Abstract
Mammalian cells respond to DNA double-strand breaks (DSBs) by recruiting DNA repair and cell-cycle checkpoint proteins to such sites. Central to these DNA damage response (DDR) events is the DNA damage mediator protein MDC1. MDC1 interacts with several DDR proteins, including the MRE11–RAD50–NBS1 (MRN) complex. Here, we show that MDC1 is phosphorylated on a cluster of conserved repeat motifs by casein kinase 2 (CK2). Moreover, we establish that this phosphorylation of MDC1 promotes direct, phosphorylation-dependent interactions with NBS1 in a manner that requires the closely apposed FHA and twin BRCT domains in the amino terminus of NBS1. Finally, we show that these CK2-targeted motifs in MDC1 are required to mediate NBS1 association with chromatin-flanking sites of unrepaired DSBs. These findings provide a molecular explanation for the MDC1–MRN interaction and yield insights into how MDC1 coordinates the focal assembly and activation of several DDR factors in response to DNA damage.
Keywords: DNA damage, CK2, MDC1, NBS1, MRN, checkpoint
Introduction
To counter the threat to the genome from DNA-damaging agents, cells use the DNA damage response (DDR): a set of events involving activation of DNA repair mechanisms and cell-cycle ‘checkpoint' signalling (Kastan & Bartek, 2004). DNA double-strand breaks (DSBs)—the most cytotoxic DNA lesions—activate the protein kinase ataxia telangiectasia mutated (ATM) to phosphorylate the carboxy-terminal tail of histone H2AX in the vicinity of the break (Stucki & Jackson, 2006). This chromatin modification is crucial for the relocalization of several proteins to sites flanking DSBs, generating ionizing radiation-induced foci (IRIF) that promote efficient repair and sustained DNA damage signalling (Fernandez-Capetillo et al, 2004; Stucki & Jackson, 2006).
A core IRIF component is the mediator of the DNA-damage checkpoint protein 1 (MDC1; Goldberg et al, 2003; Lou et al, 2003; Stewart et al, 2003; Stucki et al, 2005). MDC1 colocalizes with phospho-H2AX (γH2AX) owing to direct interactions between the C-terminal twin BRCT (BRCT2) domains of MDC1 and the γH2AX phospho-epitope (Stucki et al, 2005). MDC1 then mediates IRIF formation by other DDR factors such as p53 binding protein 1 (53BP1), breast cancer protein 1 (BRCA1) and the MRE11–RAD50–NBS1 (MRN) complex (Goldberg et al, 2003; Stewart et al, 2003). We and others have recently shown that a cluster of repeated consensus ATM phosphorylation sites in MDC1 is crucial for the recruitment of BRCA1 and 53BP1 to IRIF, but not for NBS1 focus formation (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007). On phosphorylation, these motifs are bound by the FHA domain of the ubiquitin E3 ligase ring-finger protein 8 (RNF8), which then generates ubiquitin conjugates at sites of DSBs that mediate BRCA1 and 53BP1 recruitment. Although MDC1 was shown to directly bind to MRN some years ago (Goldberg et al, 2003), the molecular basis of how MDC1 recruits MRN to IRIF has remained elusive. Here, we show that a cluster of repeated motifs in MDC1, which are phosphorylated by casein kinase 2 (CK2), are bound by MRN through phospho-dependent interactions with NBS1 that require its amino-terminal FHA and BRCT2 domains. Furthermore, we show that this MDC1–NBS1 interaction is crucial for the targeting and retention of NBS1 on chromatin-flanking DNA DSBs.
Results
Phosphorylated SDTD repeats of MDC1 bind to MRN
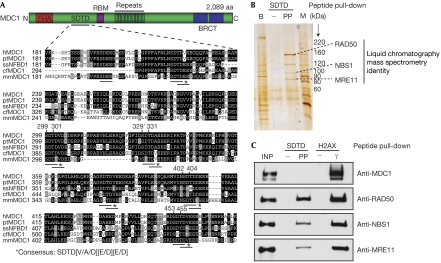
A search of published databases for the post-translational modifications of MDC1 identified many phosphorylation sites; however, two studies (Beausoleil et al, 2004; Olsen et al, 2006) identified eight sites within a cluster of short-repeat sequences that shared the consensus motif Ser-Asp-Thr-Asp (SDTD) in which both the serine and threonine are phosphorylated. This SDTD motif is repeated six times between Ser 218 and Asp 455 in human MDC1, and similar evolutionarily conserved motifs exist in MDC1 orthologues in other vertebrates (Fig 1A).
