Abstract
Although invasive plant species often have a hybrid ancestry, unambiguous evidence that hybridization has stimulated the evolution of invasive behaviors has been difficult to come by. Here, we briefly review how hybridization might contribute to the colonization of novel habitats, range expansions, and invasiveness and then describe work on hybrid sunflowers that forges a direct link between hybridization and ecological divergence. We first discuss the invasion of Texas by the common sunflower and show that the introgression of chromosomal segments from a locally adapted species may have facilitated range expansion. We then present evidence that the colonization of sand dune, desert floor, and salt marsh habitats by three hybrid sunflower species was made possible by selection on extreme or “transgressive” phenotypes generated by hybridization. This body of work corroborates earlier claims regarding the role of hybridization in adaptive evolution and provides an experimental and conceptual framework for ongoing studies in this area.
Keywords: Colonization, Helianthus, Hybridization, Introgression, Invasiveness, QTLs, Range expansion
Introduction
The enormous ecological and financial costs of invasive species (Pimental et al. 2000) have led to a recent burst of research interest in invasion dynamics. These dynamics are typically understood in the context of two sets of traits: those of the recipient community and those of the invading species (Sakai et al. 2001). For example, community species diversity has often been linked to susceptibility to invasion (Levine and D’Antonio 1999), and traits such as short generation time and rapid growth are often associated with invasive behavior of individual species (Baker and Stebbins 1965). Here, we focus on the issue of what makes one species more invasive than another, given similar community contexts.
In plants, certain life-history features are more likely to be found in invasive than in non-invasive species. They include short generation time, fast growth, general habitat requirements, self-compatibility, and small seeds with efficient dispersal mechanisms (Baker and Stebbins 1965). The correlation of characteristic life history traits with weediness is reflected in r versus K strategies (Pianka 1970) and R-C-S life history schemes, where Ruderal species maximize allocation to reproduction, Competitive species maximize allocation to growth, and Stress-tolerant species grow slowly but have high survival (Grime 1977). In a noteworthy pair of studies on life history correlates of invasiveness, four traits were found to be highly correlated with invasiveness in the woody genus Pinus: short time between large seed crops, small seeds, a short juvenile period, and high relative growth rate (Rejmanek and Richardson 1996; Grotkopp et al. 2002). However, many weedy species have alternate traits (Williamson 1996) and other species with these traits are not invasive (Erlich 1989).
In addition to identifying the attributes determining invasive success, it is important to understand how species acquire these attributes in the first place. Such knowledge could help predict which species may pose a threat and suggest strategies for the management of both demonstrated invasives and potentially invasive new introductions. While many species may arrive in a novel habitat preadapted for success, post-arrival processes may be equally important in determining invasiveness. These processes, including hybridization, in situ selection, and founder effects, have received increasing attention in an invasion context (Abbott et al. 2003; Ellstrand and Schierenbeck 2000; Lee 2002). Our goal is to briefly summarize evidence linking hybridization to the colonization of new habitats and increased invasive behavior in plants, focusing on lessons derived from a well-studied system, the annual sunflowers (Helianthus).
Hybridization and the evolution of invasive plants
Hybridization is a widespread and evolutionarily important phenomenon in plants (Stace 1975; Rieseberg and Wendel 1993; Ellstrand et al. 1996; Arnold 1997). When related species or races grow in sympatry and hybridization occurs, genes from one species or race may be added to the gene pool of the other by hybridization and backcrossing or “introgressive hybridization.” Alternatively, new hybrid gene combinations may become established through allopolyploidy, clonal or asexual reproduction, selfing, or diploid hybrid speciation (Barton 2001). Regardless of mechanism, if the hybrid gene combinations enhance growth and competitive ability in novel environments, invasive success may be increased (Harlan and De Wet 1963; Lewontin and Birch 1966; Small 1984; Stebbins 1985; Abbott 1992; Galatowitsch et al. 1999; Kim and Rieseberg 1999; Ellstrand and Schierenbeck 2000; Blumler 2003). Of course, in many instances hybridization will have no impact on invasiveness.
While other mechanisms leading to the success of invasive plants clearly occur, and numerous invasive species have no history of hybridization, interspecific gene flow may be an important factor in the success of many invasive plants, especially in closely related crop-weed complexes (Langevin et al. 1990). In a recent review, Ellstrand (2003) found that 22 of the 25 most important food crops hybridize with wild relatives in at least part of their range. One example is hybridization of maize with teosinte (Zea mexicana), a weed in agricultural fields in Mexico. Crop-wild gene flow has resulted in forms of the weed with morphological features that are typical of the crop (mimicry) (Wilkes 1977; Barrett 1983), which makes weed control extremely difficult.
Hybridization of weedy species need not be limited to crop species and may occur with other related weed species, non-weedy introduced species, and native species (Abbott 1992; Stebbins 1985; Rieseberg et al. 1990a; Ellstrand and Schierenbeck 2000; Arnold 2004). Some of the best examples come from the genus Helianthus (see below). In Britain, a novel radiate form of Senecio vulgaris apparently originated by introgressive hybridization with the radiate species S. squalidus and has become widely distributed (Abbott et al. 2003). It is unlikely that the radiate character is directly related to the success of the hybrid but may serve as a marker for other adaptive traits. Baker (1972) suggested that weedy radishes (Raphanus sativus) have arisen through repeated hybridization with cultivated forms and related weed species. Rhododendron ponticum tolerates a wider temperature range in Britain than in its native mainland Europe, perhaps as a result of hybridization with an introduced North American species (Abbott 1992; Milne and Abbott 2000). For all of these cases, however, the link between hybridization and increased invasive behavior has not been definitively established, and more work on mechanisms is clearly needed.
Hybridization may promote the persistence, aggressiveness and ecological amplitude of invasive plant populations through three distinct mechanisms.
Heterosis
Hybrid offspring may have increased vigor resulting from high levels of heterozygosity (Stebbins 1985). Possible examples are an array of Spartina hybrids currently invading several continents (Ainouche et al. 2003). In a famous case in Britain, hybridization between the native S. maritima and introduced S. alternifolia resulted in an allotetraploid (S. anglica). While heterosis has not been confirmed, the allotetraploid is a vigorous invader that has displaced the parental species in certain coastal habitats (Gray et al. 1991) and has colonized entirely new habitats (Thompson 1991). Likewise, two new Tragopogon allopolyploids have become invasive and are replacing their parents in parts of the Pacific Northwest U.S.A (Soltis et al. 2004). Other well characterized instances th at involve clonal diploid hybrids include a hybrid Tamarix genotype that now dominates many riparian areas in the western U.S. (Gaskin and Schaal 2002) and a hybrid water milfoil (Myriophyllum), which is an aggressive aquatic weed in the northeastern U.S. and upper Midwest (Moody and Les 2002). In the absence of processes that fix heterozygous genotypes, such as allopolyploidy, apomixes, and clonal spread, heterosis will be transitory. However, the high frequency of these processes in the plant kingdom suggests that hybridization has been historically important, and that heterosis can be stabilized.
Increased genetic variation
Hybridization may increase genetic variation in hybridizing populations, providing a larger pool of raw material for adaptive evolution (Anderson 1949; Anderson and Stebbins 1954; Stebbins 1959; Rattenbury 1962; Raven 1976; Sun and Corke 1992; Neuffer et al. 1999; Ainouche et al. 2003). Increased invasiveness might be due to augmented levels of genetic variability alone and/or to the fixation of novel combinations of genes and phenotypes (Stebbins 1969; Arnold 1997; Rieseberg et al. 1999a; Thompson 1991).
There are several ways in which novel gene combinations or phenotypes may arise and become established (Ellstrand and Schierenbeck 2000). For example, the new hybrid lineages may be fixed for trait values that are intermediate between the parental species, as has been reported for Carprobotus hybrids that are invading coastal habitats of California (Weber and D’Antonio 1999) and in the allopolyploid Spartina anglica (Thompson 1991). Hybrids may also recombine the traits of their parents. This appears to have happened in Iris (Burke et al. 2000; Johnston et al. 2001), in which the homoploid hybrid species Iris nelsonii (adapted to shady, freshwater habitats) appears to be a novel recombinant for ecological tolerances that differentiate its parental species (shade/saltwater and sun/freshwater). Finally, hybrids may display extreme or “transgressive” traits. A review of 171 studies of segregating plant and animal hybrids indicates that transgression is nearly ubiquitous (Rieseberg et al. 1999a). Only 15 of the 171 studies failed to report a transgressive trait, and 44% of 1,229 traits examined were transgressive. Although most transgressive hybrids will be poorly adapted to local environmental conditions, some may exhibit greater adaptation than one or the other parental species. For example, Clausen and Hiesey (1958) found that a small number of F2 progeny of intra specific hybrids of Potentilla glandulosa had significantly greater fitness than either parent over an altitudinal gradient. Patterns of herbivore resistance may also exhibit a transgressive pattern, in which some hybrids are more resistant to natural enemies than the parental species (Strauss 1994; Fritz et al. 1999). This may be a consequence of novel secondary compounds produced by hybridization (Orians 2000).
While transgression is typically associated with segregating diploid hybrids, extreme phenotypes often arise in allopolyploid lineages as a consequence of changes in genome size, heterosis (Thompson 1991; Stebbins 1985), and selection on post-polyploid genomic changes (Pires et al. 2004). As a consequence, polyploids often are able to colonize habitats that are more extreme than those of their parental species and allopolyploidy is frequently associated with invasiveness (Stebbins 1985).
Reduced genetic load
Ellstrand and Schierenbeck (2000) speculate that hybridization may provide a mechanism by which populations may purge accumulated genetic load. The rationale is that small or isolated natural populations are unlikely to become fixed for deleterious alleles at the same loci. Thus, hybridization offers a means to exchange deleterious alleles for neutral or advantageous counterparts from the alternative population or species. Possibly, the fitness boost associated with reduced load might contribute to invasiveness, but this has not been shown.
