Abstract
In many human cancers, tumor-specific chromosomal rearrangements are known to create chimeric products with the ability to transform cells. The EWS/WT1 protein is such a fusion product, resulting from a t(11;22) chromosomal translocation in desmoplastic small round cell tumors, where 265 aa from the EWS amino terminus are fused to the DNA binding domain of the WT1 tumor suppressor gene. Herein, we find that EWS/WT1 is phosphorylated in vivo on serine and tyrosine residues and that this affects DNA binding and homodimerization. We also show that EWS/WT1 can interact with, and is a substrate for, modification on tyrosine residues by c-Abl. Tyrosine phosphorylation of EWS/WT1 by c-Abl negatively regulates its DNA binding properties. These results indicate that the biological activity of EWS/WT1 is closely linked to its phosphorylation status.
Keywords: desmoplastic small round cell tumor, DNA binding, homodimerization, c-Abl
Desmoplastic small round cell tumor is an aggressive rare tumor that generally occurs in adolescence and is located to the peritoneal surfaces of the abdomen. This malignancy is associated with a recurrent translocation t(11;22)(p13;q22), which fuses the amino-terminal domain (NTD) of the EWS gene in-frame to three of the four carboxyl-terminal zinc fingers of the WT1 tumor suppressor gene (1, 2). The EWS gene is involved in several tumor-related chromosomal translocations that produce fusions with genes postulated to function as transcription factors (for a review, see ref. 3). In each case, the translocation produces chimeric molecules containing the EWS NTD fused to the DNA domain of the partner.
There are two isoforms of EWS/WT1 generated as a result of an alternative splicing event between the third and last WT1 zinc finger and resulting in the insertion or removal of three amino acids (±KTS). This event produces EWS/WT1 isoforms with distinct DNA binding properties (4). The EWS/WT1(−KTS) isoform recognizes GC-rich WT1 binding sites [5′-GCGGGGGCG-3′] with ≈10-fold higher affinity than WT1 and can transform NIH 3T3 cells (4, 5). On the other hand, EWS/WT1(+KTS) does not bind GC-rich WT1 sites and has no transforming activity. The NTD (amino acids 1–265) of EWS/WT1 is composed almost exclusively of tyrosine, glutamine, alanine, serine, threonine, glycine, and proline residues, of which some are organized in a repeated and degenerate polypeptide motif having the consensus NSYGQQS. This domain shares distant homology with the carboxyl-terminal domain (CTD) of eukaryotic RNA polymerase II (6) and is a potent transcriptional activator (4, 7, 8). One model to account for the transforming properties of EWS/WT1(−KTS) is that some of the genes normally under WT1 control are deregulated by EWS/WT1.
During our characterization of the EWS/WT1 oncogene we discovered that this product can self-associate and that the responsible region maps to the chimeric portion of the molecule, requiring both EWS and WT1 domains (see below). Herein, we report that phosphorylation of the EWS/WT1 chimeric product dramatically alters its biological properties, inhibiting both DNA recognition and homodimerization. In addition, we show that c-Abl is a candidate upstream modifier of EWS/WT1 biological activity.
Materials and Methods
Plasmid Constructions.
