Abstract
Background
A Disintegrin And Metalloprotease (ADAM) 9 has been implicated in tumour progression of various solid tumours, however, little is known about its role in renal cell carcinoma. We evaluated the expression of ADAM9 on protein and transcript level in a clinico-pathologically characterized renal cell cancer cohort.
Methods
108 renal cancer cases were immunostained for ADAM9 on a tissue-micro-array. For 30 additional cases, ADAM9 mRNA of microdissected tumour and normal tissue was analyzed via quantitative RT-PCR. SPSS 14.0 was used to apply crosstables (Fisher's exact test and χ2-test), correlations and univariate as well as multivariate survival analyses.
Results
ADAM9 was significantly up-regulated in renal cancer in comparison to the adjacent normal tissue on mRNA level. On protein level, ADAM9 was significantly associated with higher tumour grade, positive nodal status and distant metastasis. Furthermore, ADAM9 protein expression was significantly associated with shortened patient survival in the univariate analysis.
Conclusion
ADAM9 is strongly expressed in a large proportion of renal cell cancers, concordant with findings in other tumour entities. Additionally, ADAM9 expression is significantly associated with markers of unfavourable prognosis. Whether the demonstrated prognostic value of ADAM9 is independent from other tumour parameters will have to be verified in larger study cohorts.
Background
Renal cell cancer (RCC) is thought to cause 12.890 deaths in 2007 in the USA [1] and accounts for around 2–3% of cancers worldwide [2,3]. It is one of the most lethal urologic malignancies. Nodal and systemic metastasis as well as vascular invasion are important prognostic factors in this tumour entity [4]. New molecular markers are warranted to improve the classification of RCC, to provide further prognostic and predictive information and eventually to allow for an individualized cancer therapy [5-8].
In this study, we focused on ADAM9 (synonyms: MDC9, meltrin-γ), a member of the "A Disintegrin And Metalloprotease" family. Functionally, ADAMs participate in spermatogenesis, cell adhesion, myo- and neurogenesis, inflammation, cell migration and tissue remodelling [9,10]. ADAMs are membrane-anchored cell surface glycoproteins with a protease domain in addition to an adhesion domain. The structure of ADAMs was found to be related to soluble snake venom proteins which induce hemorrhage and basement membrane destruction [11,12]. The interactions of ADAMs with cell surface and extracellular matrix proteins like integrins and syndecans could be of relevance in tumour biology as these processes are vital for tumour progression defined by growth, invasion and metastasis [13-17]. Several ADAMs have been analyzed in various tumour entities and were often found to be differentially expressed, partially conveying prognostic information [18-32]. Several ADAMs have already been shown up-regulated in renal cancer on transcript level, with ADAM8 being associated with shortened survival times and distant metastasis [33,34].
ADAM9 has been proposed to be involved in the ectodomain shedding of membrane-anchored of heparin-binding epidermal growth factor-like growth factor, probably regulated by the binding protein Eve-1 [35-37]. Possible mediating effects on EGFR activity further support the notion of ADAM9 involvement in carcinogenesis and tumour progression [38,35,41]. Moreover, ADAM9 promotes cancer cell invasion by modifying or regulating e-cadherin and several types of integrins [21,42].
We evaluated the ADAM9 expression on protein and transcript level to clarify a diagnostic or prognostic value of ADAM9 in renal cell cancer. We found ADAM9 mRNA up-regulated in RCC and demonstrated a prognostic value of ADAM9 protein expression for overall survival times.
Methods
Patients (RT-PCR)
Thirty matched malignant and non-malignant kidney tissue samples were derived from patients (26 male, four female; mean age 62 years, range: 40 to 92 years) with clear cell (cc) RCC undergoing radical nephrectomy at the Department of Urology, Charité – Universitätsmedizin Berlin, between September 2003 and January 2006.
Cases used for mRNA isolation were different from the cohort used for immunohistochemistry. Thirteen of the 30 ccRCC were pT1 stage, two tumours were pT2, and 15 tumours were pT3. Histological grading: G1 (n = 3), G2 (n = 25) and G3 (n = 2). None of the patients had known nodal or distant metastasis according to preoperative screening (computed tomography of chest, abdomen and pelvis). Samples were collected immediately after surgery in tubes with RNAlater® Stabilization Reagent (Qiagen, Hilden, Germany). Until RNA isolation the tubes were stored at 4°C overnight and then at -80°C until analysis.
Patients (immunohistochemistry)
One-hundred-eight patients (83 men, 25 women) diagnosed for renal cancer at the Institute of Pathology, Charité – Universitätsmedizin Berlin between 2003 and 2005 were enclosed in this study. The study has been approved by the Charité University Ethics Committee under the title 'Retrospektive Untersuchung von Gewebeproben mittels immunhistochemischer Färbung und molekularbiologischer Methoden' ('Retrospective analysis of tissue samples by immunohistochemistry and molecular biological techniques') (EA1/06/2004) on 20 September 2004.
Patient age ranged between 28 and 92 years with a median of 62. Histological diagnosis was established according to the guidelines of the World Health Organization. Cases were selected according to tissue availability and were not stratified for any known preoperative or pathological prognostic factors. Eightysix (79.6%) patients had a clear cell RCC (ccRCC), 17 (15.7%) a papillary RCC and five (4.6%) a chromophobe RCC. Twentythree patients had systemic disease (pM1) at the time of diagnosis. Tumour specific survival times, as annually assessed, was available for all patients. The median follow-up time of all cases was 30 months, ranging from one to 47 months. Twentythree patients died from renal cancer. The pT stages: 54 (50.0%) pT1, 3 (2.8%) pT2, 47 (43.5%) pT3 and 4 (3.7%) pT4. Twelve patients (11.1%) had pathologically confirmed nodal metastases (pN1 = 3, pN2 = 9). Fiftyone (47.2%) patients had no nodal metastases (pN0). Of 45 (41.7%) patients no lymph nodes were histologically examined (pNx). Hemangiosis carcinomatosa (V1/2) was detected in 29 (26.8%) cases. Tumour grades: 11 (10.2%) G1, 75 (69.4%) G2, 18 (16.7%) G3 and 4 (3.7%) G4. The sample cohort for the immunohistochemical study was different from the sample cohort used for mRNA analysis. The grouped clinico-pathological data of the immunohistochemically analysed cases are described in the left columns of Table 1.