Figure 1.
Phosphorylated MDC1 SDTD motifs bind to MRN. (A) Domain architecture of MDC1 and alignment of the SDTD region from MDC1 proteins in human (hMDC1), chimpanzee (ptMDC1), pig (ssNFBD1), dog (cfMDC1) and mouse (mmMDC1). RBM indicates RNF8-binding motifs. Identical and similar residues are boxed in black and grey, respectively. CK2 consensus sites are underlined, and residues identified as phosphorylated (Beausoleil et al, 2004; Olsen et al, 2006) are numbered (′phosphorylated residues identified in both studies). (B) Silver-stained SDS–polyacrylamide gel of an SDTD peptide pull-down. PP, S329 T331 doubly phosphorylated peptide and −, its non-phosphorylated equivalent; B, bead-interacting proteins removed from extracts in a pre-clearing step; M, molecular weight markers. (C) Immunoblot of SDTD and histone H2AX phosphopeptide-interacting proteins. INP, input (5% of total); γ, phosphorylated H2AX peptide. CK2, casein kinase 2; MRN, MRE11–RAD50–NBS1; SDTD, Ser–Asp–Thr–Asp motif.
To investigate whether the SDTD motifs might interact with other DDR components, we generated a synthetic peptide that corresponded to MDC1 residues 325–340 bearing dual phosphorylation on Ser 329 and Thr 331. This peptide and its unphosphorylated equivalent were coupled to beads and used to retrieve interacting proteins from HeLa cell nuclear extracts. This approach identified four prominent protein bands on a silver-stained gel that bound in a phosphorylation-dependent manner (Fig 1B). Mass spectrometric analysis identified three of these as MRE11, RAD50 and NBS1. We noted that control binding assays with the γH2AX phospho-peptide retrieved MRN and MDC1 from nuclear extracts, whereas the phosphorylated SDTD peptide bound MRN only (Fig 1C). This indicates that the phosphorylated MDC1 SDTD peptide does not interact with MRN indirectly by bridging contacts with MDC1.
CK2 phosphorylation of MDC1 mediates MRN binding
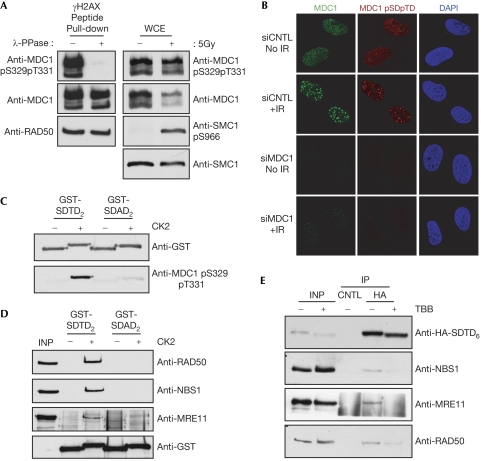
To study phosphorylation of the MDC1 SDTD region and to investigate its regulation, we raised a rabbit polyclonal antiserum against a synthetic peptide corresponding to one of the MDC1 SDTD repeats phosphorylated on Ser 329 and Thr 331 (supplementary Fig S1A online). This detected MDC1 isolated from HeLa nuclear extracts, indicating that MDC1 S329/T331 phosphorylation is present in proliferating human cells (Fig 2A, left panels). The signal from this antibody was lost when MDC1 precipitated with γH2AX peptide-coupled beads was treated with phosphatase, confirming its phospho-specificity (Fig 2A, left panels; detection of RAD50 indicates equal protein loading). Interestingly, although γ-irradiation of osteosarcoma (U2OS) cells caused little change in the amount of MDC1 S329/T331 phosphorylation, it reduced the overall levels of MDC1 (Fig 2A, right panels). This suggests that, after DNA damage, S329/T331-phosphorylated MDC1 is more stable than the unmodified protein. Consistent with these data, S329/T331-phosphorylated MDC1 was detected by immunofluorescence in both control and γ-irradiated cells (Fig 2B; note the absence of staining in cells treated with MDC1 small interfering RNA (siRNA)), confirming that SDTD-phosphorylated MDC1 is present in the absence of damage and also forms IRIF. Notably, we did not detect any cell-cycle-dependent alteration of MDC1 S329/T331 phosphorylation relative to total MDC1 protein content by using flow cytometry, which suggests that SDTD phosphorylation occurs throughout interphase (supplementary Fig S1B online).
