Abstract
Duplicated genes escape gene loss by conferring a dosage benefit or evolving diverged functions. The yeast Saccharomyces cerevisiae contains many duplicated genes encoding ribosomal proteins. Prior studies have suggested that these duplicated proteins are functionally redundant and affect cellular processes in proportion to their expression. In contrast, through studies of ASH1 mRNA in yeast, we demonstrate paralog-specific requirements for the translation of localized mRNAs. Intriguingly, these paralog-specific effects are limited to a distinct subset of duplicated ribosomal proteins. Moreover, transcriptional and phenotypic profiling of cells lacking specific ribosomal proteins reveals differences between the functional roles of ribosomal protein paralogs that extend beyond effects on mRNA localization. Finally, we show that ribosomal protein paralogs exhibit differential requirements for assembly and localization. Together, our data indicate complex specialization of ribosomal proteins for specific cellular processes, and support the existence of a ribosomal code.
Introduction
The yeast Saccharomyces cerevisiae arose from an ancient whole genome duplication followed by massive gene loss, as redundant copies were eliminated from the genome. Roughly 10% of duplicated genes were maintained, mainly through the evolution of specialized functions (Kellis et al., 2004). Remarkably, 59 of the 78 ribosomal proteins retained two genomic copies. Following the initial discovery of duplicated ribosomal protein genes, growth rates were assayed in ribosomal protein gene knockouts to determine whether paralogous genes were functionally distinct. Correspondence between fitness defects and expression levels and the fact that overexpression of one ribosomal protein rescues the growth defect from deletion of its paralog led to the conclusion that duplicated ribosomal proteins are functionally redundant, with the more highly expressed paralog playing a more significant role in the cell (Rotenberg et al., 1988).
Recent studies reveal a more complex relationship between paralogous ribosomal proteins. A study of Rps27a and Rps27b found that cells lacking Rps27a exhibited ribosomal assembly defects and deficiencies in rRNA processing despite growing at the wild-type rate (Baudin-Baillieu et al., 1997), demonstrating that growth rate does not necessarily reflect functionality. Recent high-throughput screens have suggested more subtle differences between duplicated ribosomal protein genes, including paralog-specific defects in sporulation (Enyenihi and Saunders, 2003), actin organization (Haarer et al., 2007), and bud-site selection (Ni and Snyder, 2001). Though these studies suggest functional specificity of duplicated ribosomal protein paralogs, a mechanistic role for the ribosome in these processes remains unclear.
The yeast protein Ash1 localizes exclusively to the daughter cell where it acts to suppress mating-type switching upon cell division. Protein localization is achieved through ASH1 mRNA localization, a process with a well-characterized requirement for both translation and translational regulation. Several studies have demonstrated the requirement of ongoing translation for anchoring of ASH1 mRNA at the site of growth in the emerging daughter cell (the bud tip). Mutations in ASH1 mRNA that disrupt its translation abolish bud-tip anchoring, as does inhibition of translation by cycloheximide treatment, resulting in a mis-localization of the mRNA throughout the emerging daughter cell (Gonzalez et al., 1999; Irie et al., 2002; Kruse et al., 2002). ASH1 also undergoes translational repression, and factors required for this repression are needed for its anchoring at the bud tip (Beach et al., 1999; Gu et al., 2004; Irie et al., 2002). Thus, anchoring of the ASH1 mRNA involves a complex mechanism in which both translational repression and active translation are required.
Several factors required for ASH1 localization have also been implicated in ribosomal assembly. Loc1, a strictly nuclear protein, was identified through its association with ASH1 mRNA and is required for targeting ASH1 to the bud (Long et al., 2001). Recent studies show that Loc1 associates with the 60S pre-ribosomal subunit fraction (Harnpicharnchai et al., 2001) and is required for the efficient assembly of the large ribosomal subunit (Harnpicharnchai et al., 2001) and rRNA processing (Urbinati et al., 2006). Another factor required for ASH1 localization, Puf6, has also been implicated in ribosomal assembly, having been identified among the proteins that sediment in the 60S pre-ribosomal fraction (Nissan et al., 2002). Puf6 is a member of the pumilio family that was recently shown to play a role in ASH1 localization and translational repression (Gu et al., 2004).
We sought to determine whether duplicated ribosomal proteins have distinct roles in translational regulation. We show that a specific subset of duplicated ribosomal protein genes are required for ASH1 localization, and that there is a direct correspondence between the genes required for this process and those required for bud-site selection. Transcriptional profiling of cells lacking individual genes shows additional paralog-specific differences. Analysis of phenotypic data demonstrates that functional specificity also occurs in other duplicated ribosomal protein genes and cannot simply be attributed to expression levels. Finally, we show that paralogous ribosomal proteins have different genetic requirements for their assembly, and exhibit paralog-specific aberrant localizations in certain genetic backgrounds. Together our results indicate that what was previously thought to be simple redundancy in ribosomal protein-encoding genes has significant functional consequences.
Results
Loc1 is required for the translational regulation of ASH1
The requirement for Loc1 in ASH1 localization may relate to its role in ribosomal assembly. Although Loc1 was originally implicated in the targeting of ASH1 mRNA from within the nucleus (Long et al., 2001), Loc1 is also required for ribosomal assembly (Harnpicharnchai et al., 2001; Urbinati et al., 2006). Moreover, high-throughput immunoprecipitations identified many ribosomal proteins in association with Loc1 (Collins et al., 2007), suggesting that Loc1 may play a direct role. We verified the association of Loc1 with ribosomal proteins and intermediates using both immunoprecipitation (Supplementary Figure 1) and sucrose gradient analysis (Supplementary Figure 2). Taken together with previous data implicating translation in ASH1 localization (Gonzalez et al., 1999; Irie et al., 2002; Kruse et al., 2002), these data suggest that the effect of Loc1 on ASH1 localization may be a consequence of its role in ribosomal assembly.
In order to better understand the mechanism by which Loc1 affects ASH1 localization, we used a live-cell mRNA reporter system (Brodsky and Silver, 2002) to determine its effect on individual ASH1 regulatory elements. We created reporter constructs that contain the promoter and coding sequence of yeast PGK1, an array of U1A-binding hairpins, and either PGK1’s own 3’ UTR or one of the four ASH1 localization elements (Figure 1a). Each reporter was co-expressed with U1A-GFP, which binds the hairpins and allowed us to track the localization of the reporter mRNAs in live cells (Brodsky and Silver, 2002).
Figure 1.

Loc1 is required for the translational regulation of ASH1 mRNA. (A) Reporter constructs used to assay ASH1 regulation. The reporters contain the promoter and ORF of yeast PGK1, an array of U1A hairpins, and either PGK1’s own 3’ UTR or one of ASH1’s four localization elements. Each reporter was co-expressed along with U1A-GFP, which specifically binds the U1A hairpins and allows visualization of reporter mRNA location in live cells. The fraction of cells with either bud-tip, bud-cytoplasm, or ubiquitous localization was determined (see Experimental Procedures). (B) Defective anchoring of the E3 construct in loc1 Δ cells is due to aberrant translation. Histograms of the E3 reporter construct localizations in wild-type, loc1 Δ, and wild-type cells following brief treatment with cycloheximide. Error bars represent total variation between replicate experiments. (C) Protein level of myc-Ash1 increases in loc1 Δ cells relative to actin (negative control). Western blots of myc-Ash1 and actin were performed from extracts of wild-type and loc1 Δ cells. Equal amounts of protein were loaded in each lane. (D) mRNA level of ASH1 decreases in loc1 Δ cells.