Range limits
Invasions and range expansions are qualitatively similar processes, although there are differences as well. For example, the average invasion probably has to bridge a larger change in environmental conditions than do range expansions, invasions often involve acute transitions to novel environments while colonizations may be more gradual, and invasions are less likely to be hampered by gene flow from ancestral populations than colonizations. Nonetheless, many of the factors that limit the range of a species are also likely to limit invasions, so parallels between the two processes are worth exploring.
Both ecological and genetic factors may serve to limit the geographical and/or ecological distribution of a species (Marshall 1968; Silander and Antonovics 1979; Levin and Clay 1984; Hoffmann and Blows 1994). In some cases, range is limited by dispersal. That is, appropriate habitat may exist beyond the normal range of a species, but the species may fail to occur there because of insufficient dispersal. In some instances, there may be a lack of necessary genetic variation to permit evolutionary changes allowing range expansion. Alternatively, negative genetic correlations between adaptive traits at the species margin can prevent adaptive evolution. A gain in one trait leads to a loss in another so there is no net gain. Finally, gene flow from central populations can swamp out genetic differentiation at the margin (Antonovics 1976; Kirkpatrick and Barton 1997).
In some cases the spread of plant species is related to environmental changes such as rainfall patterns or human-caused disturbances (Harper 1977), whereas in other instances range expansions may take place as a result of evolutionary changes in populations that increase adaptation to environmental conditions previously inhibitory to population persistence and growth. Hybridization may provide genetic material necessary for such evolutionary changes in range-limiting traits like competitive ability, cold or drought tolerance, photoperiodicity, and disease resistance (Prince et al. 1985; Abbott 1992). For example, hybrid violets have colonized polluted areas where neither parental species can persist (Neuffer et al. 1999). Such changes are not limited to plants. Lewontin and Birch (1966) suggested that hybridization between two fruit flies in Australia might have introduced genes increasing physiological adaptation to extreme temperatures, allowing one species to greatly increase its range over the past 120 years.
The objective of the present paper is to discuss the role of hybridization as an evolutionary mechanism leading to increased geographical range, ecological amplitude, and/or the colonization of new habitats in wild sunflower (Helianthus) species. These species represent an excellent system for investigating the role of hybridization in range expansion and adaptation to novel habitats, in part because they build on more than half a century of studies, first by Heiser (1947, 1965) and more recently by Rieseberg et al. (1988, 2003). Insights from these studies are described below.
Hybridization in sunflowers
The role of hybridization in the diversification of the North American sunflowers (Helianthus) was initially explored by Charles Heiser and his students (Heiser 1947, 1949, 1951a, b; Heiser et al. 1969). Heiser documented the occurrence of natural hybrids between the widespread, common sunflower, H. annuus and several of its congeners, including H. argophyllus (Heiser 1951a), H. bolanderi (Heiser 1949), H. debilis (Heiser 1951b), and H. petiolaris (Heiser 1947). This hybridization, Heiser argued, allowed H. annuus to acquire favorable alleles from the locally adapted species, thereby increasing its ecological amplitude and facilitating range expansion. In some instances, hybridization was believed to have contributed to the formation of introgressive races such as H. annuus ssp. texanus in Texas (Heiser 1951b) and H. bolanderi ssp. bolanderi in the California (Heiser 1949) or new species such as H. neglectus (Heiser et al. 1969).
More recently, Rieseberg and colleagues re-examined these hypotheses using a combination of molecular marker surveys and molecular phylogenetic analyses (Dorado et al. 1992; Rieseberg et al. 1988, 1990a, b, 1991a, b; Rieseberg, 1991). These studies verified much of Heiser’s work, including the occurrence of natural hybridization between H. annuus and each of the species listed above, as well as the introgressive origin of H. a. texanus. On the other hand, the molecular evidence failed to support a hybrid origin for H. neglectus or H. b. bolanderi (Rieseberg et al. 1988). Thus, while the morphological criteria employed by Heiser accurately identified contemporary hybrid swarms or zones, their utility in the detection of ancient, stabilized hybrid lineages was limited. This was not unexpected, as it has long been recognized that morphological intermediacy may sometimes result from processes other than hybridization (Gottlieb 1972).
In addition to verifying or refuting putative examples of hybrid lineage formation, molecular phylogenetic analyses detected three previously unsuspected hybrid species (Fig. 1): H. anomalus, H. deserticola, and H. paradoxus (Rieseberg et al. 1990b, 1991a; Rieseberg 1991). The three hybrids appear to be derived from the same parental species, H. annuus and H. petiolaris. However, there are subtle differences in the parental chloroplast and nuclear ribosomal DNA haplotypes found in each hybrid, suggesting that each was independently derived.
Fig. 1.
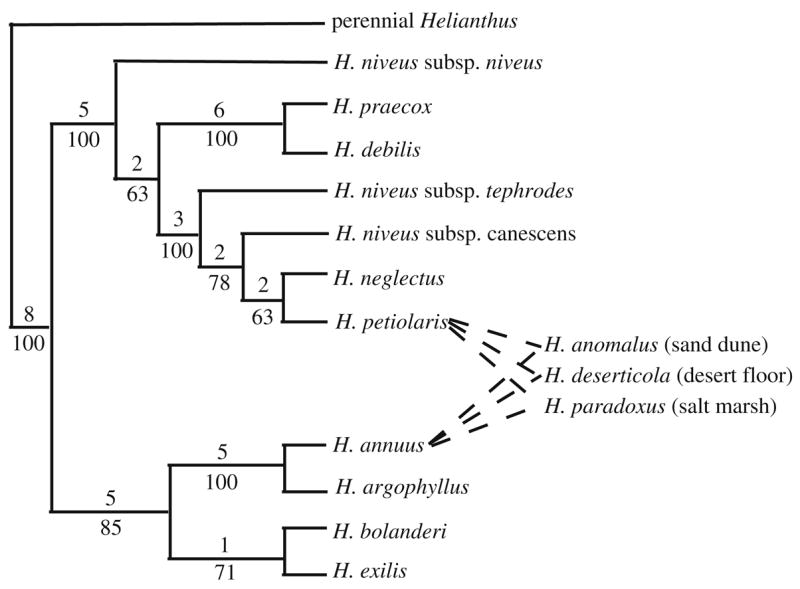
Phylogenetic tree for Helianthus section Helianthus based on combined chloroplast DNA and nuclear ribosomal DNA data (Rieseberg 1991). The number of mutations are given above and bootstrap percentages below each branch. Dashed lines indicate parentage of homoploid hybrid species
Over the past decade, these four well-established examples of hybrid lineage formation (H. a. texanus, H. anomalus, H. deserticola, and H. paradoxus) have been employed to investigate the role of hybridization in range expansion, ecological divergence, the colonization of new habits, and the evolution of invasiveness. Note that the origin of H. annuus ssp. texanus provides an example of a new geographic race formed through introgressive hybridization, whereas the remaining three hybrid lineages represent good biological species that appear to have originated through recombinational speciation (sensu Grant 1981), where chromosomal differences between the parental species are recombined to form a new homokaryotype. These two different kinds of hybrid lineage formation will be discussed separately, since the gene flow from the parental species is likely to hinder local adaptation and ecological divergence in introgressive races, but not in recombinational species that are strongly isolated by chromosomal sterility barriers (Rieseberg et al. 1995b; Lai et al. 2005).
Introgressive hybridization and range expansion of the common sunflower
The common sunflower, H. annuus, is both the progenitor of the domesticated sunflower and a major weed in corn, soybean, wheat, and sugar beets in the central U.S.A. (Al-Khatib et al. 1998). As described above, Heiser (1965) argued that H. annuus has been able to expand its ecological amplitude and geographic range by introgression with locally adapted native species. The best-studied example of this process involves the origin of H. a. texanus in Texas (Heiser 1954). This subspecies is restricted to human-disturbed areas in Texas, suggesting it was most likely recently introduced, perhaps by native Americans (Heiser 1951b). It is fully interfertile with other forms of H. annuus, but morphologically it approaches H. debilis ssp. cucumerifolius, a sunflower native to southeastern Texas. Hybridization is common when the two taxa come into contact, and while first generation hybrids are mostly sterile, averaging 6.7% viable pollen, pollen viability increases rapidly in the backcross generations (BC1: 3–67%; BC2: 20–98%; Heiser 1951b), suggesting that introgression can occur despite the highly reduced fertility of the F1 plants. Based on these observations, Heiser (1951b) postulated that H. annuus was able to colonize Texas by “capturing” advantageous alleles from H. debilis (Fig. 2).
Fig. 2.
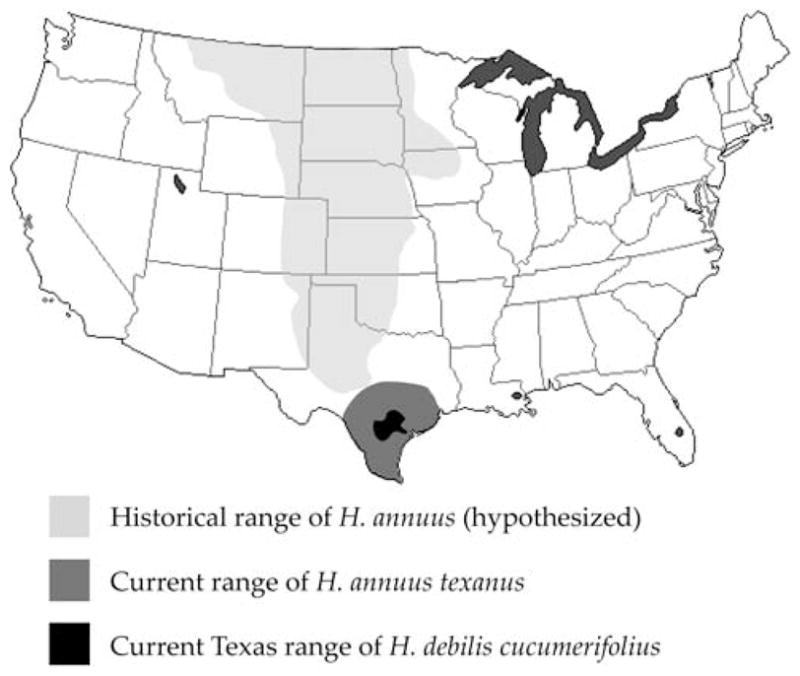
Geographic distributions of H. debilis ssp. cucumerifolius (Texas distribution only; Rogers et al. 1982) and H. annuus ssp. texanus (Heiser 1951b), and the postulated distribution of the common sunflower, H. annuus, prior to the colonization of North America by humans
Molecular data are consistent with Heiser’s hypothesis in that the introgression of presumably neutral nuclear ribosomal and chloroplast DNA markers has been documented between the species (Rieseberg et al. 1990a). However, this does not prove that the introgression was adaptive or contributed to the evolution of increased invasiveness. To address these concerns, we have conducted series of genetic and ecological studies to (1) locate chromosomal segments containing quantitative trait loci (QTLs) for important phenotypic traits and hybrid sterility; (2) assess whether any of these segments have moved from H. debilis into natural populations of H. a. texanus; and (3) test the fitness effects of these chromosomal segments in H. a. texanus habitat in southern Texas. These experiments may not only provide an answer to a very old question about the role of introgressive hybridization in the origin of invasive taxa, but they may also enable us to identify the particular trait(s) and QTLs that trigger invasions of weedy plant species following introgression with native, adapted species.