Figure 2.
Casein kinase 2-dependent phosphorylation of MDC1 SDTD motifs induces interaction with MRN. (A) Left panels: SDTD peptide interacting proteins, isolated from HeLa nuclear extracts (as in Fig 1C), were subjected to phosphatase or mock treatment, and then analysed with the indicated antibodies. Right panels: whole-cell extracts (WCE) from osteosarcoma (U2OS) cells 1 h after 5 Gy irradiation or control cells were immunoblotted with the indicated antibodies. (B) At 72 h after control (CNTL) or MDC1-targeting siRNA, control cells or cells incubated for 2 h following 5 Gy of X-rays were processed for immunofluorescence with MDC1 and MDC1 pS329pT331 antibodies. (C) Immunoblot analysis of purified GST-SDTD2 and GST-SDAD2 after in vitro phosphorylation by recombinant CK2, or mock treatment. (D) A 250 ng portion of purified GST-SDTD2 or GST-SDAD2 was mock treated or phosphorylated by CK2 in vitro and then incubated with HeLa nuclear extracts. GST fusions and interacting proteins were captured on glutathione-Sepharose beads and immunoblotted. (E) Expression plasmids encoding HA-SDTD6 were transfected into U2OS cells. After 48 h, cells were treated with 4,5,6,7-tetrabromo-benzimidazole (TBB; 75 μM; Sigma-Aldrich (Poole, Dorset, UK), T0826) or dimethyl sulphoxide (DMSO) for 6 h. Extracts were immunoprecipitated (IP) with monoclonal antibodies against HA or GFP (CNTL) and immunoblotted. CK2, casein kinase 2; DAPI, 4′,6–diamidino–2–phenylindole; GFP, green fluorescent protein; GST, glutathione S-transferase; HA, haemagglutinin; INP, input; MRN, MRE11–RAD50–NBS1; SDAD, Ser–Asp–Ala–Ser motif; SDTD, Ser–Asp–Thr–Asp motif; siRNA, small interfering RNA.
The MDC1 SDTD motif does not conform to consensus sequences targeted by ATM or other DDR kinases. Instead, both the serine and threonine residues in the SDTD sequence match the CK2 consensus motif of phosphorylated serine/threonine followed by an acidic residue at the +3 position (p[S/T]xxD/E). Consistent with this, a bacterially expressed protein (glutathione S-transferase (GST)-SDTD2) encompassing two MDC1 SDTD motifs (contained within amino-acid residues 296–340 of MDC1) was efficiently phosphorylated by recombinant CK2 in vitro, as shown by a marked decrease in electrophoretic mobility and detection by anti-MDC1 pS329/pT331 antiserum (Fig 2C, upper and lower panels, respectively). By contrast, the equivalent protein bearing alanine substitutions of Thr 301 and Thr 331 (GST-SDAD2), while being phosphorylated by CK2 to some degree (presumably on CK2 consensus serine residues retained in the SDAD motifs present in this fusion protein), was detected only weakly with the anti-MDC1 pS329/pT331 antiserum (Fig 2C). Moreover, although CK2-mediated phosphorylation of the GST-SDTD2 protein allowed it to retrieve MRN from HeLa nuclear extracts, the CK2-treated GST-SDAD2 mutant protein was unable to bind to MRN (Fig 2D).
To investigate whether CK2 phosphorylation of the MDC1 SDTD region promotes MRN binding in vivo, we transiently expressed a haemagglutinin (HA)-tagged MDC1 SDTD fragment coupled to a nuclear localization sequence in U20S cells (HA-SDTD6; supplementary Fig S1C online). Immunoblotting showed that treating these cells with the CK2 inhibitor 4,5,6,7-tetrabromo-benzimidazole increased the electrophoretic mobility of the MDC1 SDTD fragment and reduced the ability of the fragment to co-immunoprecipitate MRN (Fig 2E).
