Abstract
Formation of cancerous translocations requires the illegitimate joining of chromosomes containing double-strand breaks (DSBs). It is unknown how broken chromosome ends find their translocation partners within the cell nucleus. Here, we have visualized and quantitatively analysed the dynamics of single DSBs in living mammalian cells. We demonstrate that broken ends are positionally stable and unable to roam the cell nucleus. Immobilization of broken chromosome ends requires the DNA-end binding protein Ku80, but is independent of DNA repair factors, H2AX, the MRN complex and the cohesion complex. DSBs preferentially undergo translocations with neighbouring chromosomes and loss of local positional constraint correlates with elevated genomic instability. These results support a contact-first model in which chromosome translocations predominantly form among spatially proximal DSBs.
DSBs occur frequently in the genome through the action of DNA-damaging agents or during genome replication1,2. DSBs are hazardous to the cell because failure to properly repair them may lead to tumorigenic trans-locations3. How broken ends of different chromosomes meet in the cell nucleus to eventually form a translocation is poorly understood4. Two hypotheses have been put forth: the ‘contact-first’ model proposes that interactions between breaks on distinct chromosomes can only take place when the breaks are created in chromatid fibres that colocalize at the time of DNA damage5. In contrast, the ‘breakage-first’ hypothesis postulates that breaks formed at distant locations are able to scan the nuclear space for potential partners and come together to produce translocations6. The two models make divergent predictions as to the dynamic behaviour of broken chromosome ends. In the breakage-first model, single DSBs are required to undergo large-scale motions within the cell nucleus and must be able to roam the nuclear space in search of appropriate interaction partners. In the contact-first model, only limited local positional motion of DSBs is expected. The available experimental data is contradictory: in mammalian cells, induction of extensive chromosome damage using ultra-soft X-rays7, laser microirradiation8 or γ-irradiation8 indicates that damaged DNA is largely stationary. In contrast, α-particle irradiation leads to large-scale motion and clustering of damaged sites6. In addition, observations in Saccharomyces cerevisiae have suggested that although broken chromosome ends do not physically separate9–11, unrepaired loci are able to search the nuclear space for appropriate translocation partners and are recruited into common repair foci after damage, suggesting that they are able to undergo long range movements12,13.
To directly study the dynamics of single DSBs in living mammalian cells in vivo, we developed a cell system in which we can induce a DSB at a defined genomic site and follow the fate of each of the two damaged DNA ends in real time. NIH3T3 stable cell lines (NIH2/4) were generated containing a copy of the unique 18 nucleotide ISceI restriction site flanked by an array of 256 copies of the lac-repressor binding site and by 96 copies of the tetracycline response element (L-ISceI-T array; Fig. 1a)14. The lac- and tet-arrays were visualized simultaneously by expression of CFP–lac-repressor and YFP–tet-repressor, respectively (Fig. 1b). To temporarily control the induction of a DSB at the L-ISceI-T array in a single living cell, we took advantage of glucocorticoid-receptor chimeras that translocate from the cytoplasm to the nucleus on binding to the synthetic ligand triamcinolone acetonide (Fig. 1a)15. A chimera between the ISceI restriction endonuclease and the ligand-binding domain of the glucocorticoid receptor in frame with monomeric RFP (ISceI–GR) is cytoplasmic in the absence of triamcinolone acetonide and no DSBs were detected when cells were stained for phosphorylated H2AX (γ-H2AX; Fig. 1c). As expected, when triamcinolone acetonide was added ISceI–GR translocated to the nucleus within 2 min and lead to rapid induction of a DSB at the array, as judged by detection of a single nuclear γ-H2AX focus (Fig. 1c). More than 75% of cells showed γ-H2AX accumulation at the array within 5 min and the percentage of cells with γ-H2AX staining at the array was 85–90% after 15 min (Fig. 1d). The effect of ISceI–GR was specific for the array as no additional sites of γ-H2AX accumulation were detected. Recruitment of repair factors occurred with kinetics similar to those reported in fixed and living cells on induction of DNA damage by laser microirradiation16. The repair factor MDC1 accumulated with similar kinetics to γ-H2AX, followed by 53BP1, which associated with the array within 5 min in 30% of cells and within 15 min in 75% of cells (Fig. 1d). Concomitant staining of 35–40% of cells with the single strand binding protein RPA indicated that at least a fraction of cleaved ends was resected during that time (see Supplementary Information, Fig. S1). Cleavage kinetics were confirmed directly by ligation-mediated PCR using primers flanking the ISceI site17 (Fig. 1e). Cleavage occurred rapidly and reached a steady-state plateau of ~50% after 30 min (Fig. 1e).
Figure 1.