Quantitative trait locus studies
To identify QTLs underlying morphological differences between H. annuus and H. debilis and hybrid sterility, we generated a first generation backcross (BC1) mapping population between H. debilis and an inbred cultivated line of H. annuus (Kim and Rieseberg 1999). This allowed us to examine the segregation of H. debilis markers and traits against a homozygous H. annuus genetic background. Analysis of 226 BC1 progeny detected 56 QTLs for 15 morphological traits and two QTLs for pollen sterility; the pollen sterility QTLs presumably correspond to the two major chromosomal translocations that differentiate the species (Chandler et al. 1986).
Although the QTL data cannot tell us whether the introgression of adaptively significant genetic material has occurred, they can tell us whether it is plausible. For adaptive trait introgression to be successful, alleles contributing to an adaptation must integrate into a new genetic background before the chromosomal segment with which they are associated is eliminated by selection (Barton and Hewitt 1985). If many factors contribute to reduced hybrid fitness, then much of the genome may be resistant to introgression due to linkage. The simple genetic architecture of the sterility barrier between H. annuus and H. debilis is fully consistent with the adaptive introgression hypothesis: very few sterility QTLs were detected and only four of the 56 morphological QTLs were closely linked (<10 cM) to a sterility factor. Thus, much of genome, including most major morphological QTLs, should be able to move freely across the sterility barrier.
Introgression of QTLs
To further test the introgressive hybridization hypothesis, 14 natural populations of H. a. texanus (N = 153 individuals) were assayed for 15 mapped molecular markers specific to H. debilis (unpublished data). Our initial focus was on markers that occurred at high frequency in H. debilis and were associated with morphological and sterility QTLs (Kim and Rieseberg 1999) so that we could better explain patterns of introgression and assess whether introgression could account for the morphological convergence of H. a. texanus toward H. debilis. After adjustment for marker frequencies in pure populations of H. debilis, we asked whether all H. debilis-specific markers had introgressed into H. a. texanus at roughly the same rate, a pattern suggestive of neutral introgression, or whether certain markers were significantly over- or under-represented, a pattern most consistent with positive or negative selection, respectively, on QTLs associated with these markers (Table 1).
Table 1.
Frequency of introgressed chromosomal segments of H. debilis in natural populations of H. annuus ssp. texanus
| Linkage Group | Marker | QTLa | % PVE | Adjusted frequency | Direction of selectionb |
|---|---|---|---|---|---|
| 1 | cg.ct135 | Achene length | 8.79 | 0.01 | Negative |
| Achene shape | 11.2 | ||||
| Ligule shape | 6.9 | ||||
| 1 | cg.tg308 | Achene length | 8.79 | 0.37 | Neutral |
| Achene shape | 11.2 | ||||
| ligule shape | 6.9 | ||||
| 2 | gc.ct245 | Achene shape | 8.1 | 0.06 | Negative |
| Achene width | 7.5 | ||||
| Sterility | 38.2 | ||||
| Tooth height | 32.1 | ||||
| 2 | gc.ga140 | Achene length | 5.4 | 0.14 | Negative |
| Ligule shape | 32.7 | ||||
| Ray number | 8.7 | ||||
| 2 | ct.ct218 | Ligule width | 12.9 | 0.57 | Positive |
| 3 | gc.ct52 | Pubescence | 10.4 | 0.03 | Negative |
| 3 | gc.gc125 | Phyllary shape | 9.3 | 0.17 | Neutral |
| Phyllary width | 10.7 | ||||
| 3 | gc.cc209 | Speckled internode | 6.3 | 0.34 | Neutral |
| 7 | cg.ct92 | No QTL | 0.58 | Positive | |
| 8 | gc.ct164 | Disk diameter | 10.5 | 0.29 | Neutral |
| Phyllary shape | 7.4 | ||||
| Pubescence | 7.8 | ||||
| Speckled internode | 27.4 | ||||
| 8 | gc.tc83 | Achene shape | 5.86 | 0.12 | Negative |
| Sterility | 38.8 | ||||
| 8 | cg.gg110 | Pubescence | 7.8 | 0.41 | Neutral |
| Phyllary shape | 7.4 | ||||
| 8 | ct.ct100 | Disk diameter | 10.5 | 1.00 | Positive |
| Pubescence | 7.8 | ||||
| 11 | 1C9 | Speckled internode | 36.5 | 0.02 | Negative |
| 13 | ct.ct133 | Ligule shape | 6.7 | 0.30 | Neutral |
Quantitative trait loci (QTL) and percent variance explained (% PVE) taken from Kim and Rieseberg (1999). Bold type indicates QTL of large effect (% PVE > 25%)
The significance of frequency differences among introgressed markers was tested by generating a 95% confidence interval across all markers surveyed using a Wilcoxon sign rank test. Markers that occurred outside the confidence interval were determined to be under either positive or negative selection
We found that most (12 of 15) H. debilis markers exhibited neutral patterns of introgression or were associated with negatively selected chromosomal segments (Table 1). For example, all three markers on linkage group 3 showed low or neutral levels of introgression (Fig. 3a). Likewise, markers associated with the two pollen sterility QTLs were under-represented in H. a. texanus populations, a pattern consistent with the strong negative selection predicted for these QTLs and closely adjacent chromosomal regions (Table 1). Negative selection associated with sterility QTLs has not, however, inhibited introgression of other loosely linked markers on the same linkage group (Fig. 3b and c).
Fig. 3.
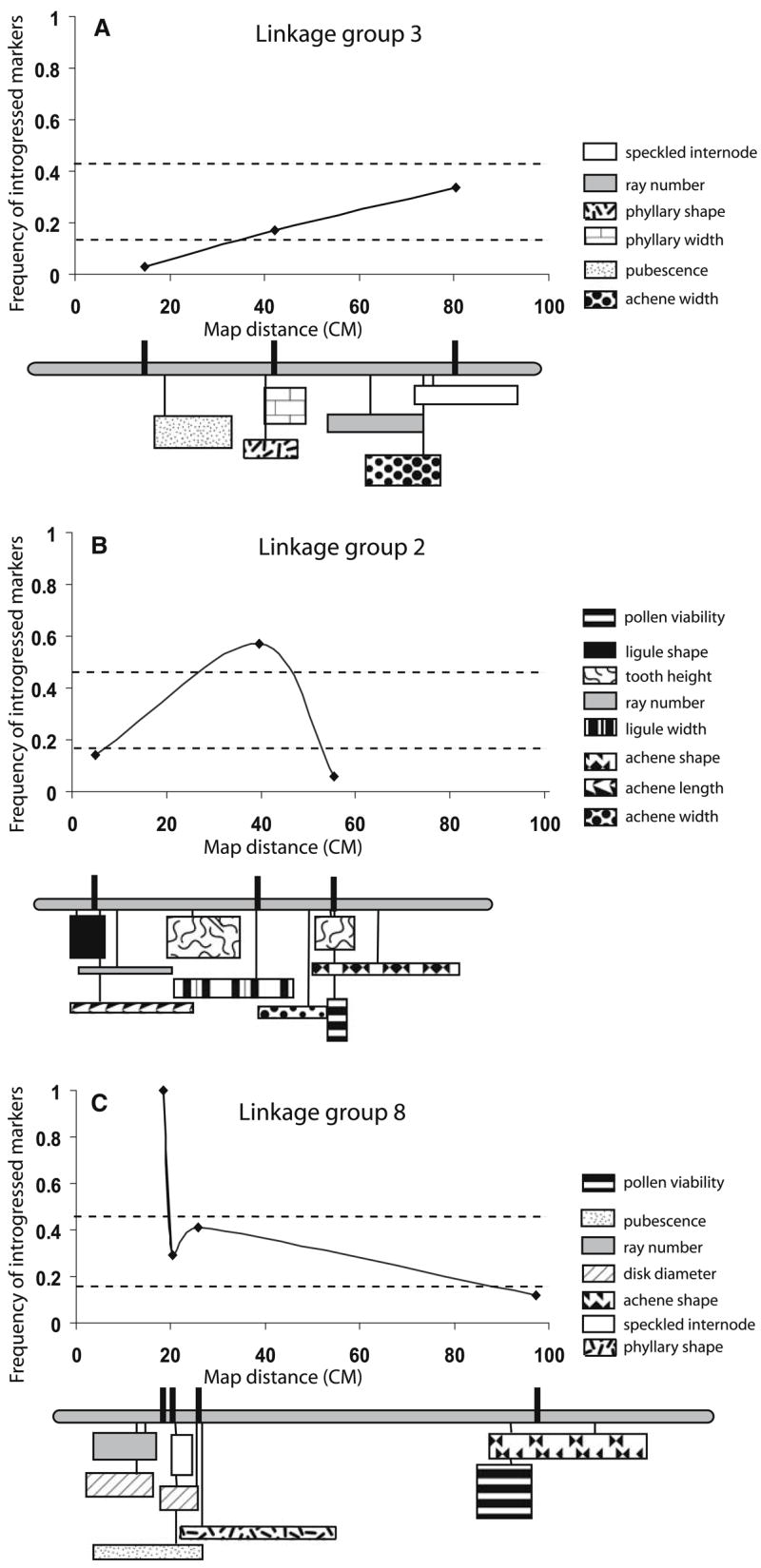
Frequency of introgressed H. debilis alleles plotted against map distance. Dashed line represents 95% confidence intervals. Linkage groups (from Kim and Rieseberg 1999) are drawn to scale below each graph. Markers surveyed are indicated with black hash marks on linkage groups. QTL positions (with one LOD support limits) and their magnitudes (indicated by height) are diagrammed below linkage groups
We also detected three markers that were over-represented in H. a. texanus and thus likely represent H. debilis chromosomal segments that have been under positive selection during the formation of H. a. texanus. This evidence of positive selection is consistent with the adaptive trait introgression hypothesis and supports Heiser’s (1951b) hypothesis regarding the role of H. debilis alleles in the formation of H. a. texanus. Moreover, two of the three positively selected markers were associated with QTLs underlying morphological traits (e.g., the size of floral parts such as ligules and the central disc) that vary in the direction of H. debilis (Table 1). This pattern is expected if the phenotypic convergence of ssp. texanus toward H. debilis were due to introgression. There are two caveats associated with these conclusions, however. First, our statistical analyses are based on comparisons of relative frequencies among introgressed markers, and it is therefore possible that we have over-estimated the number of positively selected markers. Second, introgressed chromosomal segments likely contain many genes, and the QTLs and traits we have identified may not be those actually under selection. However, both concerns may be mitigated by the selection experiments described below.