NBS1 directly binds to CK2-phosphorylated MDC1
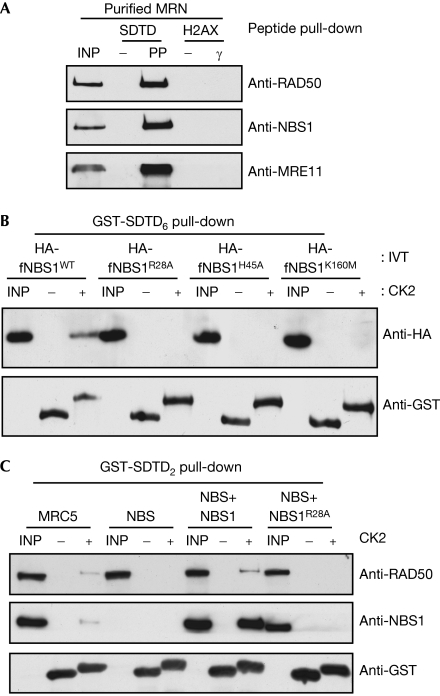
Next, we used the recombinant, purified MRN complex in peptide pull-down experiments. These showed that MRN bound to the phosphorylated but not the unphosphorylated form of the SDTD peptide—confirming the direct nature of the interaction—yet did not bind to either version of the H2AX C-terminal peptide (Fig 3A). This indicates the specificity of MRN for the phosphorylated MDC1 SDTD motif and also shows that if, as previously reported, NBS1 binds to γH2AX directly (Kobayashi et al, 2002), this interaction must be less stable than MRN binding to the phosphorylated SDTD motif.
Figure 3.
Binding of MRN to MDC1 phospho-SDTD motifs requires the NBS FHA and BRCT2 domains. (A) MRN interacts directly with phosphorylated SDTD peptides. SDTD and H2AX peptide-coupled beads were incubated with 250 ng of recombinant human MRN, and interacting proteins were analysed by western blotting. INP, input (20%); PP, MDC1 S329/T331 double phosphorylated peptide; γ, phosphorylated H2AX peptide; −, non-phosphorylated equivalent peptides. (B) A 200 ng portion of CK2-phosphorylated or mock-phosphorylated GST-SDTD6 was incubated in pull-down reactions with 25 μl of in vitro-translated (IVT) HA fusions corresponding to amino-acid residues 1–348 of NBS1 (HA-fNBS1), or analogous proteins bearing point mutations predicted to abolish either FHA (R28A/H45A) or BRCT2 (K160M) phosphorylation-dependent interactions. INP, input (20%). (C) A 2.0 mg portion of cell extracts prepared from MRC5, NBS (ILB1) or NBS (ILB1) fibroblasts stably expressing NBS1 or NBS1R28A was incubated with GST-SDAD2 treated as in (B). GST fusions were retrieved (as in Fig 2C) and interacting proteins were immunoblotted. INP, input (2.5%). CK2, casein kinase 2; GST, glutathione S-transferase; HA, haemagglutinin; MRN, MRE11–RAD50–NBS1; SDTD, Ser–Asp–Thr–Asp motif.
Within MRN, only NBS1 contains phosphorylation-specific interaction domains: an N-terminal FHA domain followed directly by a BRCT2 domain (Durocher et al, 2000; Manke et al, 2003; Yu et al, 2003; Becker et al, 2006). To investigate whether these domains bind to the phosphorylated SDTD motif of MDC1, we expressed an N-terminal fragment of NBS1 containing these motifs (residues 1–343; HA-fNBS1) in rabbit reticulocyte lysates (despite many attempts using a wide range of expression constructs, we were unable to express soluble recombinant versions of the FHA or FHA/BRCT2 domains of NBS1 in bacteria). We also generated versions of this NBS1 region in which either the FHA domain (fNBS1R28A and fNBS1H45A) or the BRCT2 region (fNBS1K160M) was mutated to abolish phospho-dependent interactions. These proteins were then tested for binding to an MDC1 fragment encompassing the SDTD repeats (GST-SDTD6) that had or had not been treated with CK2 (Fig 3B). Significantly, the wild-type NBS1 fragment bound specifically to CK2-phosphorylated—but not unphosphorylated—GST-SDTD6, whereas this binding was abolished by mutation of the FHA or BRCT2 domain of NBS1 (Fig 3B). Furthermore, CK2-phosphorylated GST-SDTD2 retrieved NBS1 and RAD50 from both MRC5- and NBS1-complemented Nijmegen breakage syndrome (NBS) cell extracts, but did not mediate such interactions when the FHA domain mutant of NBS1 was expressed in NBS cells (Fig 3C; see supplementary Fig S2 online for expression levels). Taken together, these results indicate that the integrity of both the FHA and BRCT2 domains of NBS1 is needed for binding of NBS1 to the phosphorylated SDTD region of MDC1.