Table 1.
Associations (χ2-tests/Fischers exact test) between the protein expression of ADAM9 in renal cell cancer and clinico-pathological parameters (percentages in brackets)
| Total | low ADAM9 | high ADAM9 | p-value | |
| All cases | 108 (100) | 71 (65.7) | 37 (34.3) | |
| Age | 0.225 | |||
| ≤ 62 | 59 (54.6) | 42 (71.2) | 17 (28.8) | |
| > 62 | 49 (45.4) | 29 (59.2) | 20 (40.8) | |
| Histology | < 0.001 | |||
| Clear cell | 86 (79.6) | 65 (75.6) | 21 (24.4) | |
| chromophobe | 5 (4.6) | 2 (40.0) | 3 (60.0) | |
| Papillary | 17 (15.7) | 4 (23.4) | 13 (76.5) | |
| pT-status | 0.104 | |||
| pT1 | 54 (50.0) | 40 (74.1) | 14 (25.9) | |
| pT2/3/4 | 54 (50.0) | 31 (57.4) | 23 (42.6) | |
| pN-status* | 0.022 | |||
| pN0 | 51 (47.2) | 36 (70.6) | 15 (29.4) | |
| pN1 | 12 (11.1) | 4 (33.3) | 8 (66.7) | |
| Grading | 0.002 | |||
| G 1 | 11 (10.2) | 11(100) | 0 (0) | |
| G 2 | 75 (69.4) | 50 (66.7) | 25 (33.3) | |
| G 3/4 | 22 (20.4) | 10 (45.5) | 12 (54.5) | |
| Residual tumour status# | 0.073 | |||
| R0 | 78 (72.2) | 56 (71.8) | 22 (28.2) | |
| R1/2 | 15 (13.9) | 7 (46.7) | 8 (53.3) | |
| Metastasis | 0.014 | |||
| M0 | 85 (78.7) | 61 (71.8) | 24 (28.2) | |
| M1 | 23 (21.3) | 10 (43.5) | 13 (56.5) |
* 45 cases were pNx
# 15 cases were Rx
Tissue micro array construction
A tissue micro array (TMA) was constructed as previously described [43]. Briefly, suitable areas for tissue retrieval were marked on standard haematoxylin/eosin (H&E) sections, punched out of the paraffin block and inserted into a recipient block. The tissue arrayer was purchased from Beecher Instruments (Woodland, USA). The punch diameter was 0.6 mm. The RCC array was constructed to represent 109 cases with two spots from the tumour and two spots representing matching normal tissue from the cortex region of the kidney. In four cases, the normal spots did not represent kidney tissue, leaving 105 cases with matched tumour and normal tissue, plus four cases with tumour only. The whole TMA was accomplished on three paraffin blocks.
RNA Isolation
Total RNA was isolated from 50 mg RNAlater™ stabilised kidney tissue samples using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Additionally, we introduced a DNase I (Qiagen) digestion step on the silica gel membrane of the spin column where the RNA was bound, washed and eluated. RNA was extracted with 30 μl RNase-free water and the RNA content was measured with the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, USA). The RNA integrity was validated with the RNA 6000 Nano LabChip® kit on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The Agilent 2100 Expert software generates so-called RNA Integrity Number (RIN) which is an accepted quality criterion for isolated RNA [44]. The RNA samples were stored at -80°C up to cDNA synthesis.
First Strand cDNA Synthesis
cDNA synthesis was performed with the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Penzberg, Germany) using 1 μg RNA in reaction. Kit-included random hexamer primers were applied for first strand cDNA synthesis after following procedure: 10 min at 25°C for primer annealing, 30 min at 55°C for reverse transcription step, 5 min at 85°C for inactivation of Transcriptor Reverse Transcriptase, then cooling on ice. The cDNA volume amounted to 20 μl. Tubes were stored at -20°C up to subsequent PCR. All cDNA samples were 1:5 diluted with RNase-free water for use as template in real-time PCR.