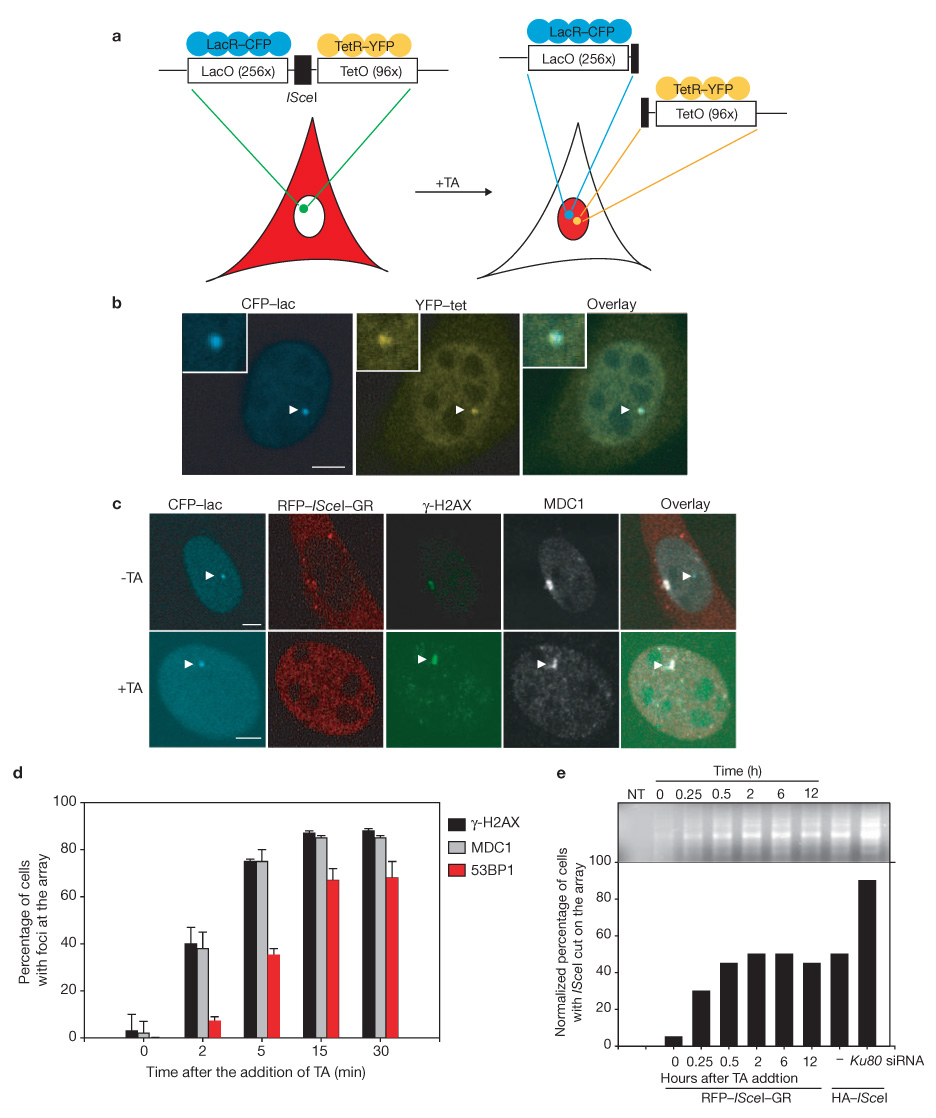
An experimental system to visualize single broken DNA ends. (a) Schematic representation of the L–ISceI–T array system. DSBs are triggered by treatment of cells with the steroid ligand triamcinolone acetonide (TA), resulting in redistribution of a RFP–ISceI–GR fusion protein (red) from the cytoplasm to the nucleus. (b) Visualization of the lac and the tet arrays in NIH2/4 cells after transient transfection of CFP–lacR and YFP–tetR. (c) Kinetics of DSBs and recruitment of DNA repair factors. Immmunofluorescence microscopy of NIH2/4 cells transiently transfected with CFP–lacR and RFP–ISceI–GR in the absence or presence of triamcinolone acetonide. Cells were stained with the indicated antibodies. On addition of triamcinolone acetonide, RFP–ISceI–GR translocates to the nucleus. In the absence of triamcinolone acetonide, RFP–ISceI–GR is cytoplasmic and no DSB recruitment of repair factors occurs. Arrowheads indicate the array. (d) Quantitative analysis of H2AX phosphorylation and recruitment kinetics of MDC1 and 53BP1 after addition of triamcinolone acetonide. Values represent averages ± s.d. (n = 100) from three independent experiments. (e) Ligation-mediated PCR in NIH2/4 cells at the indicated times after the addition of triamcinolone acetonide. NT, non-transfected cells. Cleavage levels were determined by comparison with a standard curve created by LM-PCR products from known ratios of naked DNA cleaved by ISceI in vitro to uncut DNA. Percentage of cells containing cleaved arrays was determined by normalizing for RFP–ISceI–GR and HA–ISceI transfection efficiencies. The scale bars in b and c represent 5 µm.