MDC1 SDTD motifs retain MRN at DNA DSB sites
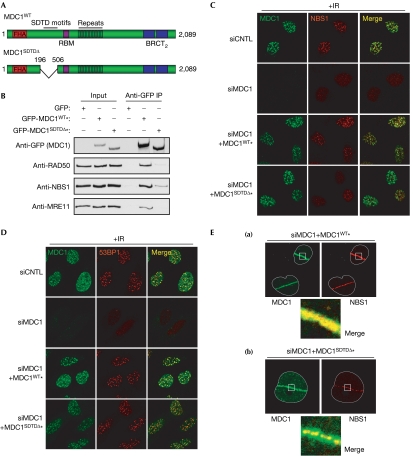
An analysis of the recruitment dynamics of fluorescently tagged proteins to laser-induced DNA damage showed similar recruitment kinetics for NBS1 and MDC1, and highlighted the importance of the FHA domain of NBS in this recruitment (Lukas et al, 2004). Therefore, we thought that the MDC1–MRN interaction, which depends on the FHA/BRCT2 domains of NBS1, might promote retention of the MRN complex in IRIF. To test this, we used siRNA to deplete endogenous MDC1 in U2OS cells stably expressing siRNA-resistant wild-type MDC1 (MDC1WT) or an MDC1 mutant without the SDTD region (MDC1SDTDΔ; Fig 4A; supplementary Fig S3A online). The wild-type MDC1 derivative efficiently co-immunoprecipitated MRN at physiological salt concentrations, whereas only low levels of MRN were recovered in immunoprecipitates of the MDC1SDTDΔ mutant (Fig 4B), consistent with the SDTD region being the principal MRN interaction interface. This residual interaction with MRN shown by the MDC1SDTDΔ mutant raises the possibility of other MDC1–MRN interactions, and would be consistent with previous findings (Goldberg et al, 2003); however, it is noteworthy that this weaker interaction was undetectable under higher salt conditions (supplementary Fig S3B online). Moreover, although both MDC1WT and MDC1SDTDΔ efficiently formed IRIF, MDC1WT, but not MDC1SDTDΔ, supported IRIF formation by NBS1 (Fig 4C; see supplementary Fig S3C online for quantification). By contrast, IRIF formation by 53BP1 and BRCA1 was normal after expression of either form of MDC1 (Fig 4D; supplementary Fig S4A online). This is consistent with 53BP1 and BRCA1 requiring the RNF8-binding motifs of MDC1 that are still intact in the MDC1SDTDΔ mutant (Fig 4A), and shows that the SDTD region of MDC1 specifically mediates IRIF formation by NBS1 by promoting efficient MDC1–MRN interactions.
Figure 4.
The MDC1 SDTD–MRN interaction recruits NBS1 to damaged chromatin. (A) SDTD deletion relative to the MDC1 domain architecture. (B) Indicated expression constructs were transfected into human embryonic kidney 293 cells. After 48 h, extracts were prepared, immunoprecipitated (IP) with GFP antibodies and immunoblotted. Immunoprecipitations and washes were performed at 150 mM salt; INP, input (5%). (C,D) Indicated osteosarcoma (U2OS) cell lines were treated with two rounds of control (CNTL) or MDC1-targeting siRNA for 72 h. Cells were then treated with 5 Gy of X-rays and processed for immunofluorescence 4 h later with MDC1, (C) NBS1 or (D) 53BP1 antibodies (non-irradiated cells are shown in supplementary Fig S4B,C online). (E) U2OS cells stably expressing siRNA-resistant MDC1wt (a) or MDC1SDTDΔ (b) were treated with siRNA against MDC1 as above, and then the nuclei were subjected to laser micro-irradiation. After 30 min, cells were pre-extracted with detergent, fixed and immunostained with antibodies against MDC1 and NBS1. Asterisks indicate resistance to MDC1 siRNA. GFP, green fluorescent protein; MRN, MRE11–RAD50–NBS1; SDTD, Ser–Asp–Thr–Asp motif; siRNA, small interfering RNA.