The construction of EWS/WT1(−KTS) has been described (4). To generate glutathione S-transferase (GST)-EWS/WT1(−KTS), which was used to produce bacterial recombinant protein, the EcoRI (Klenow blunted)–XhoI fragment from pcDNA3-EWS/WT1(−KTS) was transferred into the blunted SacI and intact XhoI sites of pGEX-RC. For expression in mammalian cells, the HincII fragment (Klenow repaired) containing the GST domain was isolated from pGEX-3T (Amersham Pharmacia) and subcloned into the EcoR V site of pcDNA3 (Invitrogen) to generate pcDNA3:GST. pcDNA3:GST-EWS/WT1(−KTS) was made by isolating an EcoNI–XhoI fragment from pGEX-RC:EWS/WT1(−KTS) and cloned into the same sites of pcDNA3:GST. To generate the amino terminally hemagglutinin (HA)-tagged EWS/WT1 [pcDNA3:HA-EWS/WT1(−KTS)], the EcoRI–XbaI fragment of pcDNA3-EWS/WT1(−KTS) was cloned into a pcDNA3-HA tag vector containing three copies of the HA peptide epitope (NH2-YPYDVPDYAG-COOH) (kindly provided by H. Imataka and N. Sonenberg, McGill University). For histidine-tagged recombinant EWS/WT1(−KTS) proteins, the EcoRI–HindIII fragment from pcDNA3-EWS/WT1(−KTS) was subcloned into the same sites of the pTrcHisB XPRESS SYSTEM vector (Invitrogen), to generate pTrcHisB-EWS/WT1(−KTS). Detailed protocols outlining plasmid constructions can be obtained from the authors on written request. All constructs involving manipulations by PCR were sequenced by using double-stranded DNA templates (9) to ensure the absence of mutations.
Cell Cultures, Transfections, Protein Purifications, and Western Blot Analyses.
COS-7 or 293T cell lines were maintained in DMEM supplemented with 10% heat-inactivated FCS (GIBCO/BRL), penicillin, and streptomycin. For phosphorylation studies, 20 μg of pcDNA3/(His)6-EWS/WT1(−KTS) was used in transient transfection assays, and cells were pretreated with sodium pervanadate, prepared fresh by mixing 6% hydrogen peroxide (Pierce) and sodium orthovanadate (Fisher Scientific) to yield a final concentration of 10 mM sodium orthopervanadate. This solution was diluted to 240 μM pervanadate with DMEM, added to the cells, and maintained at 37°C for 15 min.
For in vivo self-association experiment, 10 μg of HA-tagged EWS/WT1(−KTS) plasmid, pcDNA3:HA-EWS/WT1 (−KTS), was transfected with either 10 μg of pcDNA3:GST or pcDNA3:GST-EWS/WT1(−KTS) DNA. After 48 hr, cells were harvested in resuspension buffer (50 mM Tris⋅HCl, pH 8.0/1% NP-40/2 mM EDTA/150 mM NaCl/1 mM DTT/2 mM PMSF/1 μg/ml aprotinine/1 μg/ml leupeptine/1 μg/ml antipain) and lysed by sonication. Supernatants were collected by centrifugation at 16,000 × g for 15 min at 4°C and incubated with glutathione beads (Amersham Pharmacia) for 1 hr. Beads were collected by brief centrifugation and washed with resuspension buffer four times. The affinity-selected proteins were eluted in 1× SDS loading buffer (62.5 mM Tris⋅HCl, pH 6.9/10% glycerol/2% SDS/5% β-mercaptoethanol).
Affinity-purified proteins were separated by electrophoresis through a 10% polyacrylamide gel and transferred onto an Immobilon poly(vinylidene difluoride) (PVDF) membrane (Millipore). This filter was blocked with 5% nonfat in TBS-T (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/0.1% Tween 20) for 1 hr at room temperature, and 12CA5 (anti-HA antibody) and horseradish peroxidase-conjugated donkey anti-rabbit (Amersham Life Science) antibodies were used as primary and secondary antibodies, respectively. The blot was incubated and visualized with ECL solution according to the manufacturer’s instructions (DuPont/NEN). For antiphosphotyrosine blots, 5% BSA (Amresco, Euclid, OH) in TBS-T was used for preblocking, and the 4G10 antiphosphotyrosine antibody was used for probing.
In Vitro Transcription and Translation.
In vitro transcription and translation were performed as described (4). Briefly, pcDNA3:EWS/WT1(−KTS) was linearized with SmaI and in vitro transcriptions were performed with T7 RNA polymerase. In vitro translations were performed in the presence of [35S]methionine (NEN) in rabbit reticulocyte lysates, essentially as described by the manufacturer’s instructions (Promega). For analysis, every in vitro translation product was electrophoresed on 10% SDS/PAGE.