To directly analyse the positional motion of broken chromosome ends, the YFP–tet and CFP–lac-tagged regions flanking the ISceI site were simultaneously visualized using multicolour in vivo imaging (Fig. 2 and see Methods). To determine to what degree broken chromosome ends undergo global movements within the nucleus, complete three-dimensional stacks of the two signals were acquired every 30 s for up to 1 h after the addition of triamcinolone acetonide, and the movement of the CFP and YFP arrays was tracked in three-dimensional space (see Methods and Supplementary Information, Materials). Qualitative analysis of the position of the broken chromosome ends within the cell nucleus indicated extremely limited motion of the tagged ends and no significant change in the spatial location of either tag occurred (Fig. 2a). The CFP and YFP signals did not clearly separate, although changes in their degree of overlap were apparent during the time course (Fig. 2a and see below). The positional stability was not due to light damage, nor was it an artifact of the experimental system due to binding of the lac and tet repressors to the array as unbroken chromosome loci under-went similar local motion, as previously observed in other experimental systems (data not shown)18–20.
Figure 2.
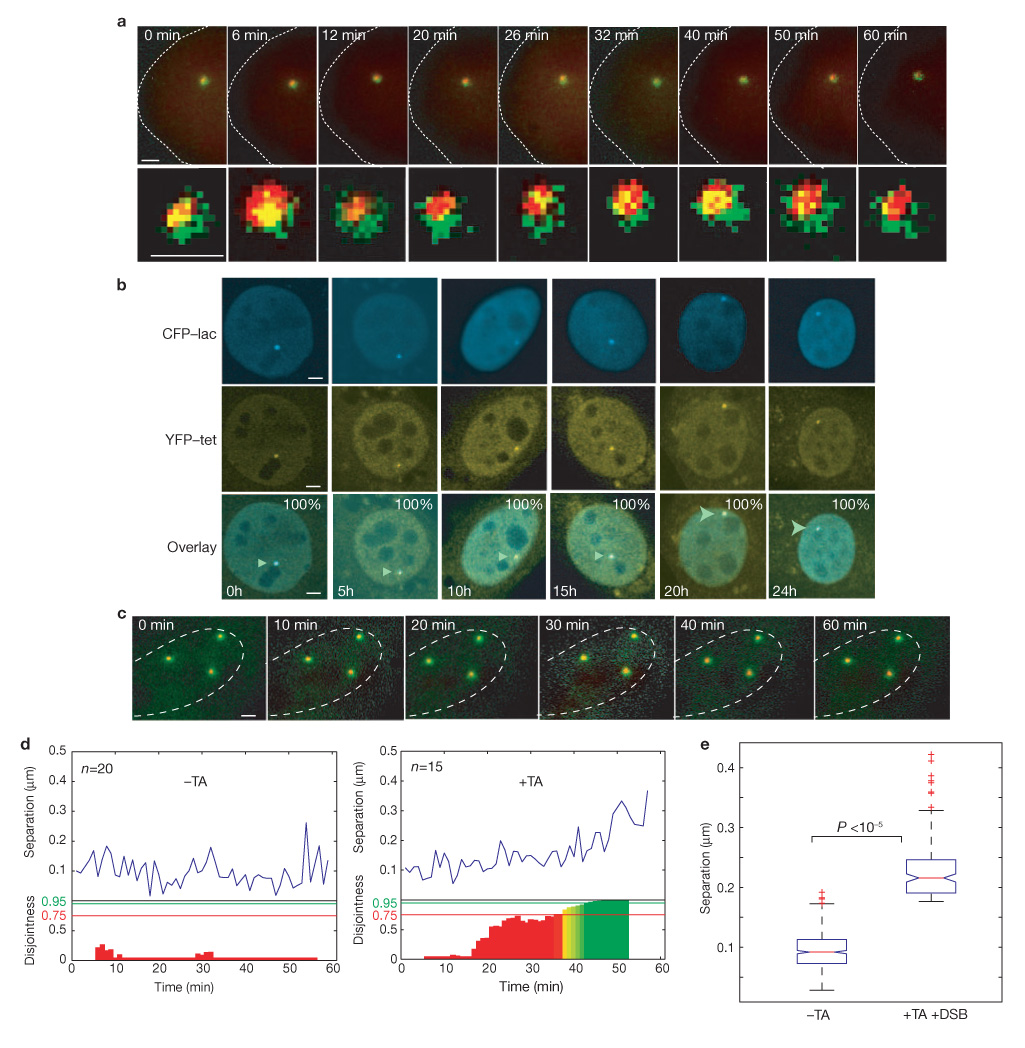
Analysis of the positional and local movement of broken DNA ends. (a) Representative time points of a time-lapse series 60 min after the addition of triamcinolone acetonide in a cell containing a single array. Each time point is a colour-combined maximum projection of three-dimensional stacks recorded in the CFP (red) and YFP (green) channels. The relative position of the two signals locally fluctuates over time (lower panel). (b) Visualization of CFP and YFP arrays several hours after the addition of triamcinolone acetonide. Arrowheads indicate the L–ISceI–T array. The percentage indicates cells with colocalized tags (n = 100 for each timepoint). (c) Time-lapse series 60 min after the addition of triamcinolone acetonide in a cell containing multiple arrays. Each time point is a colour-combined maximum projection of three-dimensional stacks recorded in the CFP (red) and YFP (green) channels. (d) Representative trajectories of CFP–YFP tag separation in the absence or presence of triamcinolone acetonide (blue line). Below the trajectories, coloured bars indicate the disjointedness probability (PD) for every 10-min window (red, PD <75%; yellow, 75 ≤ PD ≤ 95%; green, PD >95%). (e) Distribution of tag separation for control cells and cells with DSB. In the presence of DSBs, separation increases from ~0.1 µm to ~0.22 µm (P <10−5). Boxes indicate boundaries of the 25th percentile to the 75th percentile. The red line indicates the median of the data. Error bars indicate the spread of the data. Outliers are marked by red crosses, and are defined as data points that are further away than 1.5 times the width of the box. P values were obtained by t-test and represent pairwise comparisons of the indicated sample means. The scale bars represent 1 µm in a and c, and 2 µm in b. The dashed lines in a and c indicate the outline of the cells.