Real-Time PCR
Real-time PCR was performed with the LightCycler Instrument (Roche). For relative mRNA quantification of target gene (ADAM9) two stably expressed reference genes, TBP (TATA box binding protein) and PPIA (Peptidyl isomerase A) were additionally determined [45]. The PCR reaction volumes for both reference genes were 10 μl and 20 μl for the target gene. All PCR reaction mixes included 1 μl diluted cDNA. The PCR run conditions for the TBP mRNA quantification were the same as described before [46]. For the reference gene PPIA and the target gene ADAM9 the PCR methods were also described previously [45]. The used primer/probe sequences (in 5'-3' direction) for ADAM9 mRNA quantification were as follows: forward primer: ggtgacagatttggcaattgtg, reverse primer: ttgtgccttcgttaaccatcc, donor probe: acgcctagtcgaggcaccaaatgttg-6Fl and the acceptor probe: Cy5.5-gtgtggatttccagctaggatcagatgttcc-P. The cDNA amplification was performed with the ready-to-use LightCycler® FastStart DNA MasterPLUS HybProbe (Roche). Final reaction concentrations of both primers were 0.5 μmol/l and the concentration for the donor/acceptor probes were 0.2 μmol/l each. The PCR setup was: activation of FastStart Taq DNA Polymerase at 95°C for 15 min, followed by 45 cycles of denaturation at 95°C for 10 s, annealing at 62°C for 30 s and elongation at 72°C for 30 s. The temperature transition rate was in each cycle 20°C/s. The fluorescence detection was gained after each annealing step and data was evaluated by the method of second derivative maximum with the LightCycler Software 3.5 (Roche). To reduce the inter-run variability, the paired samples of non-malignant and malignant tissue areas were measured in one PCR run. Calibration curves for all three genes were generated with pooled cDNA. PCR efficiencies were calculated from cDNA dilution curves of pooled cDNA and amounted to 1.84 for PPIA, to 1.88 for TBP and to 1.95 for ADAM9. Each PCR run included a cDNA with known expression level and was used as standard for the quantification of the unknown samples calculated by LightCycler Software Version 3.5. Another pooled cDNA was used as run-to-run precision control. For relative quantification of ADAM9 mRNA the expressions were related to geometric mean of the two reference genes PPIA and TBP [47].
Immunohistochemistry
Formalin fixed paraffin embedded tissue was freshly cut (3 μm). The sections were mounted on Superfrost slides (Menzel Gläser, Braunschweig, Germany), dewaxed with xylene and gradually hydrated. Antigen retrieval was achieved by pressure cooking in 0.01 M citrate buffer for 5 min. The primary ADAM9-antibody (goat polyclonal, AF949, R&D Systems, Wiesbaden, Germany) [20] was diluted 1:50 using a background reducing dilution buffer (Dako, Glostrup, Denmark) and incubated at room temperature for 1 hour. Detection took place with the REAL™ EnVision™ System (Dako) according to the manufacturer's instructions. Diaminobenzidin (Sigma-Aldrich, Munich, Germany) served as chromogen. Afterwards the slides were briefly counterstained with haematoxylin and mounted. As negative controls a set of sequential TMA slides was processed omitting the primary antibody to exclude unspecific background. To evaluate intratumoural heterogeneity of ADAM9 expression we additionally stained 10 conventional tissue slides with RCC.
Evaluation of the immunohistochemical stainings
The immunostainings were evaluated by two genitourinary pathologists blinded for patient outcome using a multiheaded microscope. The staining intensity was evaluated with a four-tier grading system (0 = negative, 1 = weak, 2 = moderate and 3 = strong staining intensity). We used a 10% threshold to determine positivity. To delineate between low and high levels of ADAM9 expression the tumours with strong ADAM9 expression were grouped against those with none to moderate staining intensity. Additionally, we compared the difference of the ADAM9 immunoreactivity in the two tumour spots from each case.
Statistical analysis
Statistical analysis was performed using SPSS, version 14.0. Fisher's exact tests, χ2-tests and Kruskal-Wallis tests were applied to assess the statistical significance of the associations between ADAM9 expression and clinico-pathological parameters. Rank correlations were calculated according to Spearman. Univariate survival analysis was carried out according to Kaplan-Meier, differences in survival curves were assessed with the Log rank test. Cox regression model was used for multivariate survival analyses. P values < 0.05 were considered significant.
Results
RNA quality and quantitative RT-PCR
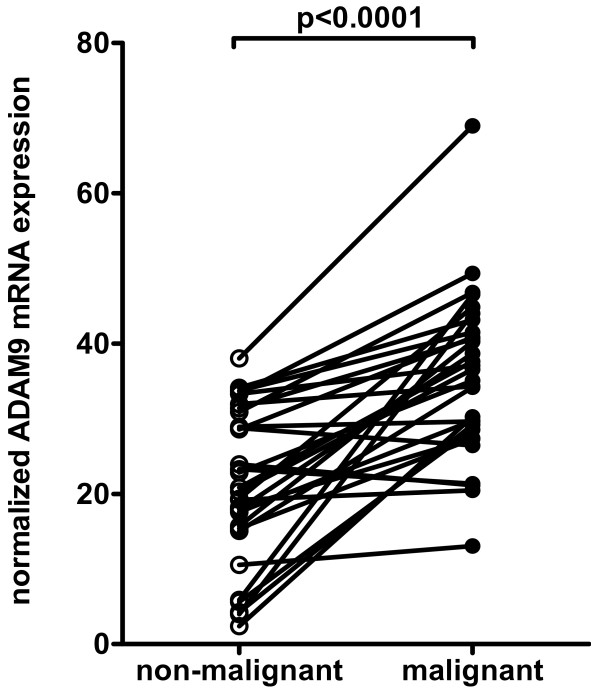
RIN values of isolated RNA from kidney tissue samples ranged from 7.0 to 10.0 (mean = 8.7, SD = 0.80) reflecting the high quality and integrity of the RNA. The normalized ADAM9 expression was significantly higher in cancer than in normal tissue samples (mean change fold 2.7, range: 0.9–12.6; p < 0.0001; Figure 1). In three out of the 30 matched pairs ADAM9 mRNA expression was higher in the normal renal tissue. Significant associations between the mRNA expression of ADAM9 in RCC and clinico-pathological parameters (tumour stage, grading, nodal status, metastasis, histologic type and residual tumour status) could not be demonstrated (all p > 0.05).
Figure 1.
ADAM9 mRNA expression. ADAM9 mRNA expression of matched pairs of non-malignant and malignant renal tissue samples was normalized to the geometric mean of the two reference genes PPIA and TBP. For most cases mRNA levels of the renal cell cancers laid above those of the normal tissue standard.