To examine further the NBS1 recruitment defect of cells expressing MDC1SDTDΔ, we used the ‘laser scissors' technique to generate DNA DSBs along defined sub-micrometre tracks in live cells (Limoli & Ward, 1993). In MDC1WT-expressing cells, the extent of NBS1 recruitment closely resembled that of MDC1 (Fig 4Ea). By contrast, although MDC1SDTDΔ recruitment was indistinguishable from that of the MDC1WT protein, the NBS1 recruitment pattern was altered. Thus, in MDC1SDTDΔ cells, although NBS1 was recruited to the laser path, its pattern no longer resembled the broad distribution of MDC1; instead, NBS1 was restricted to a discrete micro-focal pattern lying within the broader track of MDC1 staining (Fig 4Eb). This indicates that SDTD phosphorylation of MDC1 is required to retain NBS1 on γH2AX-coupled chromatin, but that NBS1 might recognize other structures induced by laser microirradiation independently of MDC1. Consistent with these results, and in line with our finding that the FHA domain of NBS1 is required for MDC1 SDTD binding, similar micro-focal patterns of NBS1 recruitment were observed in MDC1-depleted cells and in cells in which the FHA domain of NBS1 was mutated (Lukas et al, 2004).
Discussion
MDC1 binds to γH2AX flanking unrepaired DNA DSBs, and then acts as a ‘mediator' to recruit other DDR factors to such sites (Stucki & Jackson, 2006). For example, the ATM-phosphorylated TQXF motifs on MDC1 are recognized by the ubiquitin E3 ligase enzyme RNF8 (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007). RNF8 then generates ubiquitin adducts on histone H2A—and possibly other chromatin components—which lead to IRIF formation by 53BP1 and BRCA1 (Huen et al, 2007; Mailand et al, 2007). This paper describes how MDC1 facilitates the accumulation and retention of the MRN complex in IRIF, indicating a new mechanism for MDC1 mediator function. Specifically—and in contrast to the situation for 53BP1 and BRCA1—our work highlights how phospho-dependent interactions between MDC1 and MRN might precede the detection and signalling of DNA damage. This characteristic of MDC1–MRN binding might therefore allow this complex to be efficiently and rapidly recruited to sites of DNA damage, thus ensuring that the DDR is launched without delay. By contrast, the ATM- and RNF8-dependent nature of 53BP1 and BRCA1 recruitment means that it is likely to happen more slowly, possibly to ensure that the chromatin alterations and other processes triggered by these proteins are invoked only at the right time and in the correct chromatin context. This model—in which various factors are recruited with different kinetics and factor dependencies—suggests a more complex, hierarchical and highly regulated DDR than has hitherto been apparent.
Together with data obtained from proteomics screens (Beausoleil et al, 2004; Olsen et al, 2006), our findings indicate that CK2 phosphorylates several sites on MDC1, often at dual phosphorylation sites within the SDTD motif. These findings are in agreement with previous work showing that CK2 has an impact on various DDR events. For example, it was previously shown that CK2-dependent phosphorylation of the adaptor/mediator proteins XRCC1 and XRCC4 promotes the recruitment of polynucleotide kinase (PNK), Aprataxin, and Aprataxin- and PNK-like factor (APLF) to sites of chromosomal damage to facilitate the repair of DSBs and DNA single-strand breaks, respectively (Clements et al, 2004; Loizou et al, 2004; Iles et al, 2007). Interestingly, in these studies, CK2 was found to mediate factor recruitment through direct interactions between the CK2 phosphorylation sites on XRCC1/XRCC4 and the FHA domains of PNK, Aprataxin and APLF. It is noteworthy that the NBS1 recruitment defects we observed in MDC1SDTDΔ mutant cells are similar to those reported for cells containing NBS1 with mutations in its FHA domain (Lukas et al, 2004), suggesting that the FHA domain of NBS1 binds to MDC1. Although our data support this idea, the fact that mutations in either the FHA or BRCT2 domain of NBS1 impair the NBS1–MDC1 interaction suggests that both domains are intimately involved in making such contacts. It might be that these domains are orientated such that they are able to simultaneously bind to both the phosphorylated serine and the phosphorylated threonine of a single SDTD motif, or to two adjacent SDTD motifs in the MDC1 protein. This dual-interaction mode might provide a greater degree of binding selectivity and discrimination than would be possible by phospho-dependent interactions involving either domain alone. Alternatively, it is possible that the proximity of the FHA and BRCT2 domains of NBS1 makes them conformationally inter-dependent, so that mutation of one affects the structure and function of the other. The tightly interconnected FHA/BRCT2 domain architecture is conserved in virtually all known NBS1 counterparts (Becker et al, 2006), pointing to this relationship being of crucial functional importance. Resolution of the above issues will await structural determinations of the N terminus of NBS1, both alone and in a complex with the phosphorylated MDC1 SDTD motif.