To exclude the possibility that separation of DSBs occurs at later times after breakage, cell populations (n = 100) were scored for separation of the CFP–lac and YFP–tet labels at times up to 24 h after triamcinolone acetonide addition (Fig. 2b). No significant separation of the tags was observed, indicating that a sustained or repeated DSB does not lead to loss of positional stability. Furthermore, positional stability was not dependent on cell-cycle stage as no separation of tagged chromosome ends was detected in cells arrested in G0–G1, or at various times after release of cells from a G1–S-phase arrest (see Supplementary Information, Fig. S2).
As multiple DSBs in S. cerevisiae have been reported to coalesce into shared repair factories13, the dynamics of broken chromosome ends were analysed in cells with multiple lac–ISceI–tet-arrays on distinct chromosomes. Similarly to single DSBs, the broken DNA ends were positionally stable (Fig. 2c). No coalescence was observed at times up to 24 h after breakage, even between arrays separated by less than 400 nm (data not shown). We conclude that DSBs are positionally immobile within the mammalian cell nucleus.
To assess the local diffusional motion of each DNA end at the site of damage in detail, the three-dimensional position of the CFP and YFP tags was determined by high-resolution positional tracking with sub-pixel precision (see Supplementary Information, Materials). As a quantitative indicator of breakage, the distance between the fitted sub-pixel positions of the CFP tag and the YFP tag was measured (Fig. 2d, e). In more than 60% of cells (n = 15) the distance between the tags was on average at least doubled from 100 nm to 220 nm after the addition of triamcinolone acetonide (Fig. 2d, e). In contrast, no substantial increase in distance was detected in the absence of triamcinolone acetonide (Fig. 2d, e). To further support the distance analysis we evaluated the relative motion of the tags in ten-frame sliding windows and calculated a disjointedness probability (PD) defined by the tag separation combined with the relative speed of tag motion (see Supplementary Information, Materials). In more than 45% of cells (n = 15) PD was >95% after the addition of trimcinolone acetonide for 30 min (Fig. 2d, e) and more than 75% of cells had PD of >50%. In contrast, no cells had PD of >95% in the absence of triamcinolone acetonide and only 35% of cells reached a PD >50%. These data demonstrate that the local separation of broken DNA ends increases when a chromosome breaks.
To address the molecular basis of the positional stability of broken chromosome ends within the nuclear space, the repair factors Ku80, H2AX, NBS1 or the cohesin subunit SMC1 were eliminated from NIH2/4 cells using specific RNA interference (RNAi)21–26. Depletion of the targeted repair factors was confirmed by western blotting and immunocytochemistry 48 h after short interfering RNA (siRNA) trans-fection (see Supplementary Information, Fig. S3 and data not shown). When cells lacking any one of these factors were imaged using time-lapse microscopy no physical separation of broken chromosome ends was evident at times up to 2 h after induction of DSBs (see Supplementary Information, Fig. S4). Similar results were obtained in H2AX−/− mouse embryonic fibroblasts (see Supplementary Information, Fig. S5).
To determine whether the absence of repair factors leads to loss of the spatial proximity of broken ends over longer periods of time, cell populations lacking H2AX, NBS1, SMC1 or Ku80 were scored for separation of the CFP–lac and YFP–tet labels 24 h after DSB induction (Fig. 3a, b). Despite the fact that H2AX is thought to anchor or align chromosomal ends21,23 no loss of colocalization of the two signals was observed in the absence of H2AX either in RNAi knockdown cells or in H2AX−/− cells (Fig. 3b and see Supplementary Information, Fig. S5). Similarly, depletion of NBS1 or SMC1 did not impact on the proximity of the broken ends (Fig. 3b). NBS1 did not accumulate at ISceI induced DSBs in H2AX−/− cells 27 (see Supplementary Information, Fig. S5c), further supporting the notion that NBS1 is not required for positional stability. Furthermore, knockdown of the other two components of the Mre11–Rad50–Nbs1 complex did not lead to separation of cleaved ends (Fig. 3b and see Supplementary Information, Fig. S3). In striking contrast, a significant increase in the population with clearly separated broken ends (separated by >500 nm) was observed in cells lacking Ku80 (Fig. 3a, b). Separation did not require mitosis as similar numbers of separated ends were present in cells which were prevented from entering mitosis by G1–S and G2–M arrest by olomoucine (Fig. 3b). Separation of the signals was mostly due to the presence of broken, unrepaired chromosome ends, as 82% of chromosomes with aberrant fluorescent in situ hybridization (FISH) signals (22 out of 100) in metaphase spreads from Ku80-depleted cells contained the ISceI-array break site at the end of a chromosome 3 fragment (Fig. 3c, d). Eighteen percent of signals were on another chromosome, suggestive of a translocation (Fig. 3d and see below).