Assays in wild-type and loc1 Δ cells confirmed the functionality of our reporter system. We scored the fraction of large-budded cells with GFP signal enriched at the bud tip (“bud-tip”), diffuse throughout the bud cytoplasm (“bud-cytoplasm”), or evenly distributed throughout both mother and daughter cells (“ubiquitous”). As expected, in wild-type cells all four reporters bearing the ASH1 constructs exhibited bud-tip localization, whereas the reporter bearing PGK1’s own 3’ UTR was ubiquitously distributed. In contrast, the reporters bearing the E1, E2A, and E2B elements showed no bud-specific enrichment in loc1 Δ cells (Supplementary Figure 3). This result agrees with previous reports that full-length ASH1 is ubiquitously localized in loc1 Δ cells (Long et al., 2001) and indicates defective targeting of these mRNAs to the bud.
Analysis of the E3 reporter implicates Loc1 in the regulation of ASH1 translation. Unlike the other reporter constructs, the E3 reporter exhibited significant enrichment in the bud cytoplasm in loc1 Δ cells (Figure 1b). This localization has previously been shown to indicate improper translational regulation of the full-length ASH1 transcript (Beach et al., 1999; Gonzalez et al., 1999; Gu et al., 2004; Irie et al., 2002; Kruse et al., 2002). To confirm that our reporter system was similarly affected by translation defects, we examined the effect of treating wild-type cells expressing the E3 reporter with cycloheximide. As shown in Figure 1b, disrupting translation reproduces the localization observed in loc1 Δ cells, indicating that Loc1 regulates the translation of ASH1.
Analysis of Ash1 expression levels confirms a role for Loc1 in the translational regulation of ASH1 mRNA. We examined the protein and mRNA levels of ASH1 in wild-type and loc1 Δ cells. Ash1 protein expression increases approximately two-fold in cells lacking Loc1, whereas the level of actin is unaffected (Figure 1c). In contrast, ASH1 mRNA levels decrease in loc1 Δ cells (Figure 1d). We conclude that Loc1 is required for the translational regulation of ASH1.
In sum, we have further elucidated the role of Loc1 in ASH1 regulation, showing that it is not only required for targeting ASH1 mRNA to the bud tip, but also for the translational regulation of ASH1.
Genes required for bud-site selection are needed for ASH1 localization
The requirement for translational regulation in ASH1 localization suggests that Loc1 may regulate ASH1 via its role in ribosomal assembly. Intriguingly, a related phenotypic connection has been established between Loc1 and certain ribosomal protein genes: the site of daughter-cell formation is highly programmed in yeast, but cells lacking Loc1 or any of 15 specific ribosomal protein genes exhibit random bud-site positioning (Ni and Snyder, 2001). As ASH1’s E3 sequence element localizes to nascent bud sites (Beach et al., 1999), we speculated that the mechanism for ASH1 bud-tip anchoring may relate to the mechanism for regulated bud-site selection. We hypothesized that Loc1 may affect both anchoring and bud-site selection via its effects on the ribosome, and that this subset of ribosomal proteins may be directly involved.
To test this hypothesis, we compared the localizations of the E3 reporter construct in wild-type cells and in 10 strains that had been found to exhibit random bud-site selection in diploid cells: loc1 Δ, six strains that lack specific ribosomal protein genes (rpl7aΔ, rpl12bΔ, rpl14aΔ, rpl22aΔ, rps0bΔ, and rps18bΔ), and three strains lacking genes with functions unrelated to translation (CLC1, involved in protein transport and endocytosis; CWH8, required for protein N-glycosylation; and GUP1, a membrane protein involved in glycerol transport). All deletion strains exhibited defects in localization of E3-GFP (Figure 2a,b), indicating that there is a one-to-one relationship between the genes required for bud-site selection and those required for ASH1 localization.
Figure 2.
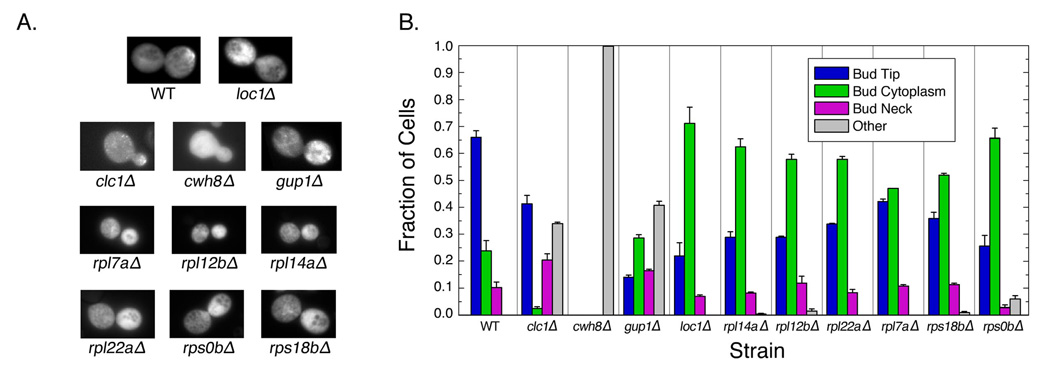
Genes required for bud-site selection in yeast are also required for ASH1 localization. (A) Representative images of cells expressing ASH1 reporter. (B) Strains that have defective bud-site selection also have defects in localization of the E3 reporter construct. Fraction of cells exhibiting bud-tip, bud-cytoplasm, bud-neck, and “other” (not bud-tip, bud-neck, or bud-cytoplasm) localizations of the E3 reporter construct in cultures lacking the genes indicated is shown. Error bars represent standard deviations of replicate experiments.
Factors unrelated to translation cause unique defects in localization of the E3 reporter construct. Strains lacking CLC1, CWH8, and GUP1 exhibited defects that were dissimilar to one another and to loc1Δ cells, but consistent with their known functions (Figure 2a). For example, Cwh8 is required for the maintenance of polarized actin cables (Bonangelino et al., 2002); since ASH1 mRNA is transported along actin filaments, an actin-assembly defect would lead to the observed ubiquitous ASH1 mRNA localization. Thus, although these factors share the bud-site selection defect observed in loc1 Δ cells, they do not necessarily have the same effect on ASH1 localization.
In contrast, each of the strains lacking ribosomal protein genes exhibited localizations indicative of defects in translation regulation. As shown in Figure 2, all six ribosomal protein knockouts had a similar phenotype to that observed in loc1Δ cells: a significantly higher fraction of cells showed bud-cytoplasmic localization of E3-GFP. This effect is directly due to the absence of the corresponding ribosomal protein, since we were able to rescue the defect by re-introducing the corresponding ribosomal protein on a plasmid (Supplementary Figure 4). Similar effects were observed for the other ASH1 reporter constructs, indicating that this defect in bud-tip anchoring is not specific to the E3 construct (data not shown).
Together, these data demonstrate a one-to-one relationship between genes required for bud-site selection and those required for ASH1 localization.