Southwestern Blot Analysis.
Protein blots for Southwestern blot analysis (10) were generated by separating EWS/WT1 protein after Ni-nitrilotriacetic acid resin purification by 10% SDS/PAGE and electroblotting onto nitrocellulose (Schleicher & Schuell). The protein was denatured 10 min with denaturation buffer (25 mM Hepes⋅KOH, pH 7.6/12.5 mM MgCl2/20% glycerol/0.1% NP-40/0.1 M KCl/10 μM ZnSO4/1 mM DTT/6 M guanidium chloride) and renatured for 10 min with equal volumes of denaturation buffer and renaturation buffer (25 mM Hepes⋅KOH, pH 7.6/12.5 mM MgCl2/20% glycerol/0.1% NP-40/0.1 M KCl/10 μM ZnSO4/1 mM DTT). These steps were repeated at least five more times. Subsequently, the blot was incubated with blocking buffer (3% nonfat dried milk in renaturation buffer) for 30 min and hybridized for 30 min with binding buffer (0.25% nonfat dried milk in renaturation buffer) containing the DNA probe. End-labeled probe was prepared as described (4), and 106 cpm/ml of probe and 0.5 mM of unlabeled ATP were used in each hybridization. The blot was washed with renaturation buffer, dried, and exposed at −80°C with intensifyng screen. Oligonucleotides containing the WT1 recognition site (in capital letters) [WTE: 5′-gagtGCGTGGGAGTagaa-3′] or, a mutant version not capable of binding WT1 [WTE(m): 5′-gagtGCGTGAGAGTagaa-3′], were used as probes.
Results
Forskolin treatment of fibroblast cells expressing WT1 leads to phosphorylation of Ser-365 and Ser-393 of zinc fingers 2 and 3, respectively, and abolishes the DNA binding activity of WT1 in vitro (12). The RNA binding activity of WT1 is not altered by phosphorylation (13). Because the EWS/WT1 contains three of the four WT1 zinc fingers, as well as several potential tyrosine phosphorylation sites, we wanted to determine whether EWS/WT1 was phosphorylated in vivo and determine whether any functional characteristics of EWS/WT1 were altered by such modification.
In Vivo Phosphorylation of EWS/WT1 Regulates Its DNA Recognition Ability.
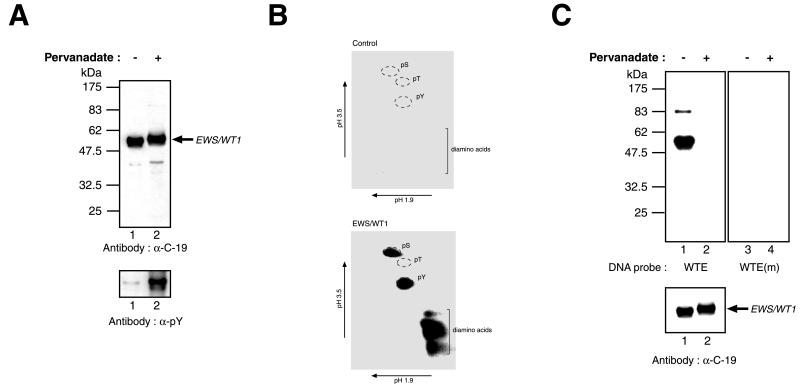
An expression vector driving the synthesis of EWS/WT1 containing six histidine residues at the amino terminus was transiently transfected into COS-7 cells. Forty-eight hours after transfection, cells were exposed for 15 min to pervanadate, a known specific inhibitor of phosphotyrosine phosphatases (14). Cell extracts probed with an antibody specific to WT1 zinc finger IV (α-C-19) revealed that the pervanadate-treated EWS/WT1 product had a slower electrophoretic mobility compared with that isolated from control cells not treated with pervanadate (Fig. 1A Upper, compare lane 2 to 1). To determine whether this decreased mobility could be caused by tyrosine phosphorylation of EWS/WT1, the same protein preparations were probed with an antiphosphotyrosine (α-pY) antibody (4G10) (Fig. 1A Lower). A significant amount of EWS/WT1 isolated from cells pretreated with pervanadate contained phosphotyrosine residues (Fig. 1A Lower, lane 2), compared with protein from untreated cells (Fig. 1A Lower, lane 1). Because Ni2+-affinity protein preparations were used in the above experiments, we interpret these results to indicate that EWS/WT1 is a substrate for (an) endogeneous protein tyrosine kinase(s).
Figure 1.
In vivo phosphorylation of EWS/WT1. (A) EWS/WT1 is a phosphotyrosine protein. Cells transfected with pcDNA3.1-(His)6EWS/WT1(−KTS) were treated with vehicle (−) or pervanadate (+) 15 min before harvesting. Western blots were performed on Ni2+-affinity-purified protein preparations prepared from cell lysates and resolved on 10% SDS-polyacrylamide gels. The nature of the antibody used in the Western blot is indicated below each panel. The arrow indicates the position of migration of the EWS/WT1 product. Molecular mass markers are provided to the left and are derived from prestained New England Biolabs (NEB) protein standards (broad range). (B) Phosphoamino acid analysis of (His)6-EWS/WT1. Transiently transfected COS cells were metabolically labeled with 0.85 mCi/ml inorganic pyrophosphate (NEN) in phosphate-free media supplemented with 10% dialyzed FBS for 4 hr. Pervanadate solution was added directly to the labeling media for 15 min, and cells were harvested and lysed by sonication in RIPA buffer (50 mM Tris⋅HCl, pH 8.0/150 mM NaCl/1.0% NP-40/0.5% sodium deoxycholate/0.1% SDS). After preclearing of extracts with rabbit preimmune serum and protein G Sepharose, immunoprecipitations were performed with the C-19 antibody (Santa Cruz Biotechnology). Bound protein was eluted under denaturing conditions (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris⋅HCl, pH 8.0), applied to Ni-nitrilotriacetic acid agarose resin (Qiagen), washed twice in wash I (8 M urea, 0.1 M sodium phosphate, 0.1 M Tris⋅HCl, pH 6.3), four times in wash II (40 mM sodium phosphate, pH 8.0, 300 mM NaCl, 30 mM immidazole), eluted with elution buffer (8 M urea/0.1 M sodium phosphate/0.01 M Tris⋅HCl, pH 6.3/100 mM EDTA), and separated on a 10% SDS/PAGE. Proteins were transferred onto Immobilon PVDF membrane and (His)6-EWS/WT1-identified by autoradiography. The same molecular mass region was excised from the PVDF membrane, which contained extract prepared from control cells transfected with empty vector. Hydrolysis was performed in 6 M HCl at 110°C for 1 hr, after which time 1 ml of water was added and samples were lyophilized. Samples then were resuspended in 10 μl electrophoresis buffer (0.58 M formic acid/1.36 M acetic acid, pH 1.9), spotted onto cellulose thin-layer plates, and analyzed by two-dimensional electrophoresis at pH 1.9 (0.58 M formic acid, 1.36 M acetic acid) and at pH 3.5 [0.87 M acetic acid, 0.5% (vol/vol) pyridine, 0.5 mM EDTA]. Nonradioactive standards were visualized by staining with ninhydrin in acetone and labeled amino acids were detected by autoradiography. (C) Southwestern blot analysis was performed on EWS/WT1(−KTS) isolated from transfected COS-7 cells treated with only vehicle (−) or with pervanadate (+) and fractionated on a 10% SDS/PAGE (Upper). After guanidine HCl denaturation/renaturation washes, the blot was probed with a 32P end-labeled oligonucleotide containing the wild-type WT1 recognition sequence (WTE) or a mutant WT1 motif [WTE(m)]. After washes, the blots were exposed to x-ray film (Kodak) at −70°C overnight with an intensifying screen. Immunoblotting with an anti-EWS/WT1 antibody (α-C-19, Santa Cruz Biotechnology) was used to confirm that equivalent levels of protein were loaded on the gels (Lower). The arrow indicates the position of migration of EWS/WT1. Molecular mass markers are provided to the left and are derived from prestained NEB protein standards (broad range).
Phosphoamino acid analysis of His-tagged EWS/WT1 used in these experiments revealed the presence of phosphotyrosine and phosphoserine residues (Fig. 1B). To assess whether in vivo phosphorylation of EWS/WT1 had a functional consequence on the DNA binding properties of EWS/WT1(−KTS), we performed Southwestern blot analysis using a 32P-labeled WT1 recognition site (called WTE) as probe (Fig. 1C). In this assay, EWS/WT1(−KTS) isolated from untreated lysates recognized the WTE probe (Fig. 1C Upper, lane 1). This binding is specific, because a mutant probe, WTE(m), containing a point mutation in the WT1 recognition site, failed to bind (Fig. 1C Upper, compare lane 3 to lane 1). Pervanadate treatment of EWS/WT1(−KTS) transfected COS-7 cells produced an isoform that no longer efficiently bound DNA (Fig. 1C Upper, compare lane 2 to 1). The differences in DNA binding are not a result of differences in protein levels, because both extracts contain equivalent amounts of EWS/WT1(−KTS) (Fig. 1C Lower). As expected, radiolabeled WTE(m) did not bind to EWS/WT1(−KTS) isolated from pervanadate-treated cells (Fig. 1C Upper, lane 4).
EWS/WT1 Is a Substrate for the c-Abl Protooncogene.
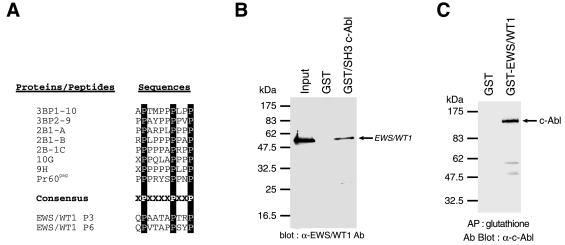
Because the CTD of RNA polymerase II is the site of multiple phosphorylation events (11), some of which are thought to be mediated by the c-Abl product (14), we decided to assess whether c-Abl resided upstream of EWS/WT1. In addition, we noted the presence of two Src homology 3 (SH3) domain interacting motifs within the EWS/WT1 NTD that corresponding to the consensus motif (PxxxxPxxP) identified in a number of c-Abl targets (Fig. 2A). Indeed, GST pull-downs of recombinant His-tagged EWS/WT1 with GST or c-Abl GST/SH3 revealed that EWS/WT1 was specifically bound to the c-Abl SH3 domain (Fig. 2B). To extend these results and demonstrate an in vivo association, EWS/WT1 and c-Abl mammalian expression vectors driving synthesis of c-Abl and either GST or GST-EWS/WT1 were introduced into COS-7 cells. Forty-eight hours after transfections, cell extracts were prepared and affinity-precipitated with glutathione affinity resin (Fig. 2C). Eluted proteins were probed by Western blotting for the presence of copurifying c-Abl protein. Clearly, c-Abl can copurify with GST-EWS/WT1, but not GST, demonstrating that EWS/WT1 and c-Abl are capable of associating in vivo (Fig. 1C), although we have yet to map the interacting domain on EWS/WT1.
Figure 2.
EWS/WT1 and c-Abl can associate. (A) Alignment of consensus interacting motif for c-Abl SH3 domain among a series of downstream c-Abl targets, including two putative SH3 domain binding sites within EWS/WT1. The c-Abl SH3 consensus binding site of PxxxxPxxP has been reported (27, 31). (B) Association of EWS/WT1 with the SH3 domain of c-Abl. Bacterially produced (His)6-EWS/WT1 protein was incubated with either GST or GST-fusion c-Abl SH3 domain. An aliquot of the input (20%) and the pellets from GST pull-down assays were analyzed by 10% SDS/PAGE, and the bound EWS/WT1 proteins were detected with an anti-WT1 antibody (C-19; Santa Cruz) and chemiluminescence (DuPont/NEN). (C) Copurification of EWS/WT1 and c-Abl from cell extracts. Forty-eight hours after cotransfection of COS-7 cells with 10 μg of pcDNA3/Abl and either 10 μg of pcDNA3/GST or pcDNA3/GST-EWS/WT1, cell extracts were prepared as described in Materials and Methods and affinity-precipitated with glutathione beads. After fractionation on an 8% SDS/PAGE and transfer of the proteins to Immobilon PVDF membrane, the filter was preblocked in 5% nonfat skim milk in TBS-T (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/0.1% Tween 20). A mAb against c-Abl [c-Abl(24–11); Santa Cruz Biotechnology] was used to interrogate the blot. Proteins were visualized with horseradish peroxidase-conjugated goat anti-mouse antibody by using an ECL solution (DuPont/NEN) according to the manufacturer’s instruction.
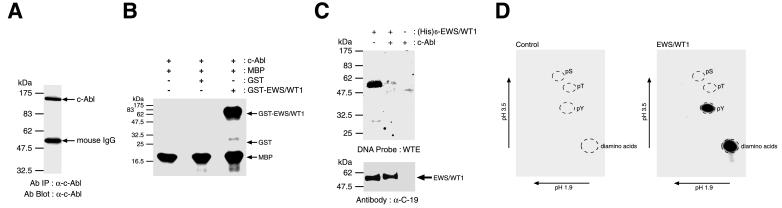
To determine whether EWS/WT1 is a substrate for phosphorylation by c-Abl, we isolated c-Abl protein from Jurkat cells by immunoprecipitation (Fig. 3A). Myelin basic protein, a nonspecific substrate, was used to normalize and monitor the immunopurified Abl kinase activity. Incubation of c-Abl with His-tagged EWS/WT1 followed by Southwestern blot analysis with WTE as a probe, abolished DNA binding (Fig. 3C). Immunohistochemical probing for EWS/WT1 on this same blot demonstrated that equivalent amounts of protein were present in both reactions and that the presence of immunoprecipitated c-Abl did not cause degradation of the substrate (Fig. 3C). Phosphoamino acid analysis of His-tagged EWS/WT1 used in these experiments revealed only the presence of phosphotyrosine residues (Fig. 3D). These results clearly demonstrate that c-Abl is capable of modifying EWS/WT1 DNA binding activity by tyrosine phosphorylation.
Figure 3.
Modification of EWS/WT1 by c-Abl. (A) Immunoprecipitation of c-Abl from Jurkat cell lysates. Human c-Abl was precipitated with anti-c-Abl antibody, c-Abl(24–11) (Santa Cruz Biotechnology) and separated on an 8% SDS/PAGE. After electrophoresis, proteins were transferred onto Immobilon PVDF membrane and a Western blot was performed with the same antibody and an horseradish peroxidase-conjugated goat anti-mouse antibody (Santa Cruz Biotechnology). Molecular mass markers are provided to the left and are derived from prestained NEB protein standards (broad range). (B) In vitro phosphorylation of EWS/WT1 by c-Abl. c-Abl immunoprecipitates were resuspended with myelin basic protein (MBP) and either recombinant GST or GST-EWS/WT1 in kinase buffer [10 mM Tris⋅HCl, pH 7.5/150 mM NaCl/10 mM MgCl2/0.5 mM DTT/50 μM ATP/10 μCi γ-32P-ATP (3,000 Ci/mmol)/240 μM pervanadate] and incubated at 37°C for 1 hr with moderate agitation. Reaction mixtures were fractionated on a 10% polyacrylamide gel, dried, and exposed to X-Omat x-ray film at −70°C for 1 hr. (C) Modification of EWS/WT1 by c-Abl abolishes DNA recognition. c-Abl immunoprecipitates were resuspended with recombinant (His)6-EWS/WT1 protein in kinase buffer (10 mM Tris⋅HCl, pH 7.5/150 mM NaCl/10 mM MgCl2/0.5 mM DTT/5 mM ATP/240 μM pervanadate) and incubated at 37°C for 1 hr with moderate agitation. Reaction mixtures were analyzed by Southwestern blotting as described in Materials and Methods. The presence of c-Abl immunoprecipitates or (His)6-EWS/WT1 in the reaction mixtures is indicated (Upper). (Lower) A Western blot analysis of the recombinant EWS/WT1 after incubation with c-Abl and probed with α-C-19 antibody. (D) Phosphoamino acid analysis of in vitro-phosphorylated (His)6-EWS/WT1 by c-Abl. Phosphorylated (His)6-EWS/WT1 was separated by 10% SDS/PAGE, transferred onto Immobilon PVDF membrane, and subjected to phosphoamino acid analysis as described in Fig. 1. The positions of the unlabeled standards, detected with ninhydrine staining are shown as pS (phosphoserine), pT (phosphothreonine), and pY (phosphotyrosine).
Inhibition of EWS/WT1 Self-Association by Tyrosine Phosphorylation.
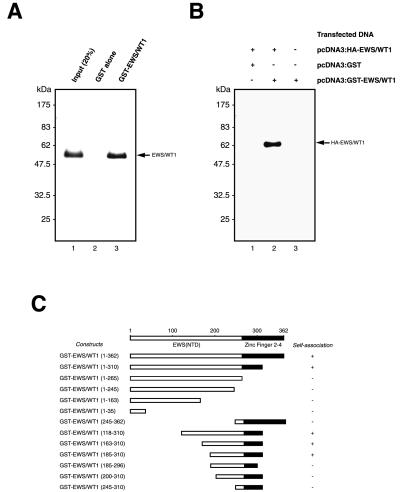
During the course of these studies, we noticed that EWS/WT1 could self-associate and that phosphorylation of EWS/WT1 also affected this property (described below). Initial experiments aimed to map the EWS/WT1 homodimerization potential using GST pull-down assays performed with in vitro-translated EWS/WT1(−KTS) and GST-EWS/WT1 recombinant protein purified from bacteria. As shown in Fig. 4A, 35S-methionine-labeled EWS/WT1(−KTS) specifically bound to GST-EWS/WT1 (lane 3) but not to GST (lane 2).
Figure 4.
Self-association of EWS/WT1. (A) Retention of EWS/WT1 by GST-EWS/WT1 recombinant protein. In vitro-transcribed and -translated 35S-methionine-labeled EWS/WT1 was incubated with either ≈2 μg of GST (lane 2) or GST-EWS/WT1 (lane 3) bound to glutathione-Sepharose beads. Approximately 20% of the 35S-methionine-labeled protein used in the experiment was loaded in lane 1 as a reference. The EWS/WT1 proteins were eluted with SDS loading buffer and analyzed by 10% SDS/PAGE. The position of the molecular mass markers are indicated on the left (NEB broad range markers) and the position of EWS/WT1 is indicated to the right. (B) EWS/WT1 self-association in vivo. 293T cells were transfected with pcDNA3:HA-EWS/WT1 and either pcDNA3:GST (lane 1) or pcDNA3:GST-EWS/WT1 (lane 2), or 293T cells were transfected with only pcDNA3:GST-EWS/WT1 (lane 3). Affinity-precipitations were performed with glutathione-Sepharose 4B beads and analyzed for the presence or absence of HA-EWS/WT1 by Western blotting with an anti-HA antibody (12CA5). + or − indicate the presence or absence, respectively, of the indicated expression vectors in the transfection mixture. The positions of molecular mass markers are indicated on the left (NEB broad range markers) and the position of HA-EWS/WT1 is indicated by an arrow. (C) The EWS/WT1 self-association domain resides between amino acids 185 and 310. A schematic diagram illustrating the EWS/WT1 deletion mutants used in this study is presented. The results from in vitro affinity selection experiments are summarized to the right. + or − indicates whether or not 35S-methionine-labeled full-length EWS/WT1 could be retained on an affinity matrix containing the corresponding GST-EWS/WT1 derivative. The amino acid numbering refers to the position of the amino acids in the fusion protein, not the numbering normally found in EWS or WT1.