Selection experiments
Currently, we are investigating whether traits promoting adaptation to the biotic and abiotic environment of south Texas have been transferred from H. debilis to H. annuus. Our approach consists of three steps. First, traits of potential ecological importance that differ between the parents were identified. Second, the fitness consequences of debilis-like trait values were quantified in field populations of hybrid individuals. Finally, QTLs for these traits are being mapped and selection coefficients for debilis-derived alleles will be calculated. We expect that trait values (and QTL alleles) conferring high fitness under current conditions would also have been advantageous to the introgressed ancestors of H. a. texanus during their Holocene colonization of south Texas (Heiser 1951b).
Several ecologically relevant traits have been identified that differ between the parental species in common gardens (unpublished data). Resistance to the seed midge Neolasioptera helianthis was significantly higher in H. debilis than in H. a. annuus. This pest has substantial fitness effects, destroying up to 90% of the seed crop of individual H. a. annuus planted in central Texas. Abiotic tolerance traits also differ; branching architecture was significantly more indeterminate in H. debilis than in H. a. annuus. Indeterminate architecture, which allows plants to add branches and flowers (and therefore seeds) in response to favorable environmental conditions that may occur late in the season, may be a key fitness trait in the unpredictable summer environment of central & southern Texas.
From field performance tests, it is clear that debilis-like trait values can be highly adaptive in hybrids. One thousand H. a. annuus × H. debilis BC1 individuals were synthesized to mimic the hybrid ancestors of H. a. texanus, and were planted in two locations in south Texas in 2003. In preliminary analyses of these populations, hybrid fitness covaried positively with both seed midge resistance and the degree of indeterminate branching. While hybrids exhibited a range of variation in these characters (and mean fitness was low relative to both parents), certain hybrid individuals were highly fit, indicating that hybridization may have increased adaptation to novel conditions and triggered the colonization of south Texas. Ongoing work will test (via QTL mapping) whether these adaptive trait values in hybrids actually result from the presence of H. debilis alleles.
Hybrid speciation and ecological divergence of annual sunflowers
Three mechanisms have been proposed by which a homoploid hybrid may become reproductively isolated from its parental species (Buerkle et al. 2000). First, the new hybrid lineage may diverge karyotypically from its parents through the sorting of chromosomal rearrangements that differentiate the parental species (Stebbins 1957; Grant 1981), and/or by the establishment of new chromosomal rearrangements induced by recombination (Rieseberg et al. 1995b; Lai et al. 2005). Second, hybrid founder events may facilitate hybrid speciation by providing initial spatial isolation for a new hybrid lineage (Charlesworth 1995). Third, a hybrid lineage may colonize a new habitat or niche and thus become ecologically isolated from its parental species (Grant 1981; Gross and Rieseberg 2005). In this section, we focus on the role of hybridization in the ecological divergence of three hybrid sunflower species: Helianthus anomalus, H. deserticola, and H. paradoxus.
The three hybrid species and their parents are self-incompatible, insect-pollinated annuals, with the same chromosome number (n = 17). All five species are native to the continental U.S. The two parental species have widespread and broadly overlapping distributions that are centered in the U.S. Great Plains (Fig. 4). They differ in soil preferences, however, with H. annuus largely restricted to heavy, clay soils, and H. petiolaris to dry, sandy soils. Nonetheless, these two habitats are often found in close proximity in the central and western U.S., resulting in the production of numerous hybrid swarms and hybrid zones. The hybrid zones are narrow, often less than 30 m, and little evidence of introgression is found outside of the hybrid zones, apparently due to the synergistic action of several reproductive barriers (Rieseberg et al. 1995a, 1999b; Schwarzbach et al. 2001; Burke et al. 2004).
Fig. 4.
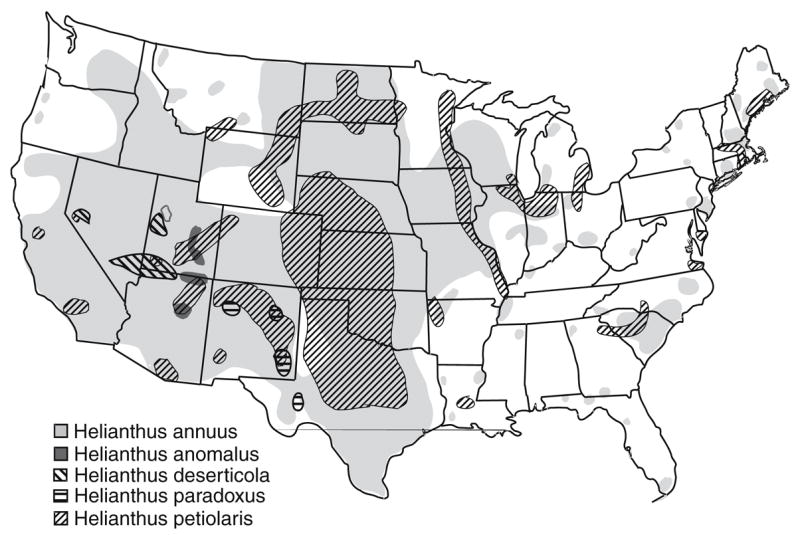
Present-day distributions of the two parental species, H. annuus and H. petiolaris, and their three hybrid derivative species, H. anomalus, H. deserticola, and H. paradoxus (based on Rogers et al. 1982)
The hybrid species are much more limited in geographic distribution than their parents (Fig. 4), with H. deserticola restricted to the Great Basin Desert in Nevada, Utah, and northern Arizona, H. anomalus to a handful of sand dune habitats in Utah and northern Arizona, and H. paradoxus to saline wetlands in western Texas and New Mexico. Despite proximity to parental populations, no natural hybrids have been reported between the three ancient hybrid species and their parents due to a strong chromosomal sterility barrier (Chandler et al. 1986; Rieseberg et al. 1995b; Rieseberg 2000; Lai et al. 2005). In contrast, the three hybrid species are almost completely allopatric to each other, but we have observed hybrids between H. anomalus and H. deserticola at the only site where they co-occur, Little Sahara Recreation Area in Central Utah.
Phylogeographic evidence implies that H. anomalus and H. deserticola have multiple independent origins (Schwarzbach and Rieseberg 2002; Gross et al. 2003), although it is difficult to rule out the alternative hypothesis of a single origin followed by local introgression with parental populations after the hybrid species had expanded its geographic distribution. In contrast, H. paradoxus clearly arose once (Welch and Rieseberg 2002). The timing of the hybrid speciation events appears to be similar between origins and between species, with estimates ranging from 63,000 and 210,000 generations ago (Schwarzbach and Rieseberg 2002; Welch and Rieseberg 2002; Gross et al. 2003).
Given the fairly ancient origin of the three hybrid species, an important question is whether hybridization has facilitated the colonization of extreme habitats, or whether most divergence occurred after speciation and hybridization was incidental to the process. To address this question, we have designed a series of selection, QTL, and comparative mapping studies, which are described below.
Selection experiments (hybrid speciation)
As discussed above, transgressive segregation is common in segregating hybrid populations and provides a potential mechanism for the generation of evolutionary novelty in hybrids. Given that H. anomalus, H. deserticola, and H. paradoxus occur in extreme habitats (sand dunes, desert floor, and salt marsh, respectively) relative to those of their parental species, it is reasonable to ask whether transgressive phenotypes generated by hybridization may have facilitated the colonization of these habitats.
There are several requirements that must be fulfilled to support the supposition that selection on transgressive traits can result in adaptation to a novel habitat. First, the natural hybrid lineage or species should possess traits that are transgressive relative to the parental species. Second, the trait variation present in synthetic hybrids should overlap the extreme traits found in the natural hybrid species. Finally, selection acting on synthetic hybrids in the field should favor the traits found in the hybrid species.
We have conducted parallel selection experiments (sensu Lande and Arnold 1983) in the habitat of all three of hybrid species to evaluate these requirements. In each case, H. annuus (parent), H. petiolaris (parent), and individuals of one of the hybrid species were planted in that hybrid species’ native habitat along with early generation hybrids between H. annuus and H. petiolaris (second generation backcrosses towards both parents; hereafter BC2Ann and BC2Pet). These early generation hybrids served as proxies for the ancestral genotypes of the hybrid species, and the experiments thus roughly recreated the birth of each hybrid species.