Translation of ASH1 mRNA requires a specific subset of duplicated ribosomal protein paralogs
Only a subset of the 137 genes encoding ribosomal proteins was implicated in bud-site selection. Of the 15 implicated genes, 14 have a duplicate within the genome. Intriguingly, although the proteins encoded by these duplicates are almost identical, only one paralog from each pair was required for bud-site selection. This suggests the existence of functional specificity between duplicated ribosomal proteins in yeast, but does not provide a mechanism. However, the role of regulated translation in ASH1 localization is well-characterized (Beach et al., 1999; Gonzalez et al., 1999; Gu et al., 2004; Irie et al., 2002; Kruse et al., 2002), thus we asked whether the paralog specificity observed in bud-site selection extends to the translational regulation of ASH1.
Ribosomal protein paralogs not required for bud-site selection are also dispensible for the translation of the ASH1 reporter. We assayed the localization of the E3 construct in cells lacking nonessential ribosomal proteins paralogous to those required for bud-site selection. Copies not implicated in bud-site selection (RPL7B, RPL12A, RPL22B, and RPS18A) had little if any effect on the anchoring of the E3 construct (Figure 3a) relative to their nearly identical counterparts (RPL7A, RPL12B, RPL22A, and RPS18B). Overexpression of the paralogous gene was unable to rescue the ASH1 defect observed when the copy implicated in bud-site selection was absent (Supplementary Figure 5). Thus, only certain ribosomal protein paralogs affect the localization and translation of ASH1 mRNA.
Figure 3.

Regulated translation of the E3 reporter construct requires a specific subset of duplicated ribosomal protein genes. (A) – (B), Error bars represent standard deviations of replicate experiments. (A) Ribosomal proteins that are required for bud-site selection have a larger defect in anchoring of the E3 reporter construct than their nearly-identical paralogs. Fraction of cells exhibiting either bud-tip or bud-cytoplasmic localization of the E3 reporter construct in cells lacking the gene is indicated. Genes that are required for bud-site selection in diploids are indicated by “*”. (B) There is a significantly greater difference in the effect on anchoring of the E3 reporter construct between pairs of duplicated ribosomal protein genes in which one copy is required for bud-site selection than for pairs in which neither copy is required for bud-site selection. The fraction of cells exhibiting bud-tip and bud-cytoplasmic localization of the E3 reporter construct was assayed in strains lacking a variety of duplicated ribosomal protein genes. The difference between the fraction of cells exhibiting bud-tip localization is plotted against the difference in the fraction exhibiting bud-cytoplasmic localization for both members of each pair.
The paralog specificity in bud-tip anchoring of E3 cannot be attributed to gene dosage effects or to paralog-specific effects on ribosomal assembly. We examined the mRNA expression level of both copies of the duplicated ribosomal proteins needed for bud-site selection. Although eight of these ribosomal protein paralogs are expressed at higher levels than their counterparts, the same is not true for the remaining four proteins (Supplementary Figure 6). Protein levels confirm that paralog-specific phenotypes are not due to gene-dosage effects; Rps18a and Rps18b expression levels are nearly identical, despite their different effects on ASH1 localization (Supplementary Figure 7). Consistent with the equal expression levels of Rps18a and Rps18b, sucrose gradient analysis shows that both paralogs have nearly identical effects on ribosomal assembly (Supplementary Figure 8a). Rpl12a and Rpl12b also have similar effects on ribosomal assembly despite different in requirements for ASH1 localization (Supplementary Figure 8b). Together these data show that the paralog specificity observed with ASH1 localization is not due to expression differences or relative contributions to ribosome assembly.
Paralog-specific effects on ASH1 localization are restricted to those proteins required for bud-site selection. We assayed E3 localization in a variety of duplicated ribosomal proteins in which neither paralog is required for bud-site selection (Supplementary Figure 9). To assess paralog-specific function, we compared the difference in the fraction of cells exhibiting bud-tip localization between paralogous genes against the difference in the fraction of cells exhibiting bud-cytoplasmic localization for the same pair of genes (Figure 3b). The difference is significantly greater between paralogous genes required for bud-site selection than between genes not required for this process (P<0.01, Mann-Whitney U-test). Thus, the differences in paralogs observed for ASH1 localization are unique to a specific subset of duplicated ribosomal proteins.
Together, these data show that a specific subset of duplicated ribosomal proteins exhibit paralog-specific requirements for the translational regulation of ASH1 mRNA.
Transcriptional profiling reveals general cellular differences between ribosomal protein paralogs
Given that certain duplicated ribosomal proteins exhibit functional specificity in the translation of ASH1 mRNA, we used transcriptional profiling to determine if the cellular roles of these paralogs differ in other respects. We analyzed the transcriptional profiles of cells lacking the eleven ribosomal proteins shown to exhibit paralog-specific roles in ASH1 localization. The resulting profiles were compared to those obtained in wild-type cells.
We examined the transcriptional profiles for evidence of functional specificity. Genes that exhibited significantly increased or decreased expression levels in each strain relative to wild-type were analyzed for significantly enriched Gene Ontology categories using Funcassociate (Berriz et al., 2003). The types of genes that were induced and repressed varied greatly between paralogous ribosomal proteins (Table 1). In some cases, there was no overlap between functional categories affected by paralogous gene deletions; for example, the absence of Rpl12a induced genes involved in amino acid metabolism and biosynthesis, while the absence of Rpl12b induced genes with products that localize to the nucleus and repressed genes involved in cell wall and RNA modification. Significantly, paralogous ribosomal protein genes also differentially affected genes involved in various aspects of ribosomal assembly. For example, deletion of Rpl12a reduced expression of genes encoding the cytosolic ribosome, while deletion of Rpl12b decreased expression of genes involved in rRNA modification (Table 1). Thus, the functional specificity of duplicated ribosomal protein genes applies not only to ASH1 translational regulation, but also to additional cellular processes, including assembly of the ribosome itself.
Table 1.
Duplicated ribosomal protein genes affect the transcription of different cellular processes. Significantly up- and down-regulated genes in the indicated gene deletions were analyzed for enriched Gene Ontology classes. Corrected P-values are shown for significantly enriched categories; positive values indicate enrichment in up-regulated genes, negative values indicate enrichment in down-regulated genes.