Figure 3.

Separation of broken DNA ends in the absence of Ku80. (a) Localization of CFP and YFP arrays in the presence and absence of Ku80. Confocal microscopy in control and in Ku80-depleted NIH2/4 cells. Two examples of separated CFP and YFP arrays with different distances are presented. Separation was defined as a tag distance of >500 nm. The scale bars indicate 5 µm in a and c. Arrow indicates ISceI array. (b) Quantification of separated CFP and YFP signals in NIH2/4 cells depleted of the indicated repair factors or in controls cells. Values represent averages ± s.d. (n = 200) from three independent experiments. (c) Partial metaphase spreads of control or Ku80-depleted NIH2/4 cells 24 h after overexpression of HA–ISceI. Localization of the array at the end of a single chromosome 3 indicates the presence of a chromosome break. Arrowheads indicate the L–ISceI–T array. Probes against the entire array were used and the Lac and Tet portions of the array can not be distinguished. (d) Quantification of L–ISceI–T array signals on broken or translocated chromosomes. One hundred metaphases were analysed per sample. (e) Representative CFP–YFP tag separation trajectory in Ku80-depleted NIH2/4 cells. Below the trajectories, coloured bars indicate the disjointness probability (PD) for every 10-min window (red, PD <75%; yellow, 75 ≤ PD ≤ 95%; green, PD >95%). (f) Distribution of relative fluorescent tag mobility. Boxes indicate boundaries of the 25th percentile to the 75th percentile. The red line indicates the median of the data. Error bars indicate the spread of the data. Outliers are marked by red crosses, and are defined as data points that are further away than 1.5 times the width of the box. P values were obtained by t-test and represent pairwise comparisons of the indicated sample means.
To determine whether the separation of broken ends on loss of Ku80 was related to the dynamic behaviour of DSBs, we analysed the local motion of broken ends in Ku80-knockdown cells using our three-dimensional tracking analysis (Fig. 3e, f). Although the average distance between the CFP and YFP tags did not increase further than in control cells (Fig. 3e and see Supplementary Information, Fig. S4), a significant increase in the ability to locally diffuse was evident on loss of Ku80 (Fig. 3f). Although broken DNA ends in control cells moved on average at 50 nm min−1, in the absence of Ku80 this speed was more than 80 nm min−1 (P <10−5; Fig. 3f). The mobility of broken ends in H2AX or NBS1-depleted cells was identical to that in control cells containing a DSB, further suggesting that these factors do not affect the motion of DSBs (Fig. 3f). We conclude that Ku80 contributes to constraining the local motion of broken chromosome ends.
The observed global positional stability of broken chromosome ends supports the contact-first model. A key prediction of this model is that translocation partners are in close spatial proximity before undergoing a rearrangement4. We directly tested this prediction by analysing the relative spatial positioning of the ISceI array and its preferential translocation partner. Using spectral karyotyping (SKY) analysis of Ku80-knockdown metaphase cells, a recurrent translocation (2 out of 20 metaphases) was identified between the dicentric chromosome 3, containing the ISceI array, and a t(8:17) present in the parental cell line giving rise to t(dic3, 8, 17) (Fig. 4a). No other translocations involving the dicentric chromosome 3 were observed. As predicted by the contact-first model, visualization of the relative positions of the translocation partners in the interphase nucleus demonstrated that dicentric chromosome 3 and t(8:17) associated significantly more frequently with each other than a non-translocating control chromosome (Fig. 4b). A cluster of at least one copy of chromosomes 3, 8 and 17 was detected in 63% of cells, and chromosomes 3 and 8 were in close spatial proximity in 72% of cells (Fig. 4a, b). In contrast, chromosome 19, which was never observed translocating with dicentric chromosome 3, associated in only 40% of cells with chromosome 3, despite the presence of four copies of chromosome 19 in the parental cells (P <0.001; Fig. 4a, b). Together with the observed positional immobility of single DSBs, these results strongly suggest that translocations preferentially occur among spatially proximal regions of the genome.
Figure 4.