Helianthus anomalus (ancient hybrid), a desert sand dune endemic, was positively transgressive for leaf succulence and negatively transgressive for leaf nitrogen (Ludwig et al. 2004). The hybrids overlapped H. anomalus for both traits, although for leaf succulence the overlap was slight and for the BC2Pet population only. Of the selective pressures calculated for transgressive traits, only selection for increased succulence in the BC2Ann population was in the direction of the H. anomalus phenotype (Table 2). Nonetheless, H. anomalus exhibited greater survivorship and reproductive biomass than either parent in the dune habitat. Likewise, the synthetic hybrids exceeded their parents in seed biomass (BC2ann only) or survivorship (BC2pet only).
Table 2.
Selection differentials (S) and gradients (β) on transgressive traits in synthetic hybrid populations of H. annuus × H. petiolaris in the habitat of the natural hybrid species (Lexer et al. 2003b; Gross et al. 2004; Ludwig et al. 2004)
| Habitat type | Trait | BC2ann
|
BC2pet
|
||
|---|---|---|---|---|---|
| S | β | S | β | ||
| H. anomalus: | Leaf succulence | 0.19*** | 0.17*** | –0.14 | –0.09 |
| Leaf N (mg/g) | 0.19* | 0.10 | 0.33*** | 0.19* | |
| H. deserticola: | Leaf area | 0.45*** | −0.32** | 0.30*** | −0.10* |
| Stem diameter | 0.80*** | 1.08*** | 0.62*** | 0.88*** | |
| Flowering date | −0.16*** | 0.23** | −0.23*** | 0.122** | |
| H. paradoxus: | Leaf succulence | – | – | +0.13*** | +0.11* |
| Ca content | – | – | +0.06 | +0.24*** | |
| Na, S, Mg, or B content | – | – | −0.25*** | −0.29*** | |
Only traits under significant selection are shown. Bold indicates selection differentials/gradients are in expected direction relative to the phenotype of the ancient hybrid species
P < 0.05;
P < 0.01;
P < 0.001
Helianthus deserticola (ancient hybrid), found in the xeric environment of the Great Basin Desert, was negatively transgressive for leaf area, stem diameter, and flowering date. Both of the BC2 populations overlapped the transgressive traits of the hybrid species, although the BC2Pet overlapped the flowering time of H. deserticola more extensively than did the BC2Ann. The selection differential for days to flowering and the selection gradient for leaf area were both in the direction of H. deserticola, although the alternate measures of selection (the selection gradient for flowering date and the selection differential for leaf area) were in the opposite direction (Table 2). Selection favored an increased stem diameter in both the BC2 populations, which contradicts the narrow stems of H. deserticola (Gross et al. 2004). Unlike H. anomalus, survivorship and reproductive biomass of H. deserticola did not exceed that of its parents, in part because the latter species is extremely sensitive to transplantation and has an abbreviated life cycle. However, BC2pet had substantially greater fitness than either parent.
Helianthus paradoxus (ancient hybrid) inhabits highly saline desert marshes, perhaps the most extreme habitat of any annual sunflower. The hybrid species was positively transgressive for sulfur and calcium content, leaf shape and leaf succulence, and negatively transgressive for boron content. The BC2Pet population overlapped the H. paradoxus phenotype for all five traits (note that the BC2Ann cross was not used in this experiment). Selection in the field favored higher leaf succulence and calcium, corresponding to the H. paradoxus phenotype (Table 2). Sulfur, boron, magnesium and sodium content were combined into a single principal component for the selection analysis due to the high degree of correlation among these elemental contents. There was strong selection against uptake of these elements, corresponding to the decreased boron content in the hybrid species and the more general expectation of sodium exclusion due to its deleterious effects (Lexer et al. 2003a). Fitness differences between the hybrid and parental species were most pronounced for H. paradoxus. None of the parental plants or synthetic hybrids survived the experiment compared to 90% survivorship for H. paradoxus. The BC2pet progeny were intermediate in fitness between the two parents.
Overall, these field experiments have shown that many of the extreme traits found in the hybrid species could have arisen via habitat-mediated selection acting on transgressive phenotypes in novel habitats (Table 2). These include high leaf succulence in H. anomalus, small leaves and early flowering in H. deserticola, and high leaf succulence, high calcium content, and low uptake of toxic elements in H. paradoxus. However, for some traits we failed to detect significant selection or selection was not in the predicted direction. Such findings might indicate that transgressive segregation did not contribute to the evolution of some traits in the hybrid species. Alternatively, they may be an artifact of the restricted spatial and temporal scale of these exploratory experiments. Given that selection fluctuates over both time and space (Grant and Grant 2002), it is not surprising that selective pressures measured over a single growing season are not fully consistent with predictions based on current phenotypes of the hybrid taxa. Thus, while informative, these experiments also illustrate the importance of replicating such trials across multiple years and multiple sites.
Genetic basis of transgressive segregation
To elucidate the genetic basis of transgressive character expression, QTL studies were performed using progeny from the same interspecific backcross populations employed above. In the most comprehensive study completed to date, analyses of close to 400 greenhouse-grown BC2Pet progeny revealed that QTLs with antagonistic effects (complementary genes) contributed to the expression of 35 of 40 phenotypic traits studied (Rieseberg et al. 2003). This genetic architecture is predicted under the “complementary gene action model”, in which QTLs with effects in opposing directions within each parent may recombine in hybrids, resulting in some hybrids having most or all QTLs with effects in the same direction (deVincente and Tanksley 1993). These individuals will have extreme (transgressive) character values, such as those observed in the phenotypic selection experiments.
Further study indicated that QTL effect sizes, expressed as a percentage of the interspecific phenotypic gap between H. annuus and H. petiolaris, were significantly larger for traits with complementary genes than for traits lacking them (164 ± 46% versus 20 ± 2%, respectively; P < 0.01; Lexer et al. 2005). This makes sense in that complementary genes represent a kind of “cryptic variation” that is not manifest in parental populations, but can be released following recombination in hybrids. Hence QTLs may be large compared to species differences.
Although the complementary gene action model assumes that QTL effects are additive, its explanatory value in sunflower hybrids is high. This is expected because non-additive gene interactions (epistasis) do not appear to contribute substantially to phenotypic differences between these species; epistatic interactions were detected for 18 of the 40 traits studied, but the sizes of the interaction effects were small (mean = 1.2 ± 0.4%; Lexer et al. 2005). Similar results have been reported by Kim and Rieseberg (2001) for another pair of Helianthus species. The situation in Helianthus, where epistatic effects tend to be small relative to main effects, is similar to that reported for most other plant and animal groups (reviewed in Tanksley 1993; Lynch and Walsh 1998), although notable exceptions are known (e.g., Doebley et al. 1995).
A second genetic study of the origin of ecological divergence in wild sunflower hybrids was conducted directly in the extreme salt marsh habitat of the hybrid species H. paradoxus, using the BC2Pet population that was employed in the phenotypic selection experiment (above). This study also detected the presence of complementary genes underlying transgressive phenotypes and showed that QTLs from both species contribute to increased fitness in the salt marsh environment (Lexer et al. 2003b). Notably, selection coefficients for individual salt tolerance QTLs of +0.126, −0.084, and −0.094, were sufficiently large to enable divergence of new diploid hybrid species in the presence of gene flow, assuming Nem estimates typical for wild annual sunflowers (Lexer et al. 2003b).
Genetic correlations
Transgressive segregation and complementary gene action provide a means by which extreme phenotypes may be generated for individual traits. For a hybrid lineage to successfully colonize a new habitat, however, all of the trait differences must be combined into a single individual or genotype, which may be difficult because of linkage and/or pleiotropy for QTLs underlying key traits. Thus, genetic correlations may limit ecological divergence through hybridization.
To determine the role of genetic correlations in the ecological divergence of the hybrid sunflower species, we asked whether closely linked or pleiotropic QTLs in the BC2 populations described above have effects that are in the same direction with respect to the hybrid species phenotype (Rieseberg et al. 2003). These positive correlations would greatly facilitate ecological divergence and hybrid speciation.
We found that closely linked QTLs were indeed positively correlated in direction of effect (P < 0.001 for all species) and that the complex, multi-trait phenotypes of the three hybrid species were recovered at low frequency in synthetic BC2 hybrids: 0.9% for H. anomalus, 6.8% for H. deserticola, and 0.2% for H. paradoxus (Rosenthal et al. 2005). Thus genetic correlations likely facilitated rather than impeded for origins of the three species. Indeed, because linkage and/or pleiotropy is so extensive among the analyzed traits, the number of ecologically relevant multi-trait phenotypes may be limited. This might partially account for why only three new hybrid species have originated from this cross.
Comparative mapping studies
Although studies of synthetic hybrids demonstrate that most of the phenotypic differences associated with each of the hybrid species could have arisen though hybridization, they fail to prove that this is what actually happened. It could be, for example, that the differences arose as a consequence of mutational divergence and that hybridization was incidental to phenotypic evolution. To distinguish between these two hypotheses, we compared the genomic composition of the three ancient hybrid species with predictions from the QTL analyses of the BC2 population of H. annuus × H. petiolaris (Rieseberg et al. 2003). That is, we asked whether the hybrid species had the predicted set of QTL alleles for producing their phenotypes. If hybridization played a key role in phenotypic divergence, then a significant correlation should be found between predicted and actual genomic composition.
Genomic composition of the ancient hybrid species did accord closely with predictions from the QTL analyses (Fig. 5). The QTL data successfully predicted the parentage of 71.8% of mapped markers in H. paradoxus, 75.6% in H. anomalus, and 79.3% in H. deserticola (P ≪ 0.0001 for all comparisons). Thus, hybridization does appear to be largely responsible for the phenotypic divergence of the three hybrid species, and by extension, the colonization of novel habitats.
Fig. 5.
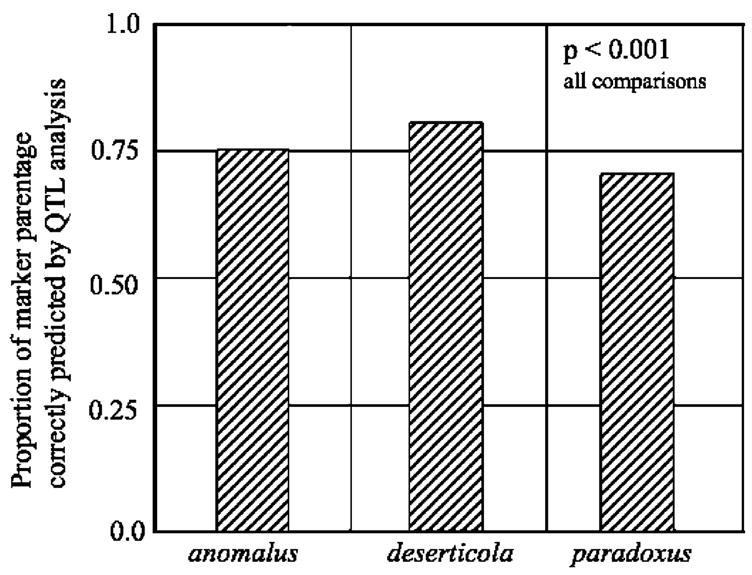
The proportion of mapped molecular markers whose parentage in the ancient hybrid species, H. anomalus, H. deserticola, and H. paradoxus was correctly predicted from a QTL analyses of 40 morphological traits in synthetic hybrids (Rieseberg et al. 2003)
Earlier studies suggested that fertility selection plays a major role in shaping hybrid genomic composition (Rieseberg et al. 1996; Rieseberg 2000), whereas the QTL comparisons described above suggest that phenotypic (presumably ecological) selection must be important as well. What is the relative importance of these two modes of selection? A preliminary analyses of the two data sets indicates that the phenotypic QTL data is a slightly better predictor of the genomic composition of the ancient hybrid species than are the products of fertility selection (P = 0.005).
Candidate genes for ecological divergence
Finally, we have assayed sequence polymorphisms for salt tolerance candidate genes in these same BC2 populations to determine whether any of the candidate genes map to QTLs of interest, as well as to identify additional genomic regions associated with salt tolerance (Lexer et al. 2004). The salt tolerance candidates were identified from an expressed sequence tag (EST) library for H. paradoxus based on homology to genes with known function. One of the 11 genes, a Ca-dependent protein kinase (CDPK), maps coincident with a previously identified QTL for mineral ion uptake. Two additional genes (an ER-type calcium ATPase and a transcriptional regulator), also exhibit a significant fitness effect in the wild. Of course, these studies are correlational only and function will have to be verified by transgenic complementation or RNAi. Nonetheless, they indicate that we soon may be able to examine the role of individual genes in ecological divergence and speciation in sunflowers.
Conclusions
The sunflower work provides compelling evidence that hybridization facilitated the colonization of novel habitats by three sunflower species, H. anomalus, H. deserticola, and H. paradoxus, and suggests a mechanism (complementary gene action) by which this occurred. Our findings also suggest that hybridization with H. debilis likely facilitated the range expansion of the common sunflower, H. annuus. These results are important because they provide perhaps the strongest link yet between hybridization, adaptive evolution, and the colonization of new habitats.
Despite the clear role hybridization played in the phenotypic evolution of the three hybrid species, this does not necessarily mean that hybridization was required for ecological divergence. It might be, for example, that gene flow from the center of parental species’ ranges, rather than limited genetic variance (Kirkpatrick and Barton 1997), prevented the parental species from colonizing novel habitats. If so, perhaps the hybrid species were able to colonize these habitats as a result of reproductive isolation from the parental species (due to karyotypic divergence) rather than the new gene combinations brought about by hybridization. We have recently initiated a long-term selection experiment to test whether the new hybrid gene combinations or reproductive isolation were most critical to ecological divergence in these species.
Our findings also demonstrate that the ability to colonize new habitats is not synonymous with range expansion or the evolution of invasiveness. While the three hybrid species have managed to colonize extreme habitats, they cannot be called invasive under any definition of the term. Rather, they are best described as rare endemics that seem unlikely to survive outside of the very restricted habitats in which they currently occur. Indeed, greenhouse experiments indicate that they grow more slowly than their parents and are less fecund (unpublished data), suggesting there may be fitness trade-offs associated with adaptation to extreme conditions (Fry 2003). This is in contrast to the common sunflower, H. annuus, which has successfully colonized most temperate regions of the world. However, with the exception of suggestive data from Texas, we do not yet know whether hybridization has facilitated these invasions.
Although much of this article has been devoted to the discussion of hybrid speciation, introgression is a more frequent outcome of hybridization and a more likely contributor to invasiveness (Buerkle et al. 2003). Hybrid speciation must be initiated in sympatry, and the development of reproductive barriers in the presence of gene flow represents a significant evolutionary challenge. In contrast, there are no theoretical reasons why introgression should not be common and frequently contribute to invasiveness. Because of dynamic nature of species ranges, most species are likely to come into contact with congeners before reproductive isolation is complete and hybridization should be common in nature. Theory also predicts that while recombinant hybrids should be less fit on average, certain hybrid genotypes should be more fit than the parents, particularly when new environments are being explored (Barton 2001). For hybridization to contribute to adaptation, however, fit hybrid genotypes must escape from “the mass of unfit recombinants” present in a hybrid population (Barton 2001). Introgression provides the simplest and most frequent means by which this occurs because the establishment of a new hybrid gene combination requires only the spread of an advantageous allele from one species into the other.
Empirical data agree well with theory. Natural hybridization is a common feature of many organismal groups (Arnold 1997; Mallet 2005), with the frequency of hybridizing species averaging 11% for plants (Ellstrand et al. 1996), 8.5% for freshwater fish (Hubbs 1955), and 9% for birds (Grant and Grant 1992). Many instances of hybridization are unlikely to be detected, however, particularly for poorly studied faunas or floras, and species that have hybridized in the past may no longer do so today (e.g., Wendel et al. 1991). On the other hand, human disturbance may have increased contemporary hybridization rates in some organismal groups (Anderson 1948; Hauser et al. 1998; Schemske 2000).
Estimates of past hybridization frequencies can be inferred from the incidence of allopolyploidy and from molecular phylogenetic studies. For example, allopolyploidy, which must be initiated by hybridization, is conservatively estimated to account for 2–4% of speciation events in flowering plants and 7% in ferns, and many asexual animal species are allopolyploids (Otto and Whitton 2000). Inferences from molecular phylogenetic studies are more difficult because incongruence between gene trees, which is often used as a diagnostic for hybridization, may arise through other processes such as the sorting of gene lineages (Avise 1994; Wendel and Doyle 1998). Nonetheless, both data sets suggest that reticulate evolution is widespread (Wang et al. 1997; Sota and Vogler 2001), particularly in plants (Rieseberg and Soltis 1991) and bacteria (Ochman et al. 2000). Thus, while it is difficult to extrapolate from contemporary to ancient frequencies of hybridization, it seems likely that hybridization has been a common feature in the evolution of many organismal groups.
Studies of hybrid fitness in natural populations (reviewed in Arnold and Hodges 1995; Barton 2001; Lexer et al. 2003a) are also in agreement with theory (above) and indicate that while average hybrid fitness typically is lower than that of both parental species, some genotypes often are more fit than either parental species (e.g., Wang et al. 1997) and fitness is frequently contingent on the environment (e.g., Grant and Grant 2002). Hybridization is less likely to be important in abundant species, where mutation may not limit adaptive divergence. As a consequence, allelic variants introduced by hybridization might be of little value (Barton 2001). However, this condition probably does not apply to many plant species, which often have small populations and/or low rates of gene flow, so that mutation may indeed limit the evolution of local populations.
While the sunflower work shows how evolutionary novelty generated by hybridization may be employed for range expansion and the colonization of new habitats, it does not address other possible contributions such as heterosis and the purging of genetic load. Heterosis, in particular, is correlated with invasiveness in many of the world’s most noxious weeds (e.g., Gray et al. 1991; Gaskin and Schaal 2002; Moody and Les 2002), but its role has not yet been verified. Even less is known about whether the purging of deleterious alleles contributes to invasiveness. Thus, much remains to be done before we can confidently describe the role of hybridization in range expansions and the evolution of invasiveness.
Acknowledgments
The research on sunflower hybridization described here has been supported by grants from the National Science Foundation, National Institutes of Health, and U.S. Department of Agriculture. This contribution was an invited paper for the 2004 Society for the Study of Evolution Symposium “All Stressed Out and Nowhere to Go: Does Evolvability Limit Adaptation in Invasive Species?”