| Gene Deletion | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GO ID | Description | Rpl7a | Rpl7b | Rpl12a | Rpl12b | Rpl14a | Rpl22a | Rpl22b | Rps0a | Rps0b | Rps18a | Rps18b |
| 0000003 | Reproduction | −0.001 | ||||||||||
| 0000051 | Urea cycle intermediate metabolism | 0.001 | 0.001 | 0.001 | ||||||||
| 0003723 | RNA binding | 0.001 | 0.001 | 0.001 | 0.001 | |||||||
| 0005618 | Cell wall | −0.004 | −0.001 | −0.011 | −0.002 | |||||||
| 0005634 | Nucleus | 0.01 | 0.023 | 0.001 | 0.001 | 0.001 | ||||||
| 0006139 | Nucleic acid metabolism | 0.001 | 0.001 | 0.001 | ||||||||
| 0006396 | RNA processing | 0.015 | 0.009 | 0.027 | ||||||||
| 0006520 | Amino acid metabolism | 0.001 | 0.001 | |||||||||
| 0006526 | Arginine biosynthesis | 0.001 | 0.001 | 0.001 | ||||||||
| 0006591 | Ornithine metabolism | 0.001 | 0.002 | |||||||||
| 0007028 | Cytoplasm organization and biogenesis | 0.003 | ||||||||||
| 0007131 | Meiotic recombination | −0.039 | ||||||||||
| 0008152 | Metabolism | 0.028 | ||||||||||
| 0008652 | Amino acid biosynthesis | 0.001 | 0.047 | 0.001 | ||||||||
| 0009165 | Nucleotide biosynthesis | −0.038 | ||||||||||
| 0009308 | Amine metabolism | 0.001 | 0.001 | |||||||||
| 0009451 | RNA modification | −0.004 | −0.028 | 0.001 | 0.001 | 0.001 | ||||||
| 0016036 | Cellular response to phosphate starvation | −0.032 | ||||||||||
| 0019752 | Carboxylic acid metabolism | 0.001 | 0.001 | |||||||||
| 0019953 | Sexual reproduction | −0.006 | −0.001 | |||||||||
| 0030312 | External encapsulating structure | −0.004 | −0.001 | −0.011 | −0.002 | |||||||
| 0030555 | RNA modification guide activity | −0.026 | −0.001 | −0.001 | 0.001 | 0.001 | 0.001 | 0.001 | ||||
| 0042221 | Response to chemical substance | −0.001 | ||||||||||
| 0043232 | Intracellular non-membrane-bound organelle | −0.037 | −0.011 | 0.001 | 0.001 | 0.001 | ||||||
| 0043412 | Biopolymer metabolism | 0.001 | 0.001 | 0.001 | ||||||||
| 0044238 | Primary metabolism | 0.001 | ||||||||||
| 0044271 | Nitrogen compound biosynthesis | 0.001 | 0.001 | |||||||||
| 0045026 | Plasma membrane fusion | −0.002 | −0.001 | |||||||||
| 0050839 | Cell adhesion molecule binding | −0.043 | ||||||||||
| 0050896 | Response to stimulus | −0.015 | ||||||||||
| 0051213 | Dioxygenase activity | −0.044 | ||||||||||
| Related to Ribosomal Assembly | ||||||||||||
| 0000154 | rRNA modification | −0.001 | −0.001 | 0.001 | 0.001 | 0.001 | 0.001 | |||||
| 0005730 | Nucleolus | −0.03 | 0.002 | 0.001 | 0.001 | 0.001 | 0.001 | |||||
| 0005830 | Cytosolic ribosome | −0.034 | ||||||||||
| 0007046 | Ribosome biogenesis | 0.018 | 0.003 | |||||||||
| 0019843 | rRNA binding | 0.019 | 0.005 | |||||||||
| 0030489 | Processing of 27S pre-rRNA | 0.04 | ||||||||||
| 0030559 | rRNA pseudouridylation guide activity | 0.003 | 0.001 | |||||||||
| 0030563 | snRNA 2’-O-ribose methylation guide | −0.002 | −0.001 | −0.001 | 0.001 | 0.001 | 0.001 | 0.001 | ||||
| 0042254 | Ribosome biogenesis and assembly | 0.003 | ||||||||||
Paralog-specific effects on expression were also observed among noncoding RNAs, such as RNAs that regulate the processing of rRNA (e.g. C/D box snoRNAs) and those involved in splicing (e.g., U1 RNA) (data not shown). Moreover, many ribosomal protein deletions exhibited misregulated expression of repetitive elements, including centromeric regions and long-terminal repeats. These data provide further support for paralog-specific cellular roles of ribosomal protein genes.
Together, our transcriptional profiling data indicate that ribosomal protein paralogs have specialized cellular roles beyond their effects on ASH1 localization.
Complex requirements for ribosomal protein paralogs that extends to other cellular processes
We next mined high-throughput datasets in order to determine if paralog-specific functional differences occur among other duplicated ribosomal protein genes. The phenotypic effects caused by the absence of all non-essential ribosomal proteins were compiled (supplementary data). A subset of this data is displayed in Table 2.
Table 2.
Paralogous ribosomal protein genes exhibit different phenotypes. Phenotypic data for all ribosomal proteins was mined from published datasets; a representative sample is shown. Sensitivity is indicated by “1” unless otherwise indicated.
| RPL7 | RPL12 | RPL13 | RPL20 | RPL27 | RPL34 | RPL41 | RPP1 | RPS4 | RPS10 | RPS14 | RPS30 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | |
| Bud-site Selection*,A | 1 | 2 | 2 | 2 | 2 | |||||||||||||||||||
| Cell Size#,B | 1 | 1 | 1 | 1 | 2 | 1 | ||||||||||||||||||
| Sporulation & MeiosisX,C | 1 | 2 | 2 | 2 | ||||||||||||||||||||
| Repressed by MMSD | 1 | 1 | 1 | |||||||||||||||||||||
| Vacuolar Protein SortingΛ,E | 2 | 2 | 2 | 1 | 1 | 2 | ||||||||||||||||||
| Wortmannin Sensitivity+,F | 2 | 4 | 3 | 2 | 2 | 1 | −2 | 2 | 3 | 1 | 2 | 2 | 2 | −3 | ||||||||||
| Caffeine Sensitivity+,G | 1 | 1 | 1 | 1 | ||||||||||||||||||||
| Cycloheximide Sensitivity+,G | 2 | 3 | ||||||||||||||||||||||
| Sulfometuron Methyl Sensitivity+,G | 1 | 2 | ||||||||||||||||||||||
| Rapamycin Sensitivity+,H | 3 | 4 | ||||||||||||||||||||||
| Abnormal Telomere Length,∇,J | −2 | −2 | −3 | 1 | 2 | 2 | ||||||||||||||||||
| Neomycin Sulfate SensitivityK | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||
| Pentamidine SensitivityK | 1 | 1 | 1 | |||||||||||||||||||||
| Hydrogen Peroxide SensitivityK | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||
| Mitomycin SensitivityK | 1 | 1 | ||||||||||||||||||||||
| Trichostatin A SensitivityK | 1 | 1 | 1 | |||||||||||||||||||||
| Benomyl SensitivityK | 1 | 1 | 1 | |||||||||||||||||||||
| Phenantroline SensitivityK | 1 | 1 | ||||||||||||||||||||||
| Hygromycin B SensitivityK | 1 | 1 | 1 | |||||||||||||||||||||
| Desipramine SensitivityK | 1 | 1 | 1 | |||||||||||||||||||||
| CG4-Theopalaumide SensitivityK | 1 | 1 | ||||||||||||||||||||||
| Caspofungin SensitivityK | 1 | 1 | 1 | |||||||||||||||||||||
| Basiliskamide SensitivityK | 1 | 1 | ||||||||||||||||||||||
| Papuamide SensitivityK | 1 | 1 | ||||||||||||||||||||||
| Geldanamycin SensitivityK | 1 | 1 | 1 | |||||||||||||||||||||
1=strong defect, 2=weak defect
1=among the smallest 5%, 2=among the largest 5%
1=low sporulation efficiency, 2=high sporulation efficiency but reduced number of spores per ascus
1=strong or moderate defect, 2=weak defect
“-“ indicates resistance; higher number indicates higher sensitivity/resistance
1=slightly long, 2=long, 3=very long, −1=slightly short, −2=short, −3=very short
L. Ni and M. Snyder, Mol. Biol. of the Cell 12(7):2147–2170, 2001.
P. Jorgensen et. al, Science 297(5580): 395–400, 2002.
A.H. Enyenihi and W.S. Saunders, Genetics 163(1): 47–54, 2003.