After DNA damage, we have consistently observed stabilization of the CK2-phosphorylated MDC1 relative to the total MDC1 pool, which is downregulated. This is consistent with a recent study showing 26S proteasome-dependent turnover of the mediator proteins topoisomerase II binding protein 1 (TOPBP1), Claspin, 53BP1 and MDC1 following DNA damage (Zhang et al, 2006). Whether CK2 phosphorylation itself, or MRN binding to the phosphorylated SDTD motifs, protects a sub-pool of MDC1 from proteolytic degradation remains to be determined. It is also noteworthy that CK2 phosphorylations on XRCC1 mediate binding to PNK, Aprataxin and APLF (Clements et al, 2004; Loizou et al, 2004; Iles et al, 2007) and, conversely, that FHA domain-containing proteins such as PNK, Aprataxin and APLF are able to bind to CK2 phosphorylation sites on both XRCC1 and XRCC4. This raises the possibility that the phosphorylated SDTD regions of MDC1 might interact with proteins in addition to NBS1, and that the FHA/BRCT2 region of NBS1 might have more than one physiological target.
Finally, a recent study of MDC1-deficient mouse cells complemented with various MDC1 domain-deletion constructs showed the central repeat region of MDC1 and FHA and BRCT2 domains are important for DSB repair by sister-chromatid recombination (Xie et al, 2007). This work also showed that MDC1-dependent recruitment of BRCA1 and 53BP1 to IRIF was not required for this repair, or for 53BP1-dependent DSB repair by non-homologous end-joining. So far, a functional role for the marked redistribution of DDR factors to regions surrounding DSBs has not been defined. Perhaps future studies using the MDC1 separation-of-function mutants created in this study will help to explain the different and possibly overlapping functional contributions made by recruiting BRCA1, 53BP1 and the MRN complex to γH2AX-associated chromatin.
Methods
See the supplementary information online for details.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Note added in proof. While this work was under revision, two related papers have been published: Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J (2008) Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol 181: 213–226; Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M (2008) Constitutive phosphorylation of MDC1 physically links the MRE11–RAD50–NBS1 complex to damaged chromatin. J Cell Biol 181: 227–240.
Supplementary Material
Supplementary Materials
Acknowledgments
We thank members of the Jackson Laboratory for input on the manuscript, M. Stucki for communications, C. Green for critical reading of the manuscript, T. Paull for purified MRN, and S. Polo and R. Hunter for assistance with laser scissors and flow cytometry, respectively. This work was supported by grants from Cancer Research UK and the European Union.
Footnotes
The authors declare that they have no conflict of interest.
References
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA 101: 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E, Meyer V, Madaoui H, Guerois R (2006) Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics 22: 1289–1292 [DOI] [PubMed] [Google Scholar]
- Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW (2004) The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair 3: 1493–1502 [DOI] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB (2000) The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell 6: 1169–1182 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of the genome. DNA Repair 3: 959–967 [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP (2003) MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421: 952–956 [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles N, Rulten S, El-Khamisy SF, Caldecott KW (2007) APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol 27: 3793–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K (2002) NBS1 localizes to γ-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol 12: 1846–1851 [DOI] [PubMed] [Google Scholar]
- Kolas NK et al. (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli CL, Ward JF (1993) A new method for introducing double-strand breaks into cellular DNA. Radiat Res 134: 160–169 [PubMed] [Google Scholar]
- Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, Caldecott KW (2004) The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 117: 17–28 [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Wu X, Chen J (2003) MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421: 957–961 [DOI] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J (2004) Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J 23: 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB (2003) BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP (2006) γH2AX and MDC1: anchoring the DNA- damage-response machinery to broken chromosomes. DNA Repair 5: 534–543 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Xie A et al. (2007) Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell 28: 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J (2003) The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ (2006) A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126: 529–542 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials