | 69.8 | 70.2 | 69.4 | 69.0 |
| 25 | 16.2 | 17.2 | 16.4 | 16.3 | G′-1 | 105.2 | 106.3 | 104.7 | 105.3 |
| 26 | 17.6 | 17.5 | 17.9 | 17.8 | -2 | 76.2 | 76.1 | 75.9 | 75.8 |
| 27 | 27.4 | 27.5 | 27.5 | 27.4 | -3 | 77.9 | 77.5 | 77.5 | 77.3 |
| 28 | 175.4 | 175.0 | 175.6 | 175.6 | -4 | 71.9 | 72.3 | 72.1 | 72.1 |
| 29 | 29.4 | 29.5 | 29.6 | 29.5 | -5 | 78.3 | 78.5 | 78.4 | 78.3 |
| 30 | 19.4 | 19.5 | 19.6 | 19.5 | -6 | 63.1 | 62.8 | 63.4 | 63.3 |
| MT-1 | 169.1 | 168.1 | 169.3 | 169.2 | F-1 | 104.0 | 103.7 | 104.1 | 104.7 |
| -2 | 129.1 | 128.9 | 129.3 | 129.2 | -2 | 82.4 | 82.6 | 81.2 | 72.4 |
| -3 | 144.0 | 142.5 | 144.2 | 144.1 | -3 | 75.4 | 75.4 | 75.7 | 75.6 |
| -4 | 24.4 | 23.9 | 24.7 | 24.5 | -4 | 72.9 | 72.9 | 72.5 | 72.5 |
| -5 | 41.0 | 40.7 | 41.2 | 41.3 | -5 | 71.7 | 71.6 | 71.4 | 72.1 |
| -6 | 81.0 | 80.1 | 81.3 | 81.2 | -6 | 16.7 | 17.7 | 16.9 | 16.9 |
| -7 | 144.1 | 144.4 | 144.3 | 144.2 | X-1 | 107.1 | 107.3 | ||
| -8 | 115.9 | 115.4 | 116.2 | 116.1 | -2 | 75.6 | 76.3 | ||
| -9 | 12.6 | 13.0 | 12.8 | 12.8 | -3 | 77.8 | 78.1 | ||
| -10 | 23.6 | 23.9 | 23.8 | 23.8 | -4 | 71.1 | 71.0 | ||
| MT′-1 | 169.6 | 168.0 | 169.8 | 169.8 | -5 | 67.3 | 67.6 | ||
| -2 | 128.4 | 128.3 | 128.6 | 128.8 | A-1 | 106.1 | |||
| -3 | 144.1 | 143.7 | 144.2 | 144.2 | -2 | 71.6 | |||
| -4 | 24.5 | 23.9 | 24.6 | 24.6 | -3 | 73.1 | |||
| -5 | 41.0 | 40.7 | 41.2 | 41.3 | -4 | 70.0 | |||
| -6 | 80.9 | 79.8 | 81.1 | 81.1 | -5 | 66.3 | |||
| -7 | 144.7 | 144.4 | 144.9 | 144.8 | G″-1 | 95.2 | 95.5 | 95.4 | 95.4 |
| -8 | 115.9 | 115.2 | 116.1 | 116.1 | -2 | 77.1 | 76.7 | 77.3 | 77.2 |
| -9 | 12.6 | 13.0 | 12.8 | 12.8 | -3 | 79.5 | 79.4 | 79.6 | 79.6 |
| -10 | 23.6 | 24.0 | 23.8 | 23.8 | -4 | 71.4 | 71.4 | 71.3 | 71.3 |
| MT″-1 | 168.8 | -5 | 79.5 | 79.4 | 79.8 | 79.6 | |||
| -2 | 128.8 | -6 | 62.2 | 62.1 | 62.4 | 62.3 | |||
| -3 | 143.8 | R-1 | 101.1 | 101.6 | 101.3 | 101.3 | |||
| -4 | 24.3 | -2 | 72.2 | 72.7 | 72.0 | 72.1 | |||
| -5 | 41.2 | -3 | 72.9 | 72.9 | 73.0 | 73.2 | |||
| -6 | 81.0 | -4 | 84.0 | 83.4 | 84.2 | 84.0 | |||
| -7 | 144.5 | -5 | 68.8 | 68.8 | 69.0 | 69.0 | |||
| -8 | 116.5 | -6 | 18.3 | 19.0 | 18.5 | 18.3 | |||
| -9 | 12.8 | X′-1 | 106.9 | 106.7 | 107.1 | 107.0 | |||
| -10 | 24.2 | -2 | 77.1 | 76.5 | 77 | 77.0 | |||
| -3 | 78.4 | 78.7 | 78.6 | 78.5 | |||||
| -4 | 71.1 | 71.2 | 71.1 | 71.0 | |||||
| -5 | 67.3 | 67.5 | 67.5 | 67.5 |
δ (ppm) 125 MHz.
The signals of the sugar carbons were assigned by HSQC-TOCSY and 13C NMR.
In CD3OD.
In C5D5N with three drops of CD3OD.
Analysis of the 1H, 13C NMR, and HSQC spectra of 1 indicated the presence of nine sugar units and additional unsaturated ester units in three fragments designated II, III, and IV. 1D TOCSY spectra were initially obtained in CD3OD to elucidate the structures of these fragments, but overlapping signals in the 1H NMR spectrum prevented complete spectroscopic interpretation. The use of C5D5N containing three drops of CD3OD as the NMR solvent was found to reduce the overlap problem, and the following discussion is based on the NMR data collected in this mixed solvent system (see Experimental Section for 1H NMR data, and Table 2 for 13C NMR data).
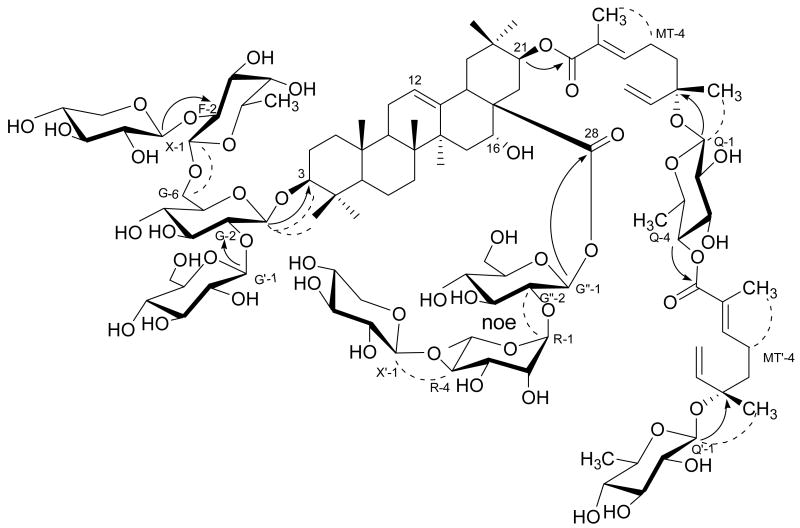
Protons H-21, H-MT-3 (δH 6.87, t, J = 7.3 Hz) and H-MT-9 (δH 1.88, s) of 1 all showed HMBC correlations to C-MT-1 (δC 168.1) (Figure 1), indicating that C-21 is esterified. The HMBC correlations between H-MT-10 (δH 1.54, s) and C-MT-5/C-MT-6/C-MT-7 (δC 40.7/80.1/144.4), and the COSY correlations between H2-MT-4 (δH 2.40, m) and H-MT-3/H2-MT-5 (δH 6.87, t, J = 7.3 Hz/1.74, m), and between H-MT-7 (δH 6.21, m) and H2-MT-8 (δH 5.44, br d, J = 15.8 Hz; δH 5.27, br d, J = 11.0 Hz) indicated that the ester unit is a 6-O-2,6-dimethylocta-2,7-dienoyl monoterpenoid moiety.10 The trisubstituted double bond in the inner monoterpenyl unit was assigned the E configuration, as evidenced by a ROESY correlation between H3-MT-9 and H2-MT-4.
Figure 1.
Key HMBC (arrows) and ROESY (dashed) correlations for compound 1
The H-Qui-1 proton (δH 4.84, d, J = 8 Hz) showed a 3J HMBC correlation to C-MT-6 (δC 80.1), establishing the connectivity from the 6-position of the inner monoterpenyl moiety to the anomeric position of the quinovopyranose unit. The spin system from the anomeric proton to the other protons of the inner quinovopyranose was clearly exhibited in a 1D TOCSY spectrum [H-Qui-1 (selected): δH 4.84, d, J = 8.0 Hz; H-Qui-2: δH 3.99, d, J = 8.0, 8.8 Hz; H-Qui-3: δH 4.17, dd, J = 8.8, 8.8 Hz; H-Qui-4: δH 5.32, dd, J = 8.8, 9.2 Hz; H-Qui-5: δH 3.66, m; H-Qui-6: δH 1.34, d, J = 6.0 Hz] and the HSQC-TOCSY spectrum (correlations from H-Qui-1 to C-Qui-1–6: C-Qui-1, δC 99.6; C-Qui-2, δC 75.8; C-Qui-3, δC 75.8; C-Qui-4, δC 77.5; C-Qui-5, δC 70.5; C-Qui-6, δC 19.1) of 1.
The outer monoterpenoid moiety (MT′) and outer quinovopyranosyl unit (Q′) of 1 were determined of being identical to the corresponding inner ones by the same methods. The HMBC correlations from H-Qui-4 to C-MT′-1, and H-Qui′-1 to C-MT′-6 established the connectivities from the 4-position of the inner quinovopyranose to the 1-position of the outer monoterpenyl moiety, and from the 6-position of the outer monoterpenyl moiety to the anomeric position of the outer quinovopyranosyl unit. H-Qui-1 and H-Qui′-1 showed ROESY correlations to H-MT-10 and H-MT′-10, respectively. The 13C NMR chemical shifts of C-MT-5/C-MT′-5 and C-MT-10/C-MT′-10 of compound 1 were nearly identical to those of C-MT-5 (δC 41.3) and C-MT-10 (δC 23.8) of the related compound kinmoonoside B, which has the S configuration at the C-MT-6 and C-MT′-6 position.14a In contrast, these chemical shifts were different from those of C-MT-5 (δC 39.5) and C-MT-10 (δC 24.5) of kinmoonoside A, which has the R configuration at the C-MT-6 and C-MT′-6 position.14a These facts indicated that compound 1 has the S stereochemistry at the 6-positions of the monoterpenoid moieties.14a These observations were used to establish the structure of fragment III.10
Starting from the anomeric and/or the sixth protons of each of the other seven sugar units, all the protons within each spin system of 1 were assigned using COSY NMR spectra with the aid of ROESY, 1D and 2D TOCSY spectra. The 13C NMR resonances of each of these seven sugar units were assigned by HSQC-TOCSY, HSQC, and HMBC spectra. One of the seven sugars was found to be a β-fucopyranosyl unit, as indicated by the presence of a methyl group at δH 1.48 (H3-F-6, d, J = 6.0 Hz). The coupling constants of H-Fuc-1 (δH 4.94, d, J = 7.8 Hz), H-Fuc-2 (δH 4.40, dd, J = 7.8, 9.6 Hz) and H-Fuc-3 (δH 4.11, dd, J = 4.0, 9.6 Hz) indicated axial positions for these three protons. The proton signal of H-Fuc-4 (δH 4.00) was broad, indicating an α-equatorial orientation. An α-axial position for H-Fuc-5 (δH 3.75, m) was required by the ROESY correlations between H-Fuc-1 and H-Fuc-3/H-Fuc-5. The carbon signal at δC 82.6 assigned to C-Fuc-2 suggested that it was a glycosidic linkage site for another sugar.15 The second of the seven sugars was identified as an α-rhamnopyranosyl unit. The proton signals of both H-Rha-1 (δH 6.36) and H-Rha-2 (δH 4.78) had small coupling constants, suggesting that both were equatorial. The coupling patterns of H-Rha-3 (δH 4.72, dd, J = 3.6, 8.8 Hz), H-Rha-4 (δH 4.43, dd, J = 8.8, 9.2 Hz), H-Rha-5 (δH 4.54, m), and H-Rha-6 (δH 1.74, d, J = 6.5 Hz) indicated a rhamnopyranosyl unit. The downfield chemical shift of C-Rha-4 (δC 83.4) indicated its connectivity to another anomeric position.16 Two of the seven sugars were identified as β-xylopyranosyl units, as evidenced by their proton and carbon chemical shifts [Xyl-1–5 (δH/δC): 5.03/107.3, 4.05/76.3, 4.05/78.1, 4.05/71.0, 3.58 and 4.45/67.6; Xyl′-1–5 (δH/δC): 5.25/106.7, 3.86-4.20/76.5, 3.86-4.20/78.7, 3.86-4.20/71.2, 3.47 and 4.23/67.5 (Experimental Section and Table 2)]. The 13C NMR chemical shifts of these two xylopyranosyl units were in good agreement with literature data.17 The final three sugars were found to be β–glucopyranosyl units. The unit at the 3-position of the aglycon was determined to be a β–glucopyranosyl moiety, the chemical shifts [C-Glc-1–6 (δH/δC): 4.87, d, J = 7.6 Hz/105.3; 4.00, dd, J = 7.6, 9.0 Hz/83.2; 4.12, dd, J = 9.0, 9.0 Hz/78.5; 4.50/71.8; 4.25/77.0; 4.40 and 4.60/70.2 of which were assigned by 1D and 2D NMR spectra (Experimental Section and Table 2)]. The axial orientations of H-Glc-3 and H-Glc-5 were determined by the observation of ROESY correlations between H-Glc-1 and H-Glc-3/H-Glc-5. The significant downfield signals for C-Glc-2 and C-Glc-6 indicated that they attached to two other sugars.18 The coupling patterns of H-Glc′-1 (δH 5.35, d, J = 7.6 Hz) and H-Glc′-2 (δH 4.08, dd, J = 7.6, 8.4 Hz), and the ROESY correlations between H-Glc′-1 and H-Glc′-3/H-Glc′-5 (δH 4.19/3.90, br d, J = 8.4 Hz) indicated a β–glucopyranosyl unit, which was supported by a set of typical carbon chemical shifts for this unit [C-Glc′-1–6 (δC): 106.3, 76.1, 77.5, 72.3, 78.5, 62.8 (Table 2)].17 Similar to the previously described β–glucopyranosyl units, the chemical shifts of a third β–glucopyranosyl unit [Glc″-1–6 (δH/δC): 6.11/95.5, 4.23/76.7, 4.23 or 3.96/79.4, 4.23 or 3.96/71.4, 4.23 or 3.96/79.4, 4.23/62.1 (Experimental Section and Table 2)] matched literature values, especially the 13C NMR chemical shifts.19
The connectivities of these seven glycosidic units in fragments II and IV were determined by analysis of HMBC and ROESY experiments (Figure 1). The anomeric proton of Glc-1 showed ROESY and HMBC correlations to H-3 and C-3, indicating a linkage of this β–glucopyranosyl unit to the 3-position of the aglycon. H-Glc′-1 correlated to C-Glc-2 in the HMBC spectrum, and H-Xyl-1 and H-Fuc-1 showed an HMBC and a ROESY correlation to C-Fuc-2 and H-Glc-6, respectively. The structure of fragment II was thus assigned as shown.10
Both H-18 and H-Glc″-1 exhibited 3J HMBC correlations to C-28 (δC 175.0), suggesting that position-28 is esterified. The interglycosidic correlations of Glc″, Xyl′, and Rha were evident from the ROESY [H-Rha-1 to H-Glc″-2] and HMBC [H-Xyl′-1 to C-Rha-4] cross-peaks. Hence, fragment IV20 and thus the final structure of 1 were determined as shown.
Compound 1 is structurally related to similar complex saponins with monoterpenoid esters at C-21 such as the julibrosides from Albizia julibrissin. Three close analogs are julibrosides I, II and J14,7,10 which differ from 1 in the nature and position of some of the sugar units. Similar compounds were also isolated from other genera of the legume family, for example the avicins from Acacia victoriae Benth.13,14 and the elliptosides from Archidendron ellipticum (Blume) I. C. Nielsen,21 all of which share the same acacic acid aglycon with monoterpenoid glycosides at the 21-position and oligosaccharides at the 3- and 28-positions.
Compound 2 was obtained as a white solid. Comparison of the NMR data (Tables 1 and 2) of 1 and 2 in CD3OD indicated that the xylopyranosyl unit at the 2-position of the fucopyranosyl unit in fragment II of 1 was replaced by an arabinopyranosyl residue in 2, while fragments I, III, and IV of 2 were the same as those of 1. The arabinopyranosyl residue was attached to the 2-position of the fucopyranosyl unit, as deduced from an HMBC correlation between H-Ara-1 (δH 4.55) and C-Fuc-2 (δC 81.2), and the 13C NMR chemical shifts of the arabinopyranosyl residue [C-Ara-1–6 (δC): 106.1, 71.6, 73.1, 70.0, 66.3 (Tables 1 and 2)] were in good agreement with the literature data,11 suggesting that fragment II of 2 is A1-2F1-6G(1-3aglycon)2-1G′. Therefore, the structure of 2 was determined as shown.
Compound 3 was also isolated as a white solid, and its aglycon was shown to be the same as that of 1 by comparison of NMR spectra. The major difference between compounds 1 and 3 was the presence of an extra monoterpenoid moiety in 3, as was evidenced by its mass spectrum and the HSQC spectrum. There were nine sugar units assigned in 3, including three quinovopyranoses, three glucopyranoses, one rhamnopyranose, one fucopyranose, and one xylopyranose. Fragments I and IV of 3 were the same as those of 1, as indicated by a comparison of their NMR spectra. There were only three sugars in fragment II [F1-6G(1-3aglycon)2-1G′]17 of 3 (Tables 1 and 2), as opposed to four in 1, but these three were the same as three of the four sugars in 1, and so this unit was identified by a comparison of NMR spectra. Protons H-Q′-4 and H-Q″-1 showed HMBC correlations to the carbons C-MT″-1 and C-MT″-6, respectively, of the additional monoterpenoid unit, indicating that fragment III is (aglycon)– 1MT6-1Q4-1MT′6-1Q′4-1MT″6-1Q″. The connectivities of these units were determined by the same methodology used in the structure elucidation of 1. The structure of 3 was thus determined as shown.
Some acacic acid-type saponins have been evaluated for their cytotoxicity. It was reported that the monoterpene-quinovopyranosyl moiety at C-21 and the oligosaccharide ester at C-28 of the acacic acid-type aglycon are crucial substituents required for the cytotoxicity of julibroside III and prosapogenins 8-10 against KB cells, and their hydroxyl group at C-16 may also be important for the cytotoxicity.6a Neither monodesmonoterpenyl elliptoside A nor any of the anatoliosides A-E (monoterpene glycosides) produced distinctive cytotoxicity in the NCI 60-cell line screen, which supported the apparent requirement for both the terminal monoterpenoid unit and the acacic acid portion of such active molecules.14, 21 It was reported that the trisaccharide unit at C-3 of kinmoonosides A-C were not crucial for their cytotoxicity,14a but oligosaccharides at 3-positions may intensify the cytotoxicity of acacic acid derivatives. The apoptotic properties of avicins from Acacia victoriae have been studied by Gutterman et al.22 The same group also reported the thioesterification of avicins by a thioester linkage between Cys-199 of OxyR and the outer monoterpene side chain; such derivatization can induce an adaptive response that protects cells against oxidative or nitrosative stress.23 In a cytotoxicity assay using the A2780 human ovarian cancer cell line, compounds 1-3 showed cytotoxicity with IC50 values of 0.8, 1.5, and 0.6 μg/mL, respectively. Compounds 1-3 possess structural features essential for cytotoxicity, similar to the cytotoxic julibrosides, prosapogenins, elliptoside, and avicins, mentioned above.