S.A. Jelinsky and L.D. Samson, PNAS 96(4): 1486–1491, 1999.
C.J. Bonangelino et. al, Mol. Biol. of the Cell 13(7): 2486–2501, 2002.
A. Zewail et. al, PNAS 100(6): 3345–3350, 2003.
A.B. Parsons et. al, Nat. Biotech. 22(1): 62–69, 2004.
N. Page et. al, Genetics 163(3): 875–894, 2003.
S.H. Askree et. al, PNAS 101(23): 8658–8663, 2004.
A.B. Parsons et. al, Cell 126(3): 611–625, 2006.
All duplicated ribosomal proteins exhibited phenotypes that differed between their nearly-identical paralogs. For example, rpl41a Δ cells are sensitive to wortmannin, neomycin sulfate, and phenantroline. In contrast, rpl41b Δ cells exhibit none of these sensitivities but instead are sensitive to benomyl, pentamidine, and hydrogen peroxide. Broad phenotypic differences are also observed when comparing all other pairs of duplicated ribosomal protein genes.
Clustering of the phenotypic data provides further support for functional diversity among duplicated ribosomal protein genes. The ribosomal protein paralogs were grouped according to phenotypic defects using hierarchical clustering (Figure 4a) (Eisen et al., 1998). In general, the two copies of each duplicated ribosomal protein clustered separately from each other. For example, as shown in Figure 4b, RPL2B clusters with RPS19A, RPP2A, and RPS11B, whereas its paralog RPL2A instead clusters with RPL43B, RPL26B, RPS25A, RPL8A, RPS29B, and RPL21B. These results indicate that paralogous ribosomal protein genes are more functionally similar to other duplicated ribosomal protein genes than to their nearly identical counterparts.
Figure 4.

Phenotypic data reveals complex functional relationships between duplicated ribosomal protein genes. (A) Hierarchical clustering analysis of phenotypic data by ribosomal protein (vertical axis) and phenotype (horizontal axis). Although many ribosomal proteins shared some phenotypes, no two proteins are required for the same set of processes, and different groups are required for each process. (B) Paralogous ribosomal proteins are not phenotypically similar. Rpl2a and Rpl2b cluster with completely different groups of genes, as indicated by the shaded boxes that correspond to (A). (C) Paralogous ribosomal proteins share no more phenotypes than non-paralogous genes. The number of shared phenotypes between all combinations of duplicated ribosomal protein genes was calculated and sorted into paralogous or non-paralogous relationships. Normalized values are displayed. (D) Phenotypic effects are not determined by expression level. mRNA expression levels of all duplicated ribosomal protein genes from transcriptional profiling data was used to determine the relative contribution of each paralog. Genes were sorted into “higher” or “lower” based on whether they contributed more or less than half of the mRNA, respectively. Error bars represent standard deviations.
Duplicated ribosomal proteins share no more phenotypes with each other than with other duplicated ribosomal proteins. We determined the number of shared phenotypes for each pairwise combination of duplicated ribosomal protein genes. As shown in Figure 4c, the distribution of the number of shared phenotypes between pairs of paralogous duplicated ribosomal proteins was highly similar to that obtained when comparing pairs of non-paralogous proteins (P = 0.71, Kolmogorov-Smirnov test). Thus, despite the high sequence similarity beween paralogous ribosomal proteins, their cellular roles are divergent.
Clustering analysis reveals additional complexity in the cellular roles of duplicated ribosomal protein genes. Although closely clustered ribosomal proteins are more similar to each other than to other ribosomal proteins, no two ribosomal proteins exhibit identical dependencies and phenotypes. This suggests that no one subset of ribosomal protein paralogs consistently acts together in various cellular processes. Instead, it implies a more complex model in which diverse combinations of ribosomal protein paralogs are required for different cellular processes. In support of this model, biochemical analysis reveals that ribosomal paralogs do not associate exclusively with specific paralogs from other duplicated ribosomal proteins. We generated strains in which two ribosomal proteins were tagged with different epitopes and assayed for co-immunoprecipitation. All duplicated ribosomal proteins we tested associate with each other, at levels corresponding to their overall expression level (Supplementary Figure 10). These data corroborate the findings in Figure 4a and provide further support for complex rules governing the associations among ribosomal paralogs (see Discussion).
The number of phenotypic defects induced by the deletion of a given ribosomal protein gene is not determined by its expression level. As shown in Table 2, the number of phenotypes observed varies among paralogs. The gene-dosage theory would predict that any observed phenotypes are a consequence of expression level, in which case the more highly expressed paralog of each pair of duplicated ribosomal protein genes would have more phenotypic defects than its counterpart. To determine if this is the case, we compared the wild-type expression level of paralogs of each duplicated pair, placing one paralog into the “higher” category and the other into the “lower” category, based on their relative expression levels. As shown in Figure 4d, when paralogs were sorted in this manner, the number of phenotypes did not differ significantly (P = 0.38, Mann-Whitney U-test). Thus, the observed phenotypic differences cannot be attributed to the relative abundance of each ribosomal protein paralog.
Together we have shown that the specificity observed between certain duplicated ribosomal proteins for ASH1 localization also applies to other duplicated ribosomal proteins and to other cellular processes. Moreover, it appears that the phenotypic relationships between duplicated ribosomal proteins are complex, such that different groups of ribosomal protein paralogs are required for different cellular processes.
Paralogous ribosomal proteins exhibit differences in their localizations and assembly requirements
The paralog-specific phenotypic effects among ribosomal proteins led us to ask whether paralogous genes also differ in their assembly requirements. As Loc1 and Puf6 have each been implicated in both ribosomal assembly (Harnpicharnchai et al., 2001; Nissan et al., 2002; Urbinati et al., 2006) and the translational regulation of ASH1 ((Gu et al., 2004) and Figure 1), we hypothesized that they may differentially affect the processing of paralogous ribosomal proteins. As a recent study had shown that improperly assembled ribosomes localize to a sub-region of the nucleolus (Dez et al., 2006), we used GFP-tagged ribosomal proteins to simultaneously assay assembly status and localization.
We tagged two pairs of duplicated ribosomal proteins with GFP. We chose Rpl7a, Rpl7b, Rps18a, and Rps18b for this analysis because these genes were implicated in ASH1 localization, the paralogs of each pair show distinct phenotypes, and together they represent both large and small ribosomal subunits. Tags were genomically integrated at the N-terminus, using the Cre/LOX system to remove markers and restore the native promoter (Gauss et al., 2005). Sucrose cushion assays demonstrated that these proteins are functional and are incorporated into ribosomes in wild-type cells (data not shown). As expected, in wild-type cells all four ribosomal proteins localize to the cytoplasm (Figure 5).
Figure 5.
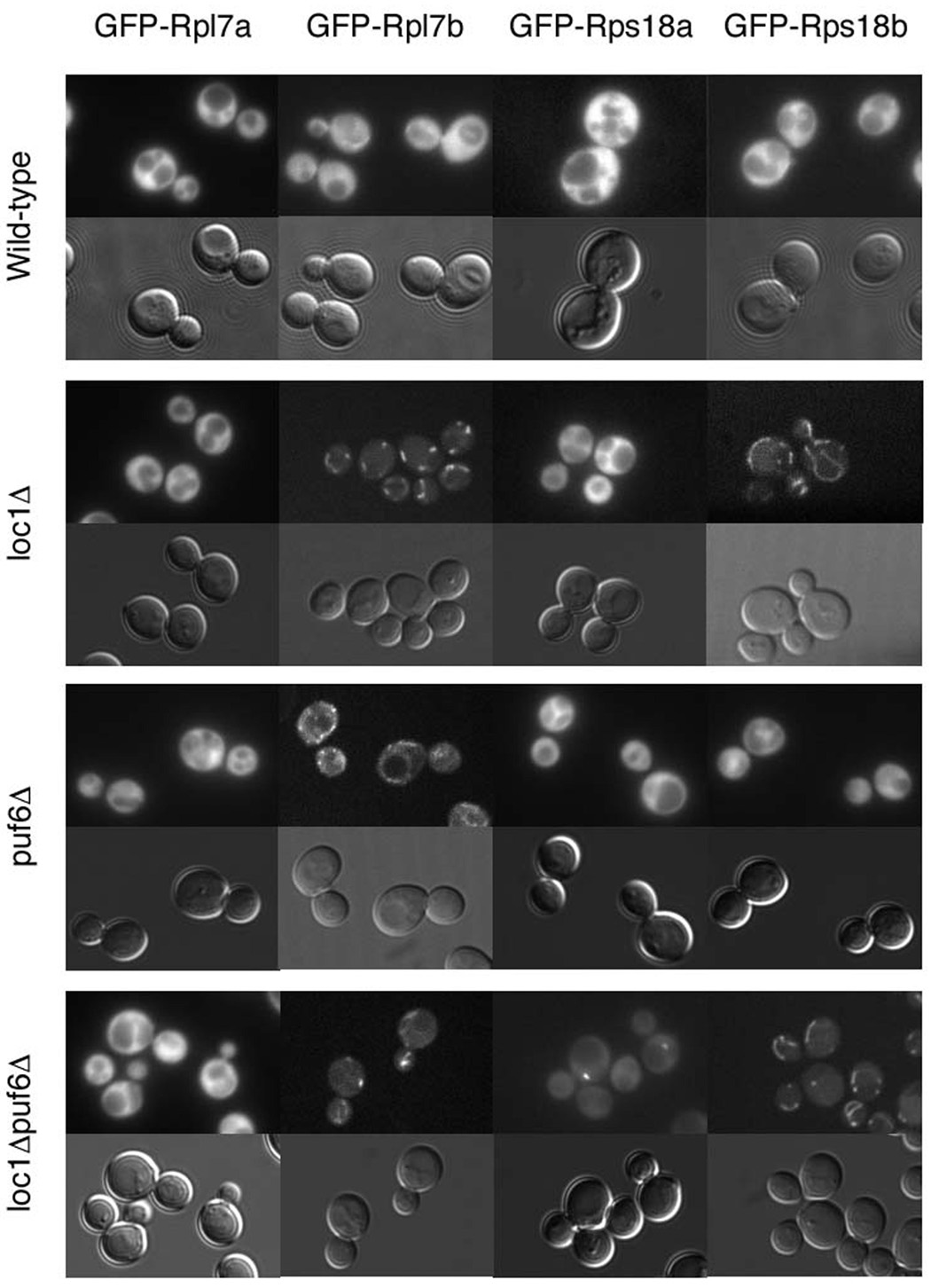
Paralogous ribosomal proteins exhibit different localizations and assembly requirements in specific genetic backgrounds. GFP-tagged Rpl7a, Rpl7b, Rps18a, and Rps18b were expressed from the genome under their own promoters in wild-type, loc1Δ , puf6Δ, and loc1Δpuf6Δ cells. Representative fluorescent (top) and nomarski (bottom) images are shown.
Intriguingly, the absence of either Loc1 or Puf6 caused paralog-specific localization defects of the GFP-tagged ribosomal proteins. As shown in Figure 5, Rpl7b and Rps18b localize to a region consistent with the endoplasmic reticulum in loc1 Δ cells, while Rpl7a and Rps18a exhibit wild-type localization. Thus, although Loc1 is required for ribosomal assembly, Rpl7b and Rps18b do not exhibit assembly defects in its absence; instead, Loc1 seems to regulate their targeting to certain cellular regions. The absence of Puf6 causes Rpl7b to exhibit a similar localization defect as observed in loc1 Δ cells but does not affect the other three paralogs (Figure 5). Together these data show that duplicated ribosomal proteins exhibit paralog-specific genetic interactions that lead to localization defects. Furthermore, as none of the ribosomal proteins exhibited nuclear or nucleolar retention, our data shows that neither Loc1 nor Puf6 is absolutely required for the assembly of these ribosomal protein paralogs.
The overlapping functions of Loc1 and Puf6 led us to ask whether they may act together in the assembly of paralogous ribosomal proteins. Specifically, we hypothesized that each ribosomal protein would still be assembled into ribosomes when only one factor was absent, but that the absence of both would have paralog-specific effects on assembly. As such, we examined the localizations of the GFP-tagged ribosomal proteins in loc1Δ puf6Δ cells. As shown in Figure 5, the localizations observed in the double deletion strain differed from both wild-type and the individual deletions. Although Rps18b exhibits the same localization observed in loc1 Δ cells, Rpl7b instead localizes to discrete cytoplasmic foci. Intriguingly, Rps18a, whose localization was unaffected in either of the single deletions, localizes to the nucleolus in loc1Δpuf6Δ cells, indicative of aberrant assembly and/or export. Thus, Loc1 and Puf6 exhibit a synthetic defect for the ribosomal assembly of Rps18a, but not Rps18b.
In sum, we have shown that paralogous ribosomal proteins require different factors for their assembly. Moreover, we have made the surprising discovery that these proteins exhibit paralog-specific aberrant localizations in the absence of certain factors.
Discussion
We have demonstrated by four criteria that paralogous ribosomal proteins, previously thought to be redundant, are functionally distinct. First, the localized translation of ASH1 mRNA requires a specific subset of duplicated ribosomal protein genes. Second, transcriptional profiling of cells lacking these same duplicated ribosomal protein genes revealed additional levels of functional divergence between paralogs. Third, the analysis of phenotypic data indicates that functional specificity occurs in all duplicated ribosomal protein genes and that no two ribosomal protein paralogs share all phenotypes. Finally, examination of paralogous ribosomal proteins revealed paralog-specific localizations and assembly defects that depend on the cell's genetic background. Together, these data indicate that duplicated ribosomal proteins are playing distinct functional roles within the cell.
Functional specificity among duplicated ribosomal protein genes
Through the analysis of ASH1, a well-characterized transcript in yeast, we have identified a new level of complexity in the regulation of gene expression. Maintenance of the bud-tip localization of ASH1 requires both translational repressors and active translation (Beach et al., 1999; Gonzalez et al., 1999; Gu et al., 2004; Irie et al., 2002; Kruse et al., 2002). We showed that Loc1, a strictly nuclear factor previously implicated in both ribosomal assembly and in the targeting of ASH1 mRNA to the bud (Harnpicharnchai et al., 2001; Long et al., 2001; Urbinati et al., 2006), is also required for the translational regulation of ASH1 (Figure 1). The bud-tip localization of ASH1 also requires a specific subset of duplicated ribosomal proteins (Figure 2), and Loc1 had previously been found to share a defect in bud-site selection with these genes (Ni and Snyder, 2001). Intriguingly, these effects are paralog-specific (Figure 3). Together these findings suggest a model in which Loc1 is required for the assembly of ribosomes containing a specific subset of duplicated ribosomal proteins, and that this “specialized” ribosome is required for the regulated translation of ASH1 mRNA.