ADAM9 immunostaining in normal and malignant renal tissues
ADAM9 was expressed in malignant and non-malignant renal epithelia (Figure 2). The generally cytoplasmic immunoreactivity of ADAM9 was accentuated in the luminal part of the non-cancerous tubular epithelia and was particularly pronounced in the proximal tubules in comparison to distal tubules, which showed a weaker and more diffuse staining pattern. Glomerula and stromal cells were negative.
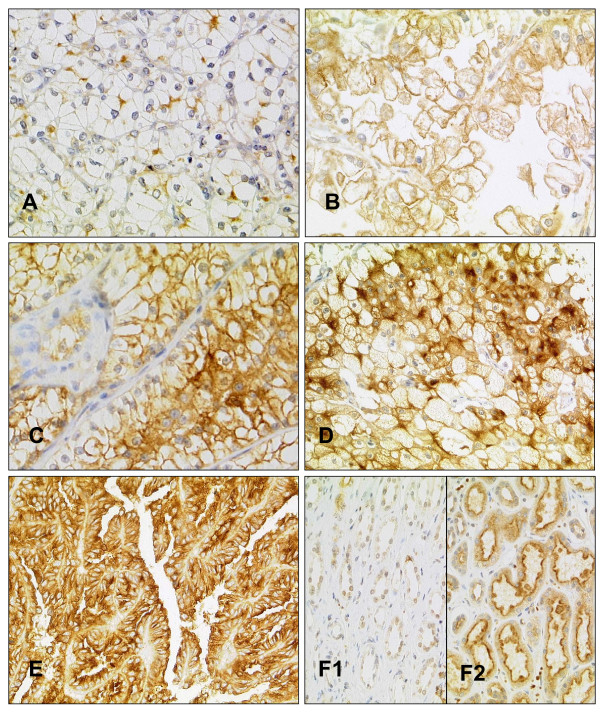
Figure 2.
ADAM9 immunohistochemistry. A-C Clear cell carcinomas with weak (A), moderate (B) and strong (C) ADAM9 protein expression. D/E Papillary (D) and chromophobe (E) renal cell carcinoma with strong ADAM9 expression. F Normal renal tissue with weak (F1) and strong (F2) ADAM9 expression.
In renal cell carcinomas, a cytoplasmic and membranous staining was seen in all tumour subtypes, with papillary carcinomas displaying the strongest cytoplasmic immunoreactivity. The distribution of the ADAM9 staining is summarized in Table 2. One case swam off the slide during immunostaining. Although strong protein expression was found much more frequently in the tumour than in the normal tissue, these differences remained insignificant according to the Wilcoxon signed rank test (p = 0.367). The median of both tissue types was 2 and the mean 1.95 (normal) and 2.04 (tumour) respectively. Matched normal tissue was available in 104 cases. It is interesting to note that none of the normal tissues and only two of the cancer cases were completely negative for ADAM9.
Table 2.
Distribution of ADAM9 staining in normal and malignant renal tissue (percentages in brackets)
| Staining | ADAM9 Normal | ADAM9 Cancer |
| Negative | 0 (0) | 2 (1.9) |
| 1+ | 9 (8.3) | 29 (26.9) |
| 2+ | 91 (84.3) | 40 (37.0) |
| 3+ | 4 (3.7) | 37 (34.3) |
Since the TMA was constructed to represent two spots of tumour from each case sampled from different tumour areas, differences of immunoreactivity between these spots can be used to estimate intratumoural heterogeneity of expression. We found ADAM9 staining completely equal both tumour spots in 91.7% (98/107; in one case only one spot had sufficient tumour tissue), which we consider a high rate of concordance. The additionally immunostained conventional tissue slides supported this estimation of a homogenous ADAM9 expression in tumour tissue. This is an important observation for it allows the use of TMAs for evaluation of ADAM9 in renal cell cancer.
ADAM9 expression, clinico-pathological correlations and disease free survival times
In bivariate Spearman's rank correlations the ADAM9 protein expression in renal cancer correlated with the tumour grade, patient age and positive tumour resection status (R1) (Table 3). Additionally performed Kruskal-Wallis tests further confirmed the non-significant associations of ADAM9 expression with M-status (p = 0.096), nodal status (p = 0.098), whereas residual tumour status was of borderline significance (p = 0.050). In the χ2-tests higher ADAM9 protein expression was significantly associated tumour grade, distant metastasis, positive nodal status and papillary as well as chromophobe histologic subtype (Table 1).
Table 3.
Correlation of ADAM9 protein expression in renal cell cancer with clinical/tumour-parameters
| ADAM9 | ADAM9 N | pT-status | Grading | pN-status | R-status | M-status | Age |
| CC | 0.107 | 0.166 | 0.239 | 0.212 | 0.206 | 0.212 | 0.219 |
| P | 0.282 | 0.085 | 0.013 | 0.096 | 0.047 | 0.076 | 0.023 |
| N | 104 | 108 | 108 | 63 | 94 | 71 | 108 |
CC = Spearman's rank correlation coefficient, p = two sided significance, N = number of cases, ADAM9 N = ADAM9 expression in normal tissue, R-status = residual tumour status (R0/R1+), M-status = Metastasis (M0/M1).
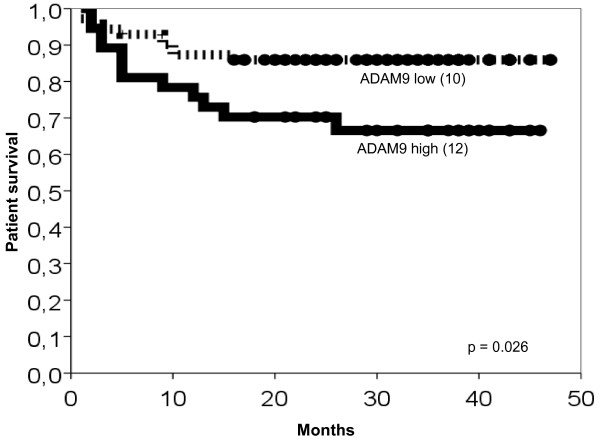
In univariate analyses (Kaplan-Meier) of patient survival the clinico-pathological characteristics pT-status, grading, nodal status, residual tumour status (R0 vs. R1) and distant metastasis (M0 vs. M1) reached statistical significance (Table 4). Histologic subtype and age were no prognosticators of patient survival (data not shown). Higher ADAM9 expression in renal cancer was significantly associated with shortened survival times (Table 4 & Fig. 3, p = 0.026). The Cox multivariate survival analysis revealed no independent prognostic value for ADAM9 expression and tumour grading whereas pT-and pM-stage remained highly significant (Table 5). The univariate prognosticators pN- and residual tumour status were not included in the Cox regression model since because of missing data this would have led to a significant drop in the number of cases for this analysis.
Table 4.
Univariate survival analysis (Kaplan-Meier)
| Characteristic | No. of cases | No. of events | Two-year survival rate (± SE) in % | p-value |
| ADAM9 expression | 0.026 | |||
| low | 71 | 10 | 85.9 ± 4.1 | |
| high | 37 | 12 | 70.3 ± 7.5 | |
| pT-status | < 0.001 | |||
| pT1 | 54 | 2 | 96.3 ± 2.6 | |
| pT2/3/4 | 54 | 20 | 64.8 ± 6.5 | |
| Grading | 0.001 | |||
| G 1 | 11 | 0 | - | |
| G 2 | 75 | 12 | 85.3 ± 4.1 | |
| G 3/4 | 22 | 10 | 54.5 ± 10.6 | |
| pN-status | 0.001 | |||
| pN0 | 51 | 10 | 80.4 ± 5.6 | |
| pN1+ | 12 | 8 | 41.7 ± 14.2 | |
| Residual tumour | < 0.001 | |||
| R0 | 78 | 10 | 87.2 ± 3.8 | |
| R1/2 | 15 | 8 | 46.7 ± 12.9 | |
| Metastasis | < 0.001 | |||
| M0 | 85 | 8 | 90.6 ± 3.2 | |
| M1 | 23 | 14 | 43.5 ± 10.3 | |
Survival times of patients with renal cancer according to clinico-pathological characteristics and ADAM9 protein expression.
Figure 3.
Kaplan-Meier survival curves for ADAM9. The number of events (deaths) is given in brackets. Tumours with high ADAM9 expression (bold line) revealed significantly shortened patient survival times if compared to those with low ADAM9 expression (dotted line).
Table 5.
Multivariate survival analysis
| Variable | Relative Risk | 95% CI | p-value |
| ADAM9 | 1.081 | 0.433–2.696 | 0.868 |
| pT-status | 6.820 | 1.462–31.827 | 0.015 |
| Grading | 1.060 | 0.434–2.589 | 0.899 |
| Metastasis | 1.496 | 1.534–13.177 | 0.006 |
Cox regression model, n = 108: ADAM9 and clinico-pathological characteristics grouped as in Table 4. CI = confidence interval.
Discussion
Several ADAMs, including ADAM9, have been described in various solid tumours on mRNA and/or protein level in and have often been associated with adverse prognostic parameters or shorter patient survival [18-21,23,25-32,48-50]. Our results are in line with this notion and demonstrate a prognostic value of ADAM9 for renal cell cancer at least in the univariate analysis. Although the whole model of the Cox analysis was significant, ADAM9 itself remained insignificant under multivariate conditions. Probably this might have been due to the strong association of ADAM9 expression with positive pM- status and higher tumour grade. Surprisingly, the histological tumour grade remained insignificant as well. This suggests that the cohort might have been not large enough to evaluate significance for these parameters, although pT-status and metastasis status could be verified as independent prognosticators. If nodal status and residual tumour status were included in the Cox analysis, all parameters would have lost their significance, probably caused by the strongly decreased number of cases in that analysis (data not shown).
ADAM9 was mainly expressed in the proximal part of the nephron. This is in line with a high expression of ADAM9 in clear cell and papillary carcinomas, which are thought to originate from that region. Interestingly also three of the five chromophobe RCC included showed a high ADAM9 expression, although this tumour type is thought to rather originate from the distal nephron. In our study we could not demonstrate a statistically significant up-regulation of ADAM9 protein expression in RCC in comparison to normal renal tissue. This is not in line with our results from the separate mRNA-sample cohort, where only 10% of the normal tissue displayed higher ADAM9 expression than the tumour. Possibly, posttranscriptional processes could be responsible for this discrepancy. On the other hand, only 4% of normal tissues showed a strong ADAM9 protein expression in comparison to 34% of the tumours, which clearly supports the notion that ADAM9 is up-regulated on protein level in a larger proportion of renal cell carcinomas.
These findings are generally coherent with results from previous studies on ADAM9 in solid tumours which evidence the role of ADAM9 in tumourigenesis and tumour progression [22,25,51,52]. Functionally, blocking of ADAM9 with specific antibodies resulted in inhibited cell growth of gastric cancer cell lines [18]. Over-expression of ADAM9 in lung cancer cell lines resulted in enhanced invasiveness and was significantly associated with brain metastases [50]. In melanoma, ADAM9 is up-regulated in vivo at the invasion front [27]. In our study, the significant associations of ADAM9 with prognostically adverse conventional tumour parameters (positive nodal status, distant metastasis, residual tumour in the resection margins and higher tumour grade) are clearly in line with these findings.
Interestingly, ADAM9 was also found in most (13 of 17) papillary renal cell carcinomas. Although the number of cases with this subtype of RCC was small in our cohort, this information might be of adjunct diagnostic use for the assessment of renal cell carcinomas.
Conclusion
In conclusion our results support the notion of ADAM9 to be associated with more aggressive tumours and unfavourable outcome. ADAM9 protein expression was significantly associated with shortened survival times but failed significance in a multivariate analysis. To further assess a possible independent prognostic role and the usefulness of ADAM9 for the sub classification of RCC, further validation using larger tumour cohorts is clearly warranted.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FRF coordinated the study, performed immunohistological and statistical analyses and wrote the paper. KW supported the immunohistological evaluation and performed statistical analyses and wrote and revised essential parts of the paper. MoJ and AT performed the RT-PCR and contributed to the study design. IK contributed in the immunohistochemical analyses and array construction. ML, MaJ and MD provided samples and clinico-pathological data. KJ coordinated the study, provided samples and contributed to the study design. GK conceived and coordinated the study, performed immunohistological and statistical analyses, wrote and revised the paper.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
We are grateful to Britta Beyer and Silvia Behnke for excellent technical assistance and the Sonnenfeld Stiftung for sponsoring the tissue micro arrayer.
Contributor Information
Florian R Fritzsche, Email: florian.fritzsche@usz.ch.
Kirsten Wassermann, Email: kirsten_wassermann@web.de.
Monika Jung, Email: monika.jung@charite.de.
Angelika Tölle, Email: angelika.toelle@charite.de.
Ilka Kristiansen, Email: i.kristiansen@gmx.de.
Michael Lein, Email: michael.lein@charite.de.
Manfred Johannsen, Email: manfred.johannsen@charite.de.
Manfred Dietel, Email: manfred.dietel@charite.de.
Klaus Jung, Email: klaus.jung@charite.de.
Glen Kristiansen, Email: glen.kristiansen@usz.ch.
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- McLaughlin JK, Lipworth L. Epidemiologic aspects of renal cell cancer. Seminars in oncology. 2000;27:115–123. [PubMed] [Google Scholar]
- Dhote R, Pellicer-Coeuret M, Thiounn N, Debre B, Vidal-Trecan G. Risk factors for adult renal cell carcinoma: a systematic review and implications for prevention. BJU international. 2000;86:20–27. doi: 10.1046/j.1464-410x.2000.00708.x. [DOI] [PubMed] [Google Scholar]
- Dall'Oglio MF, Arap MA, Antunes AA, Cury J, Leite KR, Srougi M. Impact of clinicopathological parameters in patients treated for renal cell carcinoma. The Journal of urology. 2007;177:1687–1691. doi: 10.1016/j.juro.2007.01.065. [DOI] [PubMed] [Google Scholar]
- Breda A, Konijeti R, Lam JS. Patterns of recurrence and surveillance strategies for renal cell carcinoma following surgical resection. Expert review of anticancer therapy. 2007;7:847–862. doi: 10.1586/14737140.7.6.847. [DOI] [PubMed] [Google Scholar]
- Martignoni G, Brunelli M, Gobbo S, Remo A, Ficarra V, Cossu-Rocca P, Pea M, Chilosi M, Menestrina F, Cheng L. Role of molecular markers in diagnosis and prognosis of renal cell carcinoma. Analytical and quantitative cytology and histology. Anal Quant Cytol Histol. 2007;29:41–49. [PubMed] [Google Scholar]
- Rathmell WK, Martz CA, Rini BI. Renal cell carcinoma. Current opinion in oncology. 2007;19:234–240. doi: 10.1097/CCO.0b013e3280ad4388. [DOI] [PubMed] [Google Scholar]
- Young AN, Master VA, Amin MB. Current trends in the molecular classification of renal neoplasms. The Scientific World Journal. 2006;6:2505–2518. doi: 10.1100/tsw.2006.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond ME, Fitzgibbons PL, Compton CC, Grignon DJ, Page DL, Fielding LP, Bostwick D, Pajak TF. College of American Pathologists Conference XXXV: solid tumor prognostic factors-which, how and so what? Summary document and recommendations for implementation. Cancer Committee and Conference Participants. Archives of pathology & laboratory medicine. 2000;124:958–965. doi: 10.5858/2000-124-0958-COAPCX. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Higuchi Y, Yoshiyama K, Shimizu E, Kataoka M, Hijiya N, Matsuura K. ADAM family proteins in the immune system. Immunology today. 1999;20:278–284. doi: 10.1016/s0167-5699(99)01464-4. [DOI] [PubMed] [Google Scholar]
- Bjarnason JB, Fox JW. Snake venom metalloendopeptidases: reprolysins. Methods in enzymology. 1995;248:345–368. doi: 10.1016/0076-6879(95)48023-4. [DOI] [PubMed] [Google Scholar]
- Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. The Journal of cell biology. 1995;131:275–278. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba K, Albrechtsen R, Gilpin BJ, Loechel F, Wewer UM. Cysteine-rich domain of human ADAM 12 (meltrin alpha) supports tumor cell adhesion. The American journal of pathology. 1999;154:1489–1501. doi: 10.1016/s0002-9440(10)65403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath D, Slocombe PM, Stephens PE, Warn A, Hutchinson GR, Yamada KM, Docherty AJ, Murphy G. Interaction of metargidin (ADAM-15) with alphavbeta3 and alpha5beta1 integrins on different haemopoietic cells. Journal of cell science. 1999;112:579–587. doi: 10.1242/jcs.112.4.579. [DOI] [PubMed] [Google Scholar]
- Nath D, Slocombe PM, Webster A, Stephens PE, Docherty AJ, Murphy G. Meltrin gamma(ADAM-9) mediates cellular adhesion through alpha(6)beta(1)integrin, leading to a marked induction of fibroblast cell motility. Journal of cell science. 2000;113:2319–2328. doi: 10.1242/jcs.113.12.2319. [DOI] [PubMed] [Google Scholar]
- Schwettmann L, Tschesche H. Cloning and expression in Pichia pastoris of metalloprotease domain of ADAM 9 catalytically active against fibronectin. Protein Expr Purif. 2001;21:65–70. doi: 10.1006/prep.2000.1374. [DOI] [PubMed] [Google Scholar]
- Zhou M, Graham R, Russell G, Croucher PI. MDC-9 (ADAM-9/Meltrin gamma) functions as an adhesion molecule by binding the alpha(v)beta(5) integrin. Biochem Biophys Res Commun. 2001;280:574–580. doi: 10.1006/bbrc.2000.4155. [DOI] [PubMed] [Google Scholar]
- Carl-McGrath S, Lendeckel U, Ebert M, Roessner A, Rocken C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. International journal of oncology. 2005;26:17–24. [PubMed] [Google Scholar]
- Grutzmann R, Foerder M, Alldinger I, Staub E, Brummendorf T, Ropcke S, Li X, Kristiansen G, Jesnowski R, Sipos B, et al. Gene expression profiles of microdissected pancreatic ductal adenocarcinoma. Virchows Arch. 2003;443:508–517. doi: 10.1007/s00428-003-0884-1. [DOI] [PubMed] [Google Scholar]
- Grutzmann R, Luttges J, Sipos B, Ammerpohl O, Dobrowolski F, Alldinger I, Kersting S, Ockert D, Koch R, Kalthoff H, et al. ADAM9 expression in pancreatic cancer is associated with tumour type and is a prognostic factor in ductal adenocarcinoma. British journal of cancer. 2004;90:1053–1058. doi: 10.1038/sj.bjc.6601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao T, Nanba D, Tanaka M, Ishiguro H, Kinugasa Y, Doki Y, Yano M, Matsuura N, Monden M, Higashiyama S. Overexpression of ADAM9 enhances growth factor-mediated recycling of E-cadherin in human colon cancer cell line HT29 cells. Experimental cell research. 2006;312:331–339. doi: 10.1016/j.yexcr.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Karan D, Lin FC, Bryan M, Ringel J, Moniaux N, Lin MF, Batra SK. Expression of ADAMs (a disintegrin and metalloproteases) and TIMP-3 (tissue inhibitor of metalloproteinase-3) in human prostatic adenocarcinomas. International journal of oncology. 2003;23:1365–1371. [PubMed] [Google Scholar]
- Kristiansen G, Pilarsky C, Wissmann C, Kaiser S, Bruemmendorf T, Roepcke S, Dahl E, Hinzmann B, Specht T, Pervan J, et al. Expression profiling of microdissected matched prostate cancer samples reveals CD166/MEMD and CD24 as new prognostic markers for patient survival. The Journal of pathology. 2005;205:359–376. doi: 10.1002/path.1676. [DOI] [PubMed] [Google Scholar]
- Lendeckel U, Kohl J, Arndt M, Carl-McGrath S, Donat H, Rocken C. Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. Journal of cancer research and clinical oncology. 2005;131:41–48. doi: 10.1007/s00432-004-0619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea C, McKie N, Buggy Y, Duggan C, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Expression of ADAM-9 mRNA and protein in human breast cancer. Int J Cancer. 2003;105:754–761. doi: 10.1002/ijc.11161. [DOI] [PubMed] [Google Scholar]
- Yamada D, Ohuchida K, Mizumoto K, Ohhashi S, Yu J, Egami T, Fujita H, Nagai E, Tanaka M. Increased expression of ADAM 9 and ADAM 15 mRNA in pancreatic cancer. Anticancer research. 2007;27:793–799. [PubMed] [Google Scholar]
- Zigrino P, Mauch C, Fox JW, Nischt R. Adam-9 expression and regulation in human skin melanoma and melanoma cell lines. Int J Cancer. 2005;116:853–859. doi: 10.1002/ijc.21087. [DOI] [PubMed] [Google Scholar]
- Ding X, Yang LY, Huang GW, Wang W, Lu WQ. ADAM17 mRNA expression and pathological features of hepatocellular carcinoma. World J Gastroenterol. 2004;10:2735–2739. doi: 10.3748/wjg.v10.i18.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pabic H, Bonnier D, Wewer UM, Coutand A, Musso O, Baffet G, Clement B, Theret N. ADAM12 in human liver cancers: TGF-beta-regulated expression in stellate cells is associated with matrix remodeling. Hepatology. 2003;37:1056–1066. doi: 10.1053/jhep.2003.50205. [DOI] [PubMed] [Google Scholar]
- McCulloch DR, Akl P, Samaratunga H, Herington AC, Odorico DM. Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor I, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin Cancer Res. 2004;10:314–323. doi: 10.1158/1078-0432.ccr-0846-3. [DOI] [PubMed] [Google Scholar]
- Tannapfel A, Anhalt K, Hausermann P, Sommerer F, Benicke M, Uhlmann D, Witzigmann H, Hauss J, Wittekind C. Identification of novel proteins associated with hepatocellular carcinomas using protein microarrays. The Journal of pathology. 2003;201:238–249. doi: 10.1002/path.1420. [DOI] [PubMed] [Google Scholar]
- Tian BL, Wen JM, Zhang M, Xie D, Xu RB, Luo CJ. The expression of ADAM12 (meltrin alpha) in human giant cell tumours of bone. Mol Pathol. 2002;55:394–397. doi: 10.1136/mp.55.6.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer A, Schwettmann L, Jung M, Roigas J, Kristiansen G, Schnorr D, Loening SA, Jung K, Lichtinghagen R. Increased mRNA expression of ADAMs in renal cell carcinoma and their association with clinical outcome. Oncology reports. 2004;11:529–536. [PubMed] [Google Scholar]
- Roemer A, Schwettmann L, Jung M, Stephan C, Roigas J, Kristiansen G, Loening SA, Lichtinghagen R, Jung K. The membrane proteases adams and hepsin are differentially expressed in renal cell carcinoma. Are they potential tumor markers? The Journal of urology. 2004;172:2162–2166. doi: 10.1097/01.ju.0000144602.01322.49. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Hirata M, Hasuwa H, Iwamoto R, Umata T, Miyado K, Tamai Y, Kurisaki T, Sehara-Fujisawa A, Ohno S, et al. A metalloprotease-disintegrin, MDC9/meltrin-gamma/ADAM9 and PKCdelta are involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. The EMBO journal. 1998;17:7260–7272. doi: 10.1093/emboj/17.24.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghani M, Becherer JD, Moss ML, Atherton RE, Erdjument-Bromage H, Arribas J, Blackburn RK, Weskamp G, Tempst P, Blobel CP. Metalloprotease-disintegrin MDC9: intracellular maturation and catalytic activity. The Journal of biological chemistry. 1999;274:3531–3540. doi: 10.1074/jbc.274.6.3531. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Nanba D, Mori S, Shiba F, Ishiguro H, Yoshino K, Matsuura N, Higashiyama S. ADAM binding protein Eve-1 is required for ectodomain shedding of epidermal growth factor receptor ligands. The Journal of biological chemistry. 2004;279:41950–41959. doi: 10.1074/jbc.M400086200. [DOI] [PubMed] [Google Scholar]
- Normanno N, Bianco C, Strizzi L, Mancino M, Maiello MR, De Luca A, Caponigro F, Salomon DS. The ErbB receptors and their ligands in cancer: an overview. Current drug targets. 2005;6:243–257. doi: 10.2174/1389450053765879. [DOI] [PubMed] [Google Scholar]
- Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Higashiyama S, Nanba D. ADAM-mediated ectodomain shedding of HB-EGF in receptor cross-talk. Biochimica et biophysica acta. 2005;1751:110–117. doi: 10.1016/j.bbapap.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Tanida S, Joh T, Itoh K, Kataoka H, Sasaki M, Ohara H, Nakazawa T, Nomura T, Kinugasa Y, Ohmoto H, et al. The mechanism of cleavage of EGFR ligands induced by inflammatory cytokines in gastric cancer cells. Gastroenterology. 2004;127:559–569. doi: 10.1053/j.gastro.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Mahimkar RM, Visaya O, Pollock AS, Lovett DH. The disintegrin domain of ADAM9: a ligand for multiple beta1 renal integrins. The Biochemical journal. 2005;385:461–468. doi: 10.1042/BJ20041133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen G, Yu Y, Petersen S, Kaufmann O, Schluns K, Dietel M, Petersen I. Overexpression of c-erbB2 protein correlates with disease-stage and chromosomal gain at the c-erbB2 locus in non-small cell lung cancer. Eur J Cancer. 2001;37:1089–1095. doi: 10.1016/s0959-8049(01)00096-x. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC molecular biology. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Ramankulov A, Roigas J, Johannsen M, Ringsdorf M, Kristiansen G, Jung K. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC molecular biology. 2007;8:47. doi: 10.1186/1471-2199-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl F, Jung M, Radonic A, Sachs M, Loening SA, Jung K. Identification and validation of suitable endogenous reference genes for gene expression studies of human bladder cancer. The Journal of urology. 2006;175:1915–1920. doi: 10.1016/S0022-5347(05)00919-5. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldinger I, Dittert D, Peiper M, Fusco A, Chiappetta G, Staub E, Lohr M, Jesnowski R, Baretton G, Ockert D, et al. Gene expression analysis of pancreatic cell lines reveals genes overexpressed in pancreatic cancer. Pancreatology. 2005;5:370–379. doi: 10.1159/000086537. [DOI] [PubMed] [Google Scholar]
- Shigemura K, Sung SY, Kubo H, Arnold RS, Fujisawa M, Gotoh A, Zhau HE, Chung LW. Reactive oxygen species mediate androgen receptor- and serum starvation-elicited downstream signaling of ADAM9 expression in human prostate cancer cells. The Prostate. 2007;67:722–731. doi: 10.1002/pros.20565. [DOI] [PubMed] [Google Scholar]
- Shintani Y, Higashiyama S, Ohta M, Hirabayashi H, Yamamoto S, Yoshimasu T, Matsuda H, Matsuura N. Overexpression of ADAM9 in non-small cell lung cancer correlates with brain metastasis. Cancer research. 2004;64:4190–4196. doi: 10.1158/0008-5472.CAN-03-3235. [DOI] [PubMed] [Google Scholar]
- Peduto L, Reuter VE, Shaffer DR, Scher HI, Blobel CP. Critical function for ADAM9 in mouse prostate cancer. Cancer research. 2005;65:9312–9319. doi: 10.1158/0008-5472.CAN-05-1063. [DOI] [PubMed] [Google Scholar]
- Sung SY, Kubo H, Shigemura K, Arnold RS, Logani S, Wang R, Konaka H, Nakagawa M, Mousses S, Amin M, et al. Oxidative Stress Induces ADAM9 Protein Expression in Human Prostate Cancer Cells. Cancer research. 2006;66:9519–9526. doi: 10.1158/0008-5472.CAN-05-4375. [DOI] [PubMed] [Google Scholar]