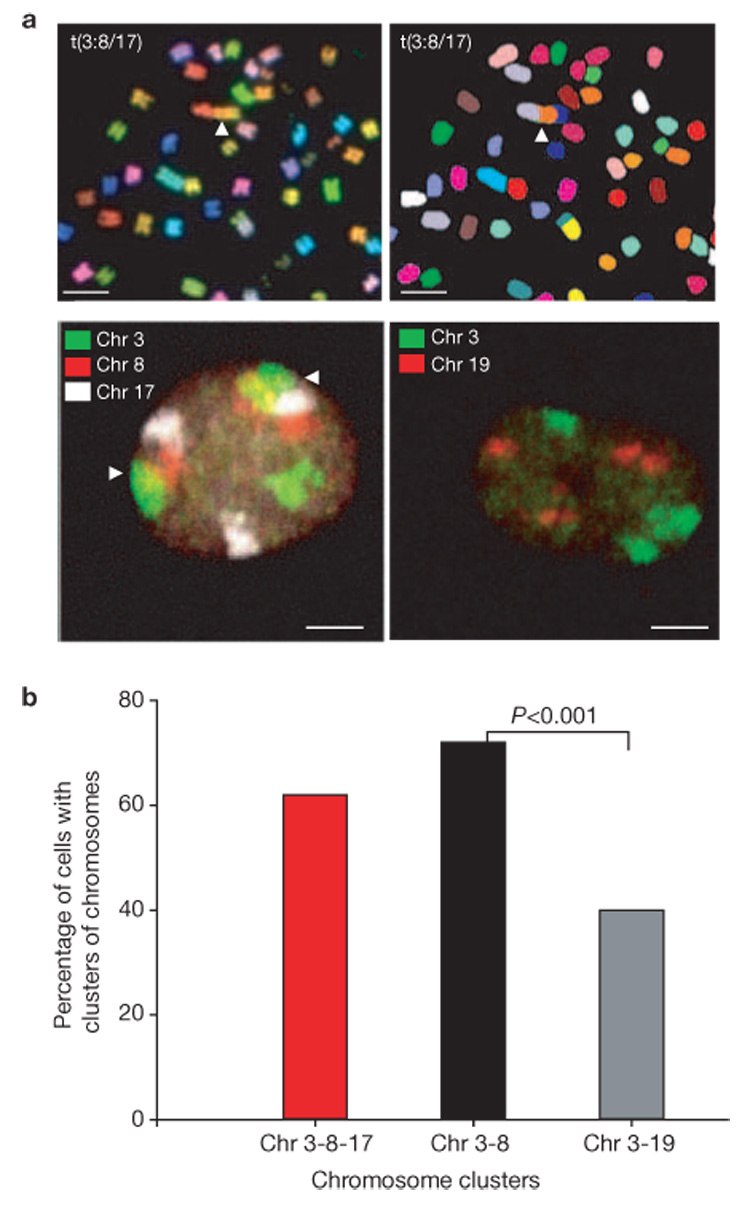
Spatial proximity of preferential translocation partners. (a) SKY analysis of Ku80-depleted NIH2/4 cells is shown in the upper panels. Arrowheads indicate the recurrent t(3:8/17). Visualization of the relative position by interphase FISH of dicentric chromosome 3 to chromosome 8 and t(8:17), or as a control to chromosome 19, is shown in the lower panels. Arrowheads indicate proximal pairs. Proximity was defined as physical touching of pixels representing distinct chromosomes. The scale bars represent 5 µm. (b) Quantification of the percentage of NIH2/4 cells containing proximal chromosome pairs. P values were obtained by x2 test.
We have developed an experimental system that allows study of single broken chromosome ends in living cells, under physiological conditions, in real time. Using this system we demonstrate that distinct broken chromosome ends are positionally stable, exhibit only small-scale local motion and undergo illegitimate joining with chromosomes in their spatial proximity. This behaviour is distinct from that observed in S. cerevisiae, where broken chromosome ends located on distinct chromosomes undergo long-range motion and coalesce into shared repair factories13. The seemingly higher mobility of broken ends in S. cerevisiae is in line with fundamental differences in higher-order genome organization compared with mammalian cells, whereby S. cerevisiae loci are able to explore relatively larger fractions of the nucleus because of the dramatically smaller-sized nuclei in yeast. In addition, the yeast genome seems to lack functionally confined chromosome territories of the type observed in mammalian cells12. The positional stability of specific DSBs observed here agrees with the behaviour of X-ray, UV- and γ-irradiated genome regions in mammalian cells7,8, but is in contrast with the large-scale motion of bulk damaged-DNA regions reported after α-particle irradiation6. Given our observations in a more physiological setting, it must be considered that previously observed large-scale motions may be the consequence of extensive DNA damage and do not reflect a physiological response.
High-precision tracking of tagged broken chromosome ends shows that broken chromosome ends separate slightly from each other, but their long-range motion is severely constrained. End separation did not merely reflect decondensation of the local chromatin around a DSB, but represents a true migration of the ends away from each other. The local mobility of broken ends was dependent on the presence of Ku80, extending into living cells the hypothesis, based on structural observations, that Ku80 forms an asymmetric ring around the two broken ends and functions to align broken chromosome termini at the site of repair24. Future studies will address what other factors or nuclear structures contribute to constraining the motion of broken ends and how their dynamic properties affect repair efficiency.