Our data indicate additional differences between duplicated ribosomal protein genes. Ribosomal protein deletions exhibit paralog-specific effects on transcription levels (Table 1) and unique phenotypes (Table 2). Additionally, paralogous genes are no more phenotypically similar to each other than they are to other duplicated ribosomal protein genes (Figure 4c). Although these other processes have not been directly linked to translation, when taken together with our data for ASH1, the extensive variation between paralogs suggests that these processes also involve “specialized” ribosomes, with each ribosome requiring different subsets of duplicated ribosomal proteins.
Our data argues against a gene-dosage model for ribosomal protein specificity. Previous characterizations of duplicated ribosomal protein genes had led to the conclusion that paralog-specific defects were due to differences in expression, with the fitness defect caused by each deletion proportional to the abundance of its transcript (Abovich and Rosbash, 1984; Herruer et al., 1987; Leer et al., 1984; Leer et al., 1985; Lucioli et al., 1988; Rotenberg et al., 1988). Analysis of ribosomal protein genes required for ASH1 localization argues against this model: several of the ribosomal protein genes required for ASH1 translation are expressed at a lower level than their corresponding paralog (Supplemental Table 1). Moreover, when we examined all duplicated ribosomal proteins and all phenotypes for which there is published data, we found no relationship between relative mRNA abundance and number of observed phenotypes (Figure 4d). Thus, paralog-specific phenotypic consequences of deleting duplicated ribosomal protein genes cannot be explained by expression level.
Findings in other organisms lend further support to the existence of specialized ribosomes. Like yeast, plants also have multiple copies of ribosomal protein genes. Many of these genes exhibit expression restricted to specific stages of development and/or specific tissues, and when mutated these genes often yield phenotypes consistent with aberrant development (Dresselhaus et al., 1999; Ito et al., 2000; Ma and Dooner, 2004; Tsugeki et al., 1996; Weijers et al., 2001; Williams and Sussex, 1995). As observed in yeast, many of these genes do not affect growth rates unless cells are exposed to genetic or environmental stresses. For example, in Arabidopsis, ARS27A is dispensable for growth in wild-type cells, but a promoter mutation leads to growth deficiencies and tumor-like structures when exposed to mutagens (Revenkova et al., 1999).
Ribosomal protein duplication also occurs in other eukaryotes. There are multiple copies of ribosomal protein genes in species as diverse as S. pombe (e.g., Rpl11-1 & -2), Drosophila (e.g., Rpl34a & b), C. elegans (e.g. rpl-11.1 & .2), and humans (Rps4X and Rps4Y). Although phenotypic data on these paralogous genes is not as extensive as in budding yeast, the conservation of ribosomal protein gene duplication among eukaryotes suggests that the functional specificity we observe in S. cerevisiae is not a special case but is instead indicative of a general phenomenon.
Other eukaryotes require ribosomal heterogeneity for mRNA localization. Drosophila, Xenopus, and Ascidians require ribosomes derived from the mitochondrion for the localized and developmentally regulated translation of maternal mRNAs (Amikura et al., 2001; Kobayashi et al., 1998; Oka et al., 1999). Intriguingly, these specialized ribosomes may only be required for the initiation of translation, after which any form of ribosome may be able to translate the regulated mRNA. This appears to be the case in Drosophila, as electron microscopy indicates that the mRNAs are translated by both mitochondrial and cytoplasmic ribosomes (Amikura et al., 2001).
Specific ribosomal protein genes have also been implicated in cancer. A recent screen in zebrafish for recessive lethal tumor suppressor genes found 11 out of the 12 tumor suppressor lines to contain ribosomal protein mutations (Amsterdam et al., 2004). In plants, the cancer-like phenomena observed following the mutation of specific ribosomal proteins (Revenkova et al., 1999) shows that ribosomal protein involvement in cancer is conserved among diverse eukaryotes.
Parallels between translational regulation and regulation of transcription: evidence for a “ribosome code”
Our data supports a model in which there are many different forms of functionally distinct ribosomes in yeast, where the functional specificity is determined by the combination of duplicated ribosomal proteins present. However, protein composition is not the only source of ribosomal heterogeneity. Many fungi express different forms of 5S rRNA, with two major species occurring in S. cerevisiae (Selker et al., 1985). Moreover, ribosomal proteins are subject to a variety of post-translational modifications, including phosphorylation, methylation, ubiquitination, and acetylation (Lee et al., 2002; Louie et al., 1996); such modifications impact the translational activity of the protein (Bachand et al., 2006; Mazumder et al., 2003). Indeed, as previously posited (Mauro and Edelman, 2002), there is a wealth of evidence for heterogeneity among ribosomes regulating the translational activity of their targets.
This model of translational regulation bears a striking resemblance to the canonical model for transcriptional regulation. The transcriptional activity of a given region of DNA is regulated by the structure of the surrounding chromatin, which is largely determined by the types of associated histones and their post-translational modifications. As with ribosomal proteins, histone genes are duplicated in yeast (Kellis et al., 2004). Moreover, several distinct forms of histones have been identified with specialized roles (Polo and Almouzni, 2006). Furthermore, as with ribosomal proteins, histones are subject to myriad post-translational modifications, and these modifications modulate the transcriptional activity of the surrounding chromatin (Kouzarides, 2007). Finally, both DNA and rRNA are subject to direct modifications (Bernstein et al., 2007; Fromont-Racine et al., 2003). In sum, the transcription state of a given region of chromatin is determined by specific combinations of histone proteins, post-translational modifications of histones, and DNA modifications; this complex relationship has been called the “histone code” (Jenuwein and Allis, 2001). Our data support a similar level of complexity for the process of translation, in which different combinations of ribosomal protein paralogs, post-translational modifications of ribosomal proteins, different forms of rRNA, and modifications to the rRNA allow calibrated translation of specific mRNAs. As with the histone code, this “ribosome code” would provide a new level of complexity in the regulation of gene expression.
Experimental Procedures
Analysis of ASH1 protein and mRNA levels
Ash1 was tagged at the N-terminus using published methods (Gauss et al., 2005). Cells were lysed, diluted to 1mg/mL total protein, and analyzed as described (Hieronymus and Silver, 2003) using c-myc A14 (Santa Cruz) and α-actin (Chemicon). ASH1 mRNA levels were determined from transcriptional profiling of loc1 Δ cells (see “Transcriptional profiling”).
Live-cell mRNA imaging
ASH1 reporter constructs were created and transformed along with pPS2035 using standard methods. Cells were grown and induced as described (Brodsky and Silver, 2002). Only large-budded cells (bud size > 75% of mother size) were counted, and each assay was repeated blind at least twice. Only cells exhibiting localized GFP expression were counted for assays performed on the E3 reporter for Figure 2 and Figure 3.