Experimental Section
General Experimental Procedures
Optical rotations were recorded on a Perkin-Elmer 241 polarimeter. IR and UV spectra were measured on MIDAC M-series FTIR and Shimadzu UV-1201 spectrophotometers, respectively. NMR spectra were obtained on a JEOL Eclipse 500 for 1H, 13C, HMQC, and HMBC and an INOVA 400 spectrometer for TOCSY, COSY, ROESY, and HSQC-TOCSY. Chemical shifts are given in δ (ppm), and coupling constants are reported in Hz. Mass spectra were obtained on a JEOL JMS-HX-110 instrument, in the positive-ion mode. HPLC was performed on a Shimadzu LC-10AT instrument with a semi-preparative C8 Varian Dynamax column (5 μm, 250 × 10 mm) and a preparative phenyl Varian Dynamax column (8 μm, 250 × 21.4 mm).
Cytotoxicity Bioassays
Cytotoxicity measurements were performed at Virginia Polytechnic Institute and State University against the A2780 ovarian cancer cell line, as described previously. The A2780 cell line is a drug-sensitive human ovarian cancer cell line.24
Plant Material
Roots of Albizia gummifera (J.F. Gmel.) C.A. Sm. var. gummifera (Fabaceae) were collected in November 2001 as collection RFA 579. The collection was made by Fidy Ratovoson et al., 3 km northwest of the village of Nosivola. The plant was growing in a dense humid forest adjacent to Zahamena National Park, in Toamasina Province, Madagascar (17. 41 01S; 48. 38. 28E, elevation 900 m). The specimen accessed was a small tree 9 m in height and trunk diameter 14 cm, with pale green sepals, white petals, and 10 dark red stamens. The vernacular name of this species in this area is “volomborona”. Duplicate voucher specimens were deposited at Centre National d'Application des Recherches Pharmaceutiques (CNARP) and the Departement des Recherches Forestieres et Piscicoles Herbarium in Antananarivo, Madagascar (TEF), at Missouri Botanical Garden, St. Louis, Missouri (MO), and the Museum National d'Histoire Naturelle in Paris, France (P).
Extraction and Isolation
Dried roots of A. gummifera (430.9 g) were ground in a hammer mill, then extracted with EtOH by percolation for 24 h at rt to give the crude extract MG 1012 (10.14 g), of which 7.44 g was shipped to Virginia Polytechnic Institute and State University (VPISU) in triplicate vials for distribution to Eisai Research Institute (2.79 g), Dow AgroSciences (2.19 g) and VPISU (2.46 g). Extract MG 1012 (1.49 g, IC50 7.2 μg/mL) was suspended in aqueous MeOH (MeOH-H2O, 9:1, 100 mL) and extracted with hexanes (3 × 100 mL portions). The aqueous layer was then diluted to 70% MeOH with H2O and extracted with CH2Cl2 (3 × 100 mL portions). The aqueous MeOH extract (1 g) was active with an IC50 less than 6.25 μg/mL, while both the hexane and CH2Cl2 extracts were inactive. The aqueous MeOH fraction was chromatographed on an open C18 column (130 × 22 mm) using H2O-MeCN (80:20 to 40:60, then 0:100) to yield the three fractions A [296 mg (polar, inactive)], B [516 mg, IC50 less than 6.25 μg/mL], and C [73 mg, nonpolar, inactive]. Fraction B furnished 15 subfractions after HPLC separation on a phenyl-bonded column (35% MeOH/H2O, 10 mL/min). HPLC of subfraction 4 (28 mg) on a C8 bonded phase column eluted with 70% MeOH/H2O (2 mL/min) yielded compounds 1 (tR 30 min, 6 mg) and 2 (tR 34 min, 3 mg). Compound 3 (tR 37 min, 3 mg) was obtained by HPLC of subfraction 14 (18 mg) also using C8 HPLC (72% MeOH/H2O, 2 mL/min).
Gummiferaoside A (1): white solid; [α]26d -15 (c 0.37, MeOH); UV (MeOH) λmax (log ε) 218 (4.6) nm; IR (film) νmax 3339, 2945, 2833, 1744, 1730, 1432, 1364, 1343, 1304, 1253, 1200, 1153, 1076, 1009, 1025 cm-1; 1H NMR (500 MHz, CD3OD), see Table 1; 13C NMR (125 MHz, CD3OD and C5D5N with three drops of CD3OD), see Table 2; 1H NMR (500 MHz, C5D5N with three drops of CD3OD): 7.02 (1H, t, J = 7.5 Hz, H-MT′-3), 6.87 (1H, t, J = 7.3 Hz, H-MT-3), 6.36 (1H, br s, H-R-1), 6.21 (1H, m, H-MT-7), 6.21 (1H, m, H-MT′-7), 6.18 (1H, m, H-21), 6.11 (1H, d, J = 7.6 Hz, H-G″-1), 5.70 (1H, br s, H-12), 5.44 (1H, br d, J = 15.8 Hz, H-MT-8a), 5.44 (1H, br d, J = 15.8 Hz, H-MT′-8a), 5.35 (1H, d, J = 7.6 Hz, H-G′-1), 5.32 (1H, dd, J = 8.8, 9.2 Hz, H-Q-4), 5.30 (1H, m, H-16), 5.27 (1H, br d, J = 11.0 Hz, H-MT-8b), 5.27 (1H, br d, J = 11.0 Hz, H-MT′-8b), 5.25 (1H, d, J = 7.0 Hz, H-X′-1), 5.03 (1H, d, J = 6.4 Hz, H-X-1), 4.94 (1H, d, J = 7.8 Hz, H-F-1), 4.87 (1H, d, J = 7.6 Hz, H-G-1), 4.84 (1H, d, J = 8.0 Hz, H-Q-1), 4.84 (1H, d, J = 8 Hz, H-Q′-1), 4.78 (1H, br s, H-R-2), 4.72 (1H, dd, J = 3.6, 8.8 Hz, H-R-3), 4.60 (1H, m, H-G-6b), 4.54 (1H, m, H-R-5), 4.50 (1H, m, H-G-4), 4.45 (1H, dd, J = 4.0, 11.4 Hz, H-X-5b), 4.43 (1H, dd, J = 8.8, 9.2 Hz, H-R-4), 4.40 (1H, dd, J = 7.8, 9.6 Hz, H-F-2), 4.40 (1H, m, H-G-6a), 4.29 (1H, m, H-G′-6b), 4.25 (1H, m, H-G-5), 4.23 (1H, m, H-X′-5b), 4.23 (1H, m, H-G″-2), 4.23 or 3.96 (2H, m, H2-G″-6), 4.23 or 3.96 (1H, m, H-G″-3), 4.23 or 3.96 (1H, m, H-G″-4) 4.23 or 3.96 (1H, m, H-G″-5), 4.22 (1H, m, H-G′-6a), 4.20-3.86 (1H, m, H-X′-2), 4.20-3.86 (1H, m, H-X′-3), 4.20-3.86 (1H, m, H-X′-4), 4.19 (1H, m, H-G′-3), 4.19 (1H, m, H-G′-4), 4.17 (1H, dd, J = 8.8, 8.8 Hz, H-Q-3), 4.12 (1H, dd, J = 9.0, 9.0 Hz, H-G-3), 4.11 (1H, dd, J = 4.0, 9.6 Hz, H-F-3), 4.08 (1H, dd, J = 7.6, 8.4 Hz, H-G′-2), 4.07 (1H, m, H-Q′-3), 4.07 (1H, m, H-Q′-4), 4.05 (1H, m, H-X-2), 4.05 (1H, m, H-X-3), 4.05 (1H, m, H-X-4), 4.00 (1H, dd, J = 7.6, 9.0 Hz, H-G-2), 4.00 (1H, br s, H-F-4), 3.99 (1H, dd, J = 8.0, 8.8 Hz, H-Q-2), 3.95 (1H, dd, J = 8.0, 8.8 Hz, H-Q′-2), 3.90 (1H, br d, J = 8.4 Hz, H-G′-5), 3.75 (1H, m, H-F-5), 3.66 (1H, m, H-Q-5), 3.66 (1H, m, H-Q′-5), 3.58 (1H, dd, J = 10.0, 11.4 Hz, H-X-5a), 3.47 (1H, dd, J = 9.6, 11.2 Hz, H-X′-5a), 3.45 (1H, m, H-3), 3.45 (1H, m, H-18), 2.91 (1H, dd, J = 14.0, 13.1 Hz, H-19a), 2.82 (1H, dd, J = 14.2, 3.9 Hz, H-22a), 2.40 (2H, m, H2-MT-4), 2.40 (2H, m, H2-MT′-4), 2.26 (1H, m, H-2a), 2.20 (1H, m, H-22b), 2.07 (1H, m, H-11a), 2.04 (2H, m, H2-15), 1.91 (1H, m, H-2b), 1.88 (3H, s, H3-MT-9), 1.86 (1H, m, H-9), 1.82 (3H, s, H3-MT′-9), 1.80 (3H, s, H3-27), 1.74 (2H, m, H2-MT-5), 1.74 (2H, m, H2-MT′-5), 1.74 (3H, d, J = 6.5 Hz, H3-R-6), 1.63 (1H, m, H-1), 1.61 (1H, m, H-11b), 1.60 (1H, m, H-6a), 1.58 (2H, m, H2-7), 1.57 (1H, d, J = 6.0 Hz, H3-Q′-6), 1.54 (3H, s, H3-MT-10), 1.54 (3H, s, H3-MT′-10), 1.48 (3H, d J = 6.0 Hz, H-F-6), 1.41 (1H, m, H-19b), 1.34 (1H, d, J = 6 Hz, H3-Q-6), 1.30 (1H, m, H-6b), 1.28 (3H, s, H3-23), 1.09 (3H, s, H3-24), 1.07 (3H, s, H3-30), 1.05 (3H, s, H3-25), 0.98 (3H, s, H3-29), 0.88 (3H, s, H3-26), 0.85 (1H, br d, J = 10.0 Hz, H-5); HRFABMS m/z 2177.9998 (calcd for C102H162O48Na, 2178.0128).
Gummiferaoside B (2): white solid; [α]26d -11 (c 0.18, MeOH); UV (MeOH) λmax (log ε) 218 (4.6) nm; IR (film) νmax 3344, 2931, 1678, 1439, 1360, 1309, 1280, 1246, 1201, 1181, 1134, 1063, 998 cm-1; 1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD), see Tables 1 and 2; HRFABMS m/z 2178.0071 (calcd for C102H162O48Na, 2178.0128).
Gummiferaoside C (3): white solid; [α]26d -24 (c 0.28, MeOH); UV (MeOH) λmax (log ε) 218 (4.7) nm; IR (film) νmax 3328, 2943, 2833, 1745, 1732, 1598, 1431, 1364, 1342, 1304, 1252, 1195, 1152, 1067, 1022, 1007 cm-1; 1H NMR (500 MHz, CD3OD) and 13C NMR (125 MHz, CD3OD), see Tables 1 and 2; HRFABMS m/z 2358.1262 (calcd for C113H178O50Na, 2358.1278).
Supplementary Material
Acknowledgments
This International Cooperative Biodiversity Group project was supported by the Fogarty International Center, the National Cancer Institute, the National Science Foundation, the National Heart, Lung and Blood Institute, the National Institute of Mental Health, the Office of Dietary Supplements, and the Office of the Director of NIH, under Cooperative Agreement U01 TW000313 from the National Institutes of Health, and this support is gratefully acknowledged. We thank Mr. B. Bebout for obtaining the mass spectra and Mr. T. Glass for assistance with the NMR spectra. Field work essential for this project was conducted under a collaborative agreement between the Missouri Botanical Garden and the Parc Botanique et Zoologique de Tsimbazaza and a multilateral agreement between the ICBG partners, including the Centre National d'Applications des Recherches Pharmaceutiques. We gratefully acknowledge courtesies extended by the Government of Madagascar (Ministère des Eaux et Forêts).
Footnotes
Dedicated to the late Dr. Kenneth Rinehart of the University of Illinois at Urbana-Champaign for his pioneering work on bioactive natural products.
References and Notes
- 1.Biodiversity Conservation and Drug Discovery in Madagascar, Part 24. For Part 23, see; Williams RB, Norris A, Miller JS, Birkinshaw C, Ratovoson F, Andriantsiferana R, Rasamison VE, Kingston DGI. J Nat Prod. 2007 doi: 10.1021/np0605034. accepted (np0605034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-Kader M, Hoch J, Berger JM, Evans R, Miller JS, Wisse JH, Mamber SW, Dalton JM, Kingston DGI. J Nat Prod. 2001;64:536–539. doi: 10.1021/np000295u. [DOI] [PubMed] [Google Scholar]
- 3.a Rukunga GM, Waterman PG. J Nat Prod. 1996;59:850–853. doi: 10.1021/np960397d. [DOI] [PubMed] [Google Scholar]; b Rukunga GM, Waterman PG. Phytochemistry. 1996;42:1211–1215. [Google Scholar]; c Ovenden SPB, Cao SG, Leong C, Flotow H, Gupta MP, Buss AD, Butler MS. Phytochemistry. 2002;60:175–177. doi: 10.1016/s0031-9422(02)00081-x. [DOI] [PubMed] [Google Scholar]
- 4.Rao YK, Reddy MVB, Rao CV, Gunasekar D, Blond A, Caux C, Bodo B. Chem Pharm Bull. 2002;50:1271–1272. doi: 10.1248/cpb.50.1271. [DOI] [PubMed] [Google Scholar]
- 5.Debella A, Haslinger E, Schmid MG, Bucar F, Michl G, Abebe D, Kunert O. Phytochemistry. 2000;53:885–892. doi: 10.1016/s0031-9422(99)00464-1. [DOI] [PubMed] [Google Scholar]
- 6.a Ikeda T, Fujiwara S, Araki K, Kinjo J, Nohara T, Miyoshi T. J Nat Prod. 1997;60:102–107. doi: 10.1021/np960556t. [DOI] [PubMed] [Google Scholar]; b Yoshikawa M, Morikawa T, Nakano K, Pongpiriyadacha Y, Murakami T, Matsuda H. J Nat Prod. 2002;65:1638–1642. doi: 10.1021/np020220l. [DOI] [PubMed] [Google Scholar]; c Haddad M, Miyamoto T, Laurens V, Lacaille-Dubois MA. J Nat Prod. 2003;66:372–377. doi: 10.1021/np020391q. [DOI] [PubMed] [Google Scholar]; d Krief S, Thoison O, Sevenet T, Wrangham RW, Lavaud C. J Nat Prod. 2005;68:897–903. doi: 10.1021/np049576i. [DOI] [PubMed] [Google Scholar]
- 7.a Haddad M, Laurens V, Lacaille-Dubois MA. Bioorg Med Chem. 2004;12:4725–4734. doi: 10.1016/j.bmc.2004.06.025. [DOI] [PubMed] [Google Scholar]; b Liang H, Tong WY, Zhao YY, Cui JR, Tu GZ. Bioorg Med Chem Lett. 2005;15:4493–4495. doi: 10.1016/j.bmcl.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 8.a Zou K, Zhao Y, Tu G, Cui J, Jia Z, Zhang R. Carbohydr Res. 2000;324:182–188. doi: 10.1016/s0008-6215(99)00294-3. [DOI] [PubMed] [Google Scholar]; b Zou K, Tong WY, Liang H, Cui JR, Tu GZ, Zhao YY, Zhang RY. Carbohydr Res. 2005;340:1329–1334. doi: 10.1016/j.carres.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 9.Rukunga GM, Waterman PG. Fitoterapia. 2001;72:140–145. doi: 10.1016/s0367-326x(00)00276-8. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda T, Fujiwara S, Kinjo J, Nohara T, Ida Y, Shoji J, Shingu T, Isobe R, Kajimoto T. Bull Chem Soc Jpn. 1995;68:3483–3490. [Google Scholar]
- 11.Haddad M, Miyamoto T, Lacaille-Dubois MA. Helv Chim Acta. 2004;87:1228–1238. [Google Scholar]
- 12.The calculator function within ChemDraw was used for this calculation.
- 13.Qiu SX, Gong Y, Cheung HTA. Phytochemistry. 1993;34:1385–1387. [Google Scholar]
- 14.a Tezuka Y, Honda K, Banskota AH, Thet MM, Kadota S. J Nat Prod. 2000;63:1658–1664. doi: 10.1021/np000347f. [DOI] [PubMed] [Google Scholar]; b Jayatilake GS, Freeberg DR, Liu Z, Richheimer SL, Blake ME, Bailey DT, Haridas V, Gutterman JU. J Nat Prod. 2003;66:779–783. doi: 10.1021/np020400v. [DOI] [PubMed] [Google Scholar]
- 15.Cao SG, Guza RC, Wisse JH, Miller JS, Evans R, Kingston DGI. J Nat Prod. 2005;68:487–492. doi: 10.1021/np049629w. [DOI] [PubMed] [Google Scholar]
- 16.Kirmizibekmez H, Calis I, Piacente S, Pizza C. Hel Chim Acta. 2004;87:1172–1179. [Google Scholar]
- 17.Kalinowski HO, Berger S, Braun S. Carbon-13 NMR Spectroscopy. John Wiley & Sons; Chichester, UK: 1988. p. 441. [Google Scholar]
- 18.Yin F, Hu L, Lou F, Pan R. J Nat Prod. 2004;67:942–952. doi: 10.1021/np0499012. [DOI] [PubMed] [Google Scholar]
- 19.Mimaki Y, Harada H, Sakuma C, Haraguchi M, Yui S, Kudo T, Yamazaki M, Sashida Y. Helv Chim Acta. 2004;87:851–865. [Google Scholar]
- 20.Li TZ, Zhang WD, Yang GJ, Liu WY, Chen HS, Shen YH. J Nat Prod. 2006;69:591–594. doi: 10.1021/np050439a. [DOI] [PubMed] [Google Scholar]
- 21.Beutler JA, Kashman Y, Pannell LK, Cardellina JH, II, Alexander MRA, Balaschak MS, Prather TR, Shoemaker RH, Boyd MR. Bioorg Med Chem. 1997;5:1509–1517. doi: 10.1016/s0968-0896(97)00098-9. [DOI] [PubMed] [Google Scholar]
- 22.Haridas V, Higuchi M, Jayatilake GS, Bailey D, Mujoo K, Blake ME, Arntzen CJ, Gutterman JU. Proc Natl Acad Sci USA. 2001;98:5821–5826. doi: 10.1073/pnas.101619098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haridas V, Kim SO, Nishimura G, Hausladen A, Stamler JS, Gutterman JU. Proc Natl Acad Sci USA. 2005;102:10088–10093. doi: 10.1073/pnas.0504430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louie KG, Behrens BC, Kinsella TJ, Hamilton TC, Grotzinger KR, McKoy WM, Winker MA, Ozols RF. Cancer Res. 1985;45:2110–2115. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.