References
- Abbott R. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol Evol. 1992;7:401–405. doi: 10.1016/0169-5347(92)90020-C. [DOI] [PubMed] [Google Scholar]
- Abbott RJ, James JK, Milne RI, Gillies ACM. Plant introductions, hybridization and gene flow. Phil Trans R Soc B. 2003;358:1123–1132. doi: 10.1098/rstb.2003.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainouche ML, Baumel A, Salmon A, Yannic G. Hybridization, polyploidy and speciation in Spartina (Poaceae) New Phytol. 2003;161:165–172. [Google Scholar]
- Al-Khatib K, Baumgartner JR, Peterson DE, Currie RS. Imazethapyr resistance in common sunflower (Helianthus annuus) Weed Sci. 1998;46:403–407. [Google Scholar]
- Anderson E. Hybridization of the habitat. Evolution. 1948;2:1–9. [Google Scholar]
- Anderson E. Introgressive hybridization. John Wiley and Sons; New York: 1949. [Google Scholar]
- Anderson E, Stebbins GL. Hybridization as an evolutionary stimulus. Evolution. 1954;8:378–388. [Google Scholar]
- Antonovics J. The nature of limits to natural selection. Ann MO Bot Gard. 1976;63:224–247. [Google Scholar]
- Arnold ML. Natural hybridization and evolution. Oxford Univ. Press; Oxford: 1997. [Google Scholar]
- Arnold ML. Transfer and origin of adaptations through natural hybridization: were Anderson and Stebbins right? Plant Cell. 2004;16:562–570. doi: 10.1105/tpc.HistPersp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML, Hodges SA. Are natural hybrids fit or unfit relative to their parents? Trends Ecol Evol. 1995;10:67–71. doi: 10.1016/S0169-5347(00)88979-X. [DOI] [PubMed] [Google Scholar]
- Avise JC. Molecular markers, natural history, and evolution. Chapman & Hall; New York: 1994. [Google Scholar]
- Baker HG. Migration of weeds. In: Valentine DH, editor. Taxonomy, phytogeography and evolution. Academic Press; London: 1972. pp. 327–347. [Google Scholar]
- Baker HG, Stebbins GL. The genetics of colonizing species. Academic Press; New York: 1965. [Google Scholar]
- Barrett SCH. Crop mimicry in weeds. Econ Bot. 1983;37:255–282. [Google Scholar]
- Barton NH. The role of hybridisation in evolution. Mol Ecol. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Ann Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- Blumler MA. Introgression as a spatial phenomenon. Phys Geogr. 2003;24:414–432. [Google Scholar]
- Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH. The likelihood of homoploid hybrid speciation. Heredity. 2000;84:441–451. doi: 10.1046/j.1365-2540.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- Buerkle CA, Wolf DE, Rieseberg LH. The origin and extinction of species through hybridization. In: Brigham CA, Schwartz MW, editors. Population viability in plants. Springer Verlag; Berlin: 2003. pp. 117–141. [Google Scholar]
- Burke JM, Bulger MR, Wesselingh RA, Arnold ML. Frequency and spatial patterning of clonal reproduction in Louisiana iris hybrid populations. Evolution. 2000;54:137–144. doi: 10.1111/j.0014-3820.2000.tb00014.x. [DOI] [PubMed] [Google Scholar]
- Burke JM, Lai Z, Salmaso M, Nakazato T, Tang S, Heesacker A, Knapp SJ, Rieseberg LH. Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics. 2004;167:449–457. doi: 10.1534/genetics.167.1.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JM, Jan C, Beard BH. Chromosomal differentiation among the annual Helianthus species. Syst Bot. 1986;11:353–371. [Google Scholar]
- Charlesworth D. Evolution under the microscope. Curr Biol. 1995;5:835–836. doi: 10.1016/s0960-9822(95)00166-7. [DOI] [PubMed] [Google Scholar]
- Clausen J, Hiesey WM. IV. Genetic structure of ecological races. Carnegie Institute of Washington; Washington, DC: 1958. Experimental studies on the nature of species. [Google Scholar]
- deVicente MC, Tanksley SD. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics. 1993;134:585–596. doi: 10.1093/genetics/134.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorado O, Rieseberg LH, Arias D. Chloroplast DNA introgression in southern California sunflowers. Evolution. 1992;46:566–572. doi: 10.1111/j.1558-5646.1992.tb02061.x. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C. Teosinte Branched1 and the origin of maize – evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC. Dangerous liaisons? When cultivated plants mate with their wild relatives. John Hopkins University Press; Balitmore, MD: 2003. [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC, Whitkus R, Rieseberg LH. Distribution of spontaneous plant hybrids. Proc Natl Acad Sci USA. 1996;93:5090–5093. doi: 10.1073/pnas.93.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich PR. Attributes of invaders and the invading process: vertebrates. In: Drake JA, Mooney HA, di Castri F, Groves RH, Kruger FJ, Rejmanek M, Williamson M, editors. Biological invasions: a global perspective. John Wiley; London: 1989. pp. 315–328. [Google Scholar]
- Fritz RS, Moulia C, Newcombe G. Resistance of hybrid plants and animals to herbivores, pathogens, and parasites. Annu Rev Ecol Syst. 1999;30:565–591. [Google Scholar]
- Fry JD. Detecting ecological trade-offs using selection experiments. Ecology. 2003;84:1672–1678. [Google Scholar]
- Galatowitsch SM, Anderson NO, Ascher PD. Invasiveness in wetland plants in temperate North America. Wetlands. 1999;19:733–755. [Google Scholar]
- Gaskin JF, Schaal BA. Hybrid Tamarix widespread in US invasion and undetected in native Asian range. Proc Natl Acad Sci USA. 2002;99:11256–11259. doi: 10.1073/pnas.132403299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb LD. Levels of confidence in the analysis of hybridization in plants. Ann MO Bot Gard. 1972;59:435–446. [Google Scholar]
- Grant V. Plant speciation. Columbia Univ. Press; New York: 1981. [Google Scholar]
- Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin’s finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- Gray AJ, Marshall DF, Raybould AF. A century of evolution in Spartina anglica. Adv Ecol Res. 1991;21:1–62. [Google Scholar]
- Grime JP. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am Nat. 1977;111:1169–1194. [Google Scholar]
- Gross BL, Rieseberg LH. The ecological genetics of homoploid hybrid speciation. J Hered. 2005;96:241–252. doi: 10.1093/jhered/esi026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Schwarzbach AE, Rieseberg LH. Origin(s) of the diploid hybrid species Helianthus deserticola (Asteraceae) Am J Bot. 2003;90:1708–1719. doi: 10.3732/ajb.90.12.1708. [DOI] [PubMed] [Google Scholar]
- Gross BL, Kane NC, Lexer C, Ludwig F, Rosenthal DM, Donovan LA, Rieseberg LH. Reconstructing the origin(s) of Helianthus deserticola: survivorship and selection on the desert floor. Am Nat. 2004;164:145–156. doi: 10.1086/422223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotkopp E, Rejmanek M, Rost TL. Toward a causal explanation of plant invasiveness: seedling growth and life-history strategies of 29 pine (Pinus) species. Am Nat. 2002;159:396–419. doi: 10.1086/338995. [DOI] [PubMed] [Google Scholar]
- Harlan JR, De Wet JMJ. The compilospecies concept. Evolution. 1963;17:497–501. [Google Scholar]
- Harper JL. Population biology of plants. Academic Press; New York: 1977. [Google Scholar]
- Hauser L, Carvalho GR, Pitcher TJ, Ogutu-Ohwayo R. Genetic affinities of an introduced predator: Nile perch in Lake Victoria, East Africa. Mol Ecol. 1998;7:849–857. [Google Scholar]
- Heiser CB. Hybridization between the sunflower species Helianthus annuus and H. petiolaris. Evolution. 1947;1:249–262. [Google Scholar]
- Heiser CB. Study in the evolution of the sunflower species Helianthus annuus and H. bolanderi. Univ Calif Publ Bot. 1949;23:157–196. [Google Scholar]
- Heiser CB. Hybridization in the annual sunflowers: Helianthus annuus × H. argophyllus. Am Nat. 1951a;85:64–72. [Google Scholar]
- Heiser CB. Hybridization in the annual sunflowers: Helianthus annuus × H. debilis var. cucumerifolius. Evolution. 1951b;5:42–51. [Google Scholar]
- Heiser CB. Variation and subspeciation in the common sunflower, Helianthus annuus. Am Midl Nat. 1954;51:287–305. [Google Scholar]
- Heiser CB. Sunflowers, weeds, and cultivated plants. In: Barker HG, Stebbins GL, editors. The genetics of colonizing species. Academic Press; Orlando, FL: 1965. pp. 391–401. [Google Scholar]
- Heiser CB, Smith DM, Clevenger S, Martin WC. The North American sunflowers (Helianthus) Mem Torrey Bot Club. 1969;22:1–218. [Google Scholar]
- Hoffmann AA, Blows MW. Species borders: ecological and evolutionary perspectives. Trends Ecol Evol. 1994;9:223–227. doi: 10.1016/0169-5347(94)90248-8. [DOI] [PubMed] [Google Scholar]
- Hubbs CL. Hybridization between fish species in nature. Syst Zool. 1955;4:1–20. [Google Scholar]
- Johnston JA, Grise DJ, Donovan LA, Arnold ML. Environment-dependent performance and fitness of Iris brevicaulis, I. fulva (Iridaceae), and hybrids. Am J Bot. 2001;88:933–938. [PubMed] [Google Scholar]
- Kim S-C, Rieseberg LH. Genetic architecture of species differences in annual sunflowers: implications for adaptive trait introgression. Genetics. 1999;153:965–977. doi: 10.1093/genetics/153.2.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-C, Rieseberg LH. The contribution of epistasis to species differences in annual sunflowers. Mol Ecol. 2001;10:683–690. doi: 10.1046/j.1365-294x.2001.01203.x. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton NH. Evolution of a species’ range. Am Nat. 1997;150:1–23. doi: 10.1086/286054. [DOI] [PubMed] [Google Scholar]
- Lai Z, Nakazato T, Salmaso M, Burke JM, Tang S, Knapp SJ, Rieseberg LH. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics. 2005;171:1933–1940. doi: 10.1534/genetics.105.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Langevin SA, Clay K, Grace JB. The incidence and effects of hybridization between cultivated rice and its related weed red rice (Oryza sativa L.) Evolution. 1990;44:1000–1008. doi: 10.1111/j.1558-5646.1990.tb03820.x. [DOI] [PubMed] [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends Ecol Evol. 2002;17:386–391. [Google Scholar]
- Levine JM, D’Antonio CM. Elton revisited: a review of evidence linking diversity and invasibility. Oikos. 1999;87:15–26. [Google Scholar]
- Lewontin RC, Birch LC. Hybridization as a source of variation for adaptation to new environments. Evolution. 1966;20:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. [DOI] [PubMed] [Google Scholar]
- Levin DA, Clay K. Dynamics of synthetic Phlox drummondii populations at the species margin. Am J Bot. 1984;71:1040–1050. [Google Scholar]
- Lexer C, Welch M, Durphy JL, Rieseberg LH. Natural selection for salt tolerance QTL in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a homoploid hybrid species. Mol Ecol. 2003a;12:1225–1235. doi: 10.1046/j.1365-294x.2003.01803.x. [DOI] [PubMed] [Google Scholar]
- Lexer C, Welch M, Raymond O, Rieseberg LH. The origins of ecological divergence in Helianthus paradoxus (Asteraceae): selection on transgressive characters in a novel hybrid habitat. Evolution. 2003b;57:1989–2000. doi: 10.1111/j.0014-3820.2003.tb00379.x. [DOI] [PubMed] [Google Scholar]
- Lexer C, Rosenthal DM, Raymond O, Donovan L, Rieseberg LH. Genetics of species differences in the wild annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 2005;169:2225–2239. doi: 10.1534/genetics.104.031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer C, Lai Z, Rieseberg LH. Candidate gene polymorphisms associated with salt tolerance in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. New Phytol. 2004;161:225–233. doi: 10.1046/j.1469-8137.2003.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig F, Rosenthal D, Johnston JA, Kane N, Gross BL, Lexer C, Dudley SA, Rieseberg LH, Donovan LA. Selection on leaf ecophysiological traits in a hybrid Helianthus species and early generation hybrids in a desert dune habitat. Evolution. 2004;58:2682–2692. doi: 10.1111/j.0014-3820.2004.tb01621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and the analysis of quantitative traits. Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Marshall JK. Factors limiting the survival of Corynephorus canescens (L.) Beauv. in Great Britain at the northern edge of its distribution. Oikos. 1968;19:206–216. [Google Scholar]
- Milne RI, Abbott RJ. Origin and evolution of invasive naturalized material of Rhododendron ponticum L. in the British Isles. Mol Ecol. 2000;9:541–556. doi: 10.1046/j.1365-294x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- Moody ML, Les DH. Evidence of hybridity in invasive watermilfoil (Myriophyllum) populations. Proc Natl Acad Sci USA. 2002;99:14867–14871. doi: 10.1073/pnas.172391499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer B, Auge H, Mesch H, Amarell U, Brandl R. Spread of violets in polluted pine forests: morphological and molecular evidence for the ecological importance of inter-specific hybridization. Mol Ecol. 1999;8:365–377. [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Orians CM. The effects of hybridization in plants on secondary chemistry: implications for the ecology and evolution of plant–herbivore interactions. Am J Bot. 2000;87:1749–1756. [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Pianka ER. On r and K selection. Am Nat. 1970;102:592–597. [Google Scholar]
- Pimental D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the United States. BioScience. 2000;50:53–65. [Google Scholar]
- Pires JC, Zhao JW, Schranz ME, Leon EJ, Quijada PA, Lukens LN, Osborn TC. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Biol J Linn Soc. 2004;82:675–688. [Google Scholar]
- Prince SD, Carter RN, Dancy KJ. The geographical distribution of prickly lettuce (Lactuca serriola) II. Characteristics of populations near its distribution limit in Britain. J Ecol. 1985;73:39–48. [Google Scholar]
- Rattenbury JA. Cyclic hybridization as a survival mechanism in the New Zealand forest flora. Evolution. 1962;16:348–363. [Google Scholar]
- Raven PH. The bases of angiosperm phylogeny: cytology. Ann MO Bot Gard. 1976;62:409–451. [Google Scholar]
- Rejmanek M, Richardson DM. What attributes make some plant species more invasive? Ecology. 1996;77:1655–1660. [Google Scholar]
- Rieseberg LH. Homoploid reticulate evolution in Helianthus: evidence from ribosomal genes. Am J Bot. 1991;78:1218–1237. [Google Scholar]
- Rieseberg LH. Crossing relationships among ancient and experimental sunflower hybrid lineages. Evolution. 2000;54:859–865. doi: 10.1111/j.0014-3820.2000.tb00086.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Soltis DE. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Plants. 1991;5:65–84. [Google Scholar]
- Rieseberg LH, Wendel J. Introgression and its consequences in plants. In: Harrison R, editor. Hybrid zones and the evolutionary process. Oxford University Press; New York: 1993. pp. 70–114. [Google Scholar]
- Rieseberg LH, Soltis DE, Palmer JD. A molecular reexamination of introgression between Helianthus annuus and H. bolanderi. Evolution. 1988;42:227–238. doi: 10.1111/j.1558-5646.1988.tb04127.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Beckstrom-Sternberg S, Doan K. Helianthus annuus ssp. texanus has chloroplast DNA and nuclear ribosomal RNA genes of Helianthus debilis ssp. cucumerifolius. Proc Natl Acad Sci USA. 1990a;87:593–597. doi: 10.1073/pnas.87.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Carter R, Zona S. Molecular tests of the hypothesized hybrid origin of two diploid Helianthus species (Asteraceae) Evolution. 1990b;44:1498–1511. doi: 10.1111/j.1558-5646.1990.tb03841.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Beckstrom-Sternberg S, Liston A, Arias D. Phylogenetic and systematic inferences from chloroplast DNA and isozyme variation in Helianthus sect. Helianthus. Syst Bot. 1991a;16:50–76. [Google Scholar]
- Rieseberg LH, Choi H, Ham D. Differential cytoplasmic versus nuclear gene flow in Helianthus. J Hered. 1991b;82:489–493. [Google Scholar]
- Rieseberg LH, Desrochers A, Youn SJ. Interspecific pollen competition as a reproductive barrier between sympatric species of Helianthus (Asteraceae) Am J Bot. 1995a;82:515–519. [Google Scholar]
- Rieseberg LH, Van Fossen C, Desrochers A. Hybrid speciation accompanied by genomic reorganization in wild sunflowers. Nature. 1995b;375:313–316. [Google Scholar]
- Rieseberg LH, Sinervo B, Linder CR, Ungerer M, Arias DM. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science. 1996;272:741–745. doi: 10.1126/science.272.5262.741. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. Transgressive segregation, adaptation, and speciation. Heredity. 1999a;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two wild sunflower species. Genetics. 1999b;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C. Major ecological transitions in annual sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Rogers CE, Thompson TE, Seiler GJ. Sunflower species of the United States. National Sunflower Association; Bismarck, ND: 1982. [Google Scholar]
- Rosenthal DM, Rieseberg LH, Donovan LA. Recreating ancient hybrid species’ complex multi-trait phenotypes from early generation synthetic hybrids: three examples using wild sunflowers. Am Nat. 2005;166:26–41. doi: 10.1086/430527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughmann S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SC. The population biology of invasive species. Annu Rev Ecol Syst. 2001;32:305–332. [Google Scholar]
- Schemske DW. Understanding the origin of species. Evolution. 2000;54:1069–1073. [Google Scholar]
- Schwarzbach AE, Rieseberg LH. Likely multiple origins of a diploid hybrid sunflower species. Mol Ecol. 2002;11:1703–1717. doi: 10.1046/j.1365-294x.2002.01557.x. [DOI] [PubMed] [Google Scholar]
- Schwarzbach AE, Donovan LA, Rieseberg LH. Transgressive character expression in a hybrid sunflower species. Am J Bot. 2001;88:270–277. [PubMed] [Google Scholar]
- Silander JA, Antonovics J. The genetic basis of the ecological amplitude of Spartina patens. 1. Morphometric and physiological traits. Evolution. 1979;33:1114–1127. doi: 10.1111/j.1558-5646.1979.tb04766.x. [DOI] [PubMed] [Google Scholar]
- Small E. Hybridization in the domesticated-weed-wild complex. In: Grant WF, editor. Plant biosystematics. Academic Press; Toronto: 1984. pp. 195–210. [Google Scholar]
- Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol J Linn Soc. 2004;82:485–501. [Google Scholar]
- Sota T, Vogler AP. Incongruence of mitochondrial and nuclear gene trees in Carabid beetles Ohomopterus. Syst Biol. 2001;50:39–59. [PubMed] [Google Scholar]
- Stace CA. Hybridization and the Flora of the British Isles. Cambridge University Press; Cambridge: 1975. [Google Scholar]
- Stebbins GL. The hybrid origin of microspecies in the Elymus glaucus complex. Cytolo Suppl. 1957;36:336–340. [Google Scholar]
- Stebbins GL. The role of hybridization in evolution. Proc Am Phil Soc. 1959;103:231–251. [Google Scholar]
- Stebbins GL. The significance of hybridization for plant taxonomy and evolution. Taxon. 1969;18:26–35. [Google Scholar]
- Stebbins GL. Polyploidy, hybridization, and the invasion of new habitats. Ann MO Bot Gard. 1985;72:824–832. [Google Scholar]
- Strauss SY. Levels of herbivory and parasitism in host hybrid zones. Trends Ecol Evol. 1994;9:209–214. doi: 10.1016/0169-5347(94)90245-3. [DOI] [PubMed] [Google Scholar]
- Sun M, Corke H. Population genetics of colonizing success of weedy rye in Northern California. Theor Appl Genet. 1992;83:321–329. doi: 10.1007/BF00224278. [DOI] [PubMed] [Google Scholar]
- Tanksley SD. Mapping polygenes. Annu Rev Genet. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]
- Thompson JD. The biology of an invasive plant – what makes Spartina anglica so successful? Bioscience. 1991;41:393–401. [Google Scholar]
- Wang H, McArthur ED, Sanderson SC, Graham JH, Freeman DC. Narrow hybrid zone between two subspecies of big sagebrush (Artemisia tridentata): Asteraceae. IV: reciprocal transplant experiments. Evolution. 1997;51:95–102. doi: 10.1111/j.1558-5646.1997.tb02391.x. [DOI] [PubMed] [Google Scholar]
- Wang RL, Wakeley J, Hey J. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics. 1997;147:1091–1106. doi: 10.1093/genetics/147.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, D’Antonio CM. Phenotypic plasticity in hybridizing Carpobrotus spp. (Aizoaceae) from coastal California and its role in plant invasion. Can J Bot/Rev Can Bot. 1999;77:1411–1418. [Google Scholar]
- Welch ME, Rieseberg LH. Patterns of genetic variation suggest a single, ancient origin for the diploid hybrid species Helianthus paradoxus. Evolution. 2002;56:2126–2137. doi: 10.1111/j.0014-3820.2002.tb00138.x. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Doyle JJ. Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II: DNA sequencing. Kluwer Academic Publishers; Boston: 1998. pp. 256–296. [Google Scholar]
- Wendel JF, Stewart JM, Rettig JH. Molecular evidence for homoploid reticulate evolution among Australian species of Gossypium. Evolution. 1991;45:694–671. doi: 10.1111/j.1558-5646.1991.tb04339.x. [DOI] [PubMed] [Google Scholar]
- Wilkes HG. Hybridization of maize and teosinte in Mexico and Guatemala and the improvement of maize. Econ Bot. 1977;31:254–293. [Google Scholar]
- Williamson M. Biological invasions. Chapman & Hall; London: 1996. [Google Scholar]