Our observations have direct implications for understanding how translocations form in vivo and they support the contact-first theory that translocations generally occur between chromosomes that are in spatial proximity at the time of breakage. These results are consistent with the emerging notion that non-random higher-order spatial organization of chromosomes contributes to determining the formation of recurrent translocations4. In support of this hypothesis, several frequent translocation partners including Myc–Igh, Bcl–Abl and RAR–PML have been found to be preferentially positioned in close spatial proximity relative to each other in normal cells before formation of translocations4. Our observation of spatial proximity of a preferred translocation partner after induction of a defined DSB confirms and extends those correlative studies. The formation of chromosomal translocations through misjoining of proximally positioned genome regions is also in agreement with the observed correlation between the degree of intermingling of neighbouring chromosomes and translocation frequencies28. The observation of positional immobility of broken chromosome ends explains why these proximally positioned gene loci are frequent partners in chromosomal translocations and suggests that the non-random spatial positioning of genomes is a significant contributor to determining translocation frequencies.
METHODS
Cell culture and transfections
To generate the NIH2/4 stable cell line or the H2AX−/− lac–ISceI–tet containing stable cells line, NIH 3T3 and H2AX−/− MEFs cells were transfected using Lipofectamine 2000 with the lac–ISceI–tet plasmid and the pTRE2hyg vector (BD Bioscience, San Jose, CA) at a ratio of 10:1. The cells were selected with 400 µg ml-1 hygromycin (BD Biosciences) for 2 weeks. Stable lines were retained in the same concentration of selection medium. siRNAs targeted against NBS1, Ku80, H2AX, SMC1, MRE11 and RAD50 (Dharmacon smart pools; Dharmacon, Lafayette, CO; see Supplementary Information, Methods for sequences) were transfected using Lipofectamine 2000 and 48 h later the cells were further transfected with the appropriate DNA plasmids using the Nucleofector kit for immortalized cell lines (Amaxa, Gaithersburg, MD). Transfection with the RFP–ISceI–GR chimera was performed in cells cultured in DMEM supplemented with 10% stripped serum (Tet system approved; Clontech, Mountain View, CO) and the concentration of triamcinolone acetonide (Sigma, St Louis, MO) added was 10−7 M.
Immunofluorescence and confocal microscopy
Indirect immunofluorescence microscopy was performed as previously described29. After fixation in 4% buffered paraformaldehyde in PBS for 10 min at room temperature, cells were immunolabelled using specific antibodies against γH2AX (anti-phospho H2AX S139, clone JBW103; Upstate, Charlottesville, VA), MDC1 (ref. 30), H2AX (Novus, Littleton, CO), 53BP1 (NB100-304A-1, Novus), Ku80 (M-20, sc-1485; Santa Cruz, Santa Cruz, CA), NBS1 (ref. 27), SMC1 (100-2004, Novus), RAD51 (PC130, Oncogen, Cambridge, MA), MRE11 (ref. 31), RPA (LabVision, Freemont, CA) and RAD50 (MA1-23269; ABR, Princeton, NJ). Cells were mounted using Vectashield and observed on a LSM 510 META confocal microscope using a 100× 1.4NA objective.
Ligation-mediated (LM)-PCR
Genomic DNA (1 µg) purified from NIH2/4 cells was ligated with 100 pmol of an asymmetric adaptor (GCATCACTACGATGTAGGATG and CATCCTACATCGTAGTGATGCTTAT) at 15 °C for 14 h. PCR was performed with the specific adaptor primer LM–ISCE: CATCCTACATCGTAGTGATGC and a primer from the lac repeats Lac-R: TGTGGAATTGTGAGGGGATA. The PCR products were normalized to a standard curve created by the LM-PCR products from known ratios of naked DNA cleaved by ISceI in vitro to uncut DNA (100%, 50/50%, 25/75%, 10/90%, 1/99%), as previously described17. In vivo cleavage frequencies were determined by normalizing for transfection efficiency.
Cell-cycle synchronization
NIH2/4 cells were synchronized at the G1–S boundary by double-thymidine block and in G0–G1 by serum starvation. For thymidine block, cells were incubated in 2 mM thymidine for 14 h, washed extensively with PBS and released for 8 h followed by a second thymidine block for 14 h and released. For serum starvation, cells were plated in medium containing 0.1% serum for 72 h. Transfection was performed, when indicated, before the second thymidine block and 48 h after the serum deprivation. To prevent cells from progressing to mitosis, NIH2/4 cells were arrested in G1 and G2 by treatement with 100 µm olomoucine (Calbiochem, San Diego, CA) for 48 h. Cell-cycle arrest was confirmed by FACS analysis. Transfection was performed 24 h after the addition of olomoucine.
BrdU incorporation assay
NIH2/4 cells on coverslips in G0–G1 or released from G1–S arrest were incubated with 1000× BrdU (Amersham, Piscataway, NJ) for 30 mins and were fixed in 4% PFA. Coverslips were subsequently denatured in 70% formamide with 2× SSC at pH 7 for 2 min at 80 °C followed by immunofluorescence microscopy with the anti-BrdU antibody (Axyll, Bethesda, MD).