Transcriptional profiling
Transcriptional profiling of Loc1 was performed as described (Casolari et al., 2004) on four independent cultures each for wild-type (PSY3259) versus loc1 Δ (PSY3262), with two arrays for each fluor orientation. RNA was prepared similarly from two independent cultures for ribosomal knockouts and wild-type cells and hybridized to Affymetrix Yeast 98 arrays; mRNAs were considered significantly changed if their ratios differed more than two-fold from wild-type.
Clustering analysis
Phenotypic data was clustered using hierarchical clustering with complete linkage and visualized using published software (Eisen et al., 1998; Saldanha, 2004).
Ribosomal protein imaging
GFP-tagged ribosomal proteins were imaged in live cells in mid-log phase (2–8×107 cells/mL); a subset of these images were obtained using a Nikon TE2000U inverted microscope with PerkinElmer ultraview spinning disk confocal.
Supplementary Material
Acknowledgements
The authors thank Joe Salas-Marco, John Tsang, Guillaume Adelement, Fred Winston, Rebecca Ward, and Dale Muzzey for helpful discussions and critical evaluation of the manuscript, the Dana-Farber Cancer Institute microarray facility for assistance with Affymetrix arrays, and Paul Grosu, Reddy Gali, and the Bauer Center for Genomics Research (Harvard University) for assistance with ORF microarray analysis. S.K. would like to thank Ana Forrest, Eve Kodiak, Luella Earley and Lydia Knutson for assistance. This work was supported by a fellowship from NSERC (to S.K.), the Harvard Biophysics Graduate Program, Milton Fund of Harvard University, NIH grant HG003224 (to F.P.R.), an institutional grant to Harvard Medical School from the HHMI, and grants from the NIH to P.A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers Microarray data are available at http://ncbi.nih.gov/geo under the accession numbers GSE8761 and GSE8765.
Bibliography
- Abovich N, Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984;4:1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikura R, Kashikawa M, Nakamura A, Kobayashi S. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc Natl Acad Sci U S A. 2001;98:9133–9138. doi: 10.1073/pnas.171286998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachand F, Lackner DH, Bahler J, Silver PA. Autoregulation of ribosome biosynthesis by a translational response in fission yeast. Mol Cell Biol. 2006;26:1731–1742. doi: 10.1128/MCB.26.5.1731-1742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Tollervey D, Cullin C, Lacroute F. Functional analysis of Rrp7p, an essential yeast protein involved in pre-rRNA processing and ribosome assembly. Mol Cell Biol. 1997;17:5023–5032. doi: 10.1128/mcb.17.9.5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Silver PA. Identifying proteins that affect mRNA localization in living cells. Methods. 2002;26:151–155. doi: 10.1016/S1046-2023(02)00017-8. [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. Embo J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T, Cordts S, Heuer S, Sauter M, Lorz H, Kranz E. Novel ribosomal genes from maize are differentially expressed in the zygotic and somatic cell cycles. Mol Gen Genet. 1999;261:416–427. doi: 10.1007/s004380050983. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi AH, Saunders WS. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Gauss R, Trautwein M, Sommer T, Spang A. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast. 2005;22:1–12. doi: 10.1002/yea.1187. [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Buonomo SB, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B, Viggiano S, Hibbs MA, Troyanskaya OG, Amberg DC. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev. 2007;21:148–159. doi: 10.1101/gad.1477507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Herruer MH, Mager WH, Woudt LP, Nieuwint RT, Wassenaar GM, Groeneveld P, Planta RJ. Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res. 1987;15:10133–10144. doi: 10.1093/nar/15.24.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- Irie K, Tadauchi T, Takizawa PA, Vale RD, Matsumoto K, Herskowitz I. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. Embo J. 2002;21:1158–1167. doi: 10.1093/emboj/21.5.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Kim GT, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22:257–264. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Amikura R, Mukai M. Localization of mitochondrial large ribosomal RNA in germ plasm of Xenopus embryos. Curr Biol. 1998;8:1117–1120. doi: 10.1016/s0960-9822(98)70466-x. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kruse C, Jaedicke A, Beaudouin J, Bohl F, Ferring D, Guttler T, Ellenberg J, Jansen RP. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J Cell Biol. 2002;159:971–982. doi: 10.1083/jcb.200207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, Shen Y, Zhao R, Smith RD. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci U S A. 2002;99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer RJ, van Raamsdonk-Duin MM, Mager WH, Planta RJ. The primary structure of the gene encoding yeast ribosomal protein L16. FEBS Lett. 1984;175:371–376. doi: 10.1016/0014-5793(84)80771-1. [DOI] [PubMed] [Google Scholar]
- Leer RJ, van Raamsdonk-Duin MM, Molenaar CM, Witsenboer HM, Mager WH, Planta RJ. Yeast contains two functional genes coding for ribosomal protein S10. Nucleic Acids Res. 1985;13:5027–5039. doi: 10.1093/nar/13.14.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Gu W, Meng X, Gonsalvez G, Singer RH, Chartrand P. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J Cell Biol. 2001;153:307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie DF, Resing KA, Lewis TS, Ahn NG. Mass spectrometric analysis of 40 S ribosomal proteins from Rat-1 fibroblasts. J Biol Chem. 1996;271:28189–28198. doi: 10.1074/jbc.271.45.28189. [DOI] [PubMed] [Google Scholar]
- Lucioli A, Presutti C, Ciafre S, Caffarelli E, Fragapane P, Bozzoni I. Gene dosage alteration of L2 ribosomal protein genes in Saccharomyces cerevisiae: effects on ribosome synthesis. Mol Cell Biol. 1988;8:4792–4798. doi: 10.1128/mcb.8.11.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Dooner HK. A mutation in the nuclear-encoded plastid ribosomal protein S9 leads to early embryo lethality in maize. Plant J. 2004;37:92–103. doi: 10.1046/j.1365-313x.2003.01942.x. [DOI] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci U S A. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. Embo J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Amikura R, Kobayashi S, Yamamoto H, Nishida H. Localization of mitochondrial large ribosomal RNA in the myoplasm of the early ascidian embryo. Dev Growth Differ. 1999;41:1–8. doi: 10.1046/j.1440-169x.1999.00409.x. [DOI] [PubMed] [Google Scholar]
- Polo SE, Almouzni G. Chromatin assembly: a basic recipe with various flavours. Curr Opin Genet Dev. 2006;16:104–111. doi: 10.1016/j.gde.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Masson J, Koncz C, Afsar K, Jakovleva L, Paszkowski J. Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. Embo J. 1999;18:490–499. doi: 10.1093/emboj/18.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg MO, Moritz M, Woolford JL., Jr. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Selker EU, Stevens JN, Metzenberg RL. Heterogeneity of 5S RNA in fungal ribosomes. Science. 1985;227:1340–1343. doi: 10.1126/science.2579431. [DOI] [PubMed] [Google Scholar]
- Tsugeki R, Kochieva EZ, Fedoroff NV. A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 1996;10:479–489. doi: 10.1046/j.1365-313x.1996.10030479.x. [DOI] [PubMed] [Google Scholar]
- Urbinati CR, Gonsalvez GB, Aris JP, Long RM. Loc1p is required for efficient assembly and nuclear export of the 60S ribosomal subunit. Mol Genet Genomics. 2006;276:369–377. doi: 10.1007/s00438-006-0151-7. [DOI] [PubMed] [Google Scholar]
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- Williams ME, Sussex IM. Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 1995;8:65–76. doi: 10.1046/j.1365-313x.1995.08010065.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