Chromosome paint and FISH in metaphase spreads
NIH2/4 cells were incubated in colcemid (KaryoMAX; Gibco, Carlsbad, CA) for 3 h, swollen in pre-warmed 50 mM KCl for 30 min at room temperature, fixed in methanol:acetic acid (3:1), and air dried on slides overnight. Dual colour FISH was performed with a chromosome-painting probe specific for mouse chromosome 3 and lac–ISceI–tet plasmid DNA. The latter was isolated and labelled with biotin–dUTP after standard nick translation. The chromosome-painting probe for mouse chromosome 3 was labelled with spectrum orange. The chromosome-painting probe and the FISH probe were coprecipitated in 50% formamide, 20% Dextran sulphate and 2× SSC, and hybridized to slides containing metaphase spreads for 48 h at 37 °C. For detection, slides were washed three times in 50% formamide in 2× SSC at 45 °C, three times in 0.1× SSC at 60 °C, and incubated for 45 min at 37 °C with avidin–FITC (1:200) diluted in 4× SSC, 0.1% Tween20. The slides were washed three times in 4× SSC, 0.1% Tween20 at 45 °C, stained with DAPI for 5 min and preserved in mounting media. The imaging was done on an upright Nikon Eclipse E800 using a 100× Apo 1. 4NA objective.
Time-lapse microscopy
Live-cell imaging was performed using a TE300 Nikon Eclipse optical sectioning microscope with a 100× 1.4NA objective, a Cool Snap Camera (Photometrics, Tuscon, AZ) and CFP (430/25, 470/30) and YFP (500/20, 535/30) filter sets (86002v2; Chroma Technology, Rockingham, VT). Three-dimensional image stacks of 16 optical slices separated by 0.3 µm were collected every 30 s for 1–2 h, controlled by Metamorph software. Exposure time was 100–500 ms (using 10% ND filter) for each channel and each frame. Three-dimensional time-lapse sequences were analysed with custom-written spot-tracking software, as described previously32.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to: T. Karpova and M. Kruhlak for help with the microscopy; E. Martinez for providing reagents; S. Mabon for technical assistance; E. Brunet for the help with LMPCR; T. Voss, K. Meaburn and all members of the Misteli laboratory for discussions. Imaging was performed at the National Cancer Institute (NCI) Fluorescence Imaging Facility. E.S. was supported by a fellowship from the Human Frontiers Science Program (HFSP). J.D. is a fellow of the Roche Research Foundation. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), NCI, Center for Cancer Research and by the NIH grant GM 68956.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Note: Supplementary Information is available on the Nature Cell Biology website.
References
- 1.Kanaar R, Hoeijmakers JH, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 2.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 3.Elliott B, Jasin M. Double-strand breaks and translocations in cancer. Cell. Mol. Life Sci. 2002;59:373–385. doi: 10.1007/s00018-002-8429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin. Cancer Biol. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikiforova MN, et al. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science. 2000;290:138–141. doi: 10.1126/science.290.5489.138. [DOI] [PubMed] [Google Scholar]
- 6.Aten JA, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 7.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 8.Kruhlak MJ, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisby M, Antunez de Mayolo A, Mortensen UH, Rothstein R. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle. 2003;2:479–483. [PubMed] [Google Scholar]
- 10.Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome frag-mentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 11.Kaye JA, et al. DNA breaks promote genomic instability by impeding proper chromo-some segregation. Curr. Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 12.Haber JE, Leung WY. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc. Natl Acad. Sci. USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nature Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 14.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez ED, Rayasam GV, Dull AB, Walker DA, Hager GL. An estrogen receptor chimera senses ligands by nuclear translocation. J. Steroid Biochem. Mol. Biol. 2005;97:307–321. doi: 10.1016/j.jsbmb.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 16.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villalobos MJ. Detection of DNA double-strand breaks and chromosome translocations using ligation-mediated PCR and inverse PCR. Methods Mol. Biol. 2006;314:109–121. doi: 10.1385/1-59259-973-7:109. [DOI] [PubMed] [Google Scholar]
- 18.Vazquez J, Belmont AS, Sedat JW. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 2001;11:1227–1239. doi: 10.1016/s0960-9822(01)00390-6. [DOI] [PubMed] [Google Scholar]
- 19.Gerlich D, et al. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 20.Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys. J. 1999;77:2871–2886. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Celeste A, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassing CH, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 23.Franco S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nature Rev. Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 25.Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- 26.Wyman C, Kanaar R. Chromosome organization: reaching out to embrace new models. Curr. Biol. 2002;12:R446–R448. doi: 10.1016/s0960-9822(02)00941-7. [DOI] [PubMed] [Google Scholar]
- 27.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 28.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soutoglou E, et al. The nuclear import of TAF10 is regulated by one of its three histone fold domain-containing interaction partners. Mol. Cell Biol. 2005;25:4092–4104. doi: 10.1128/MCB.25.10.4092-4104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee AC, Fernandez-Capetillo O, Pisupati V, Jackson SP, Nussenzweig A. Specific association of mouse MDC1/NFBD1 with NBS1 at sites of DNA-damage. Cell Cycle. 2005;4:177–182. doi: 10.4161/cc.4.1.1354. [DOI] [PubMed] [Google Scholar]
- 31.Difilippantonio S, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nature Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 32.Thomann D, Dorn J, Sorger PK, Danuser G. Automatic fluorescent tag localization II: Improvement in super-resolution by relative tracking. J. Microsc. 2003;211:230–248. doi: 10.1046/j.1365-2818.2003.01223.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