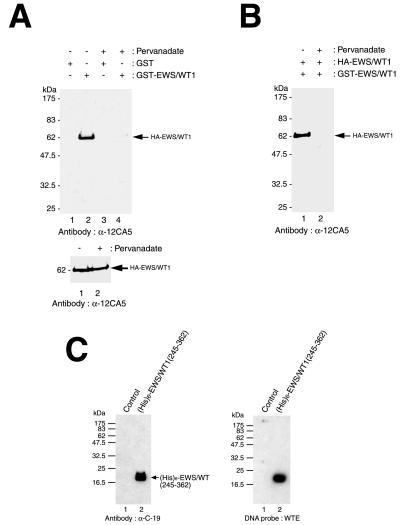
To demonstrate that self-association was not restricted to our in vitro binding conditions, affinity coprecipitation experiments were performed directly from transfected cells (Fig. 4B). 293T cells were cotransfected with pcDNA3:HA-EWS/WT1 and pcDNA3:GST-EWS/WT1 or pcDNA3:GST, and lysates were prepared 48 hr after transfection. GST or GST-EWS/WT1 were affinity-precipitated with glutathione-Sepharose beads, and subsequent immunoblotting performed by using an anti-HA antibody (12CA5) revealed that HA-EWS/WT1 is present only in extracts containing GST-EWS/WT1 (Fig. 4B).
To identify the domain(s) within EWS/WT1 that mediate self-association, in vitro GST pull-down assays were performed with a series of EWS/WT1 deletion mutants (Fig. 4C). Amino-terminal and carboxyl-terminal deletions of the EWS(NTD) were generated and fused in-frame to the GST domain. After purification of the recombinant protein, in vitro-translated EWS/WT1 was assessed for its ability to interact with the GST-EWS/WT1 derivatives. These experiments map the interaction domain to a 126-aa region (amino acids 185–310), encompassing both a region of the WT1 zinc fingers and the EWS sequences immediately upstream of the fusion junction (Fig. 4C). We were unable to refine the definition of this region because a series of finer deletions [e.g., GST-EWS/WT1(200–310) or GST-EWS/WT1(185–296)] failed to self-associate.
To determine the effect of in vivo phosphorylation on dimerization of EWS/WT1, we transfected COS-7 cells with pcDNA3:HA-EWS/WT1 and 48 hr later prepared cell extracts. These were used in affinity-selection assays with immobilized GST or GST-EWS/WT1 and revealed that HA-EWS/WT1 isolated from pervanadate-treated cells could not self-associate, whereas EWS/WT1 from untreated cells could (Fig. 5A, compare lane 4 to 2). The differences in self-association behavior between phosphorylated or underphosphorylated EWS/WT1 species (Fig. 5A, compares lanes 4 and 2) cannot be attributed to differences in protein levels (Fig. 5A Lower). A possible alternative interpretation to our data is that phosphorylation of HA-EWS/WT1 increases self-association, such that no monomers remain when extracts are challenged with immobilized GST-EWS/WT1. To test this hypothesis, COS-7 cells were transfected with HA- and GST-tagged EWS/WT1. After treatment with vehicle or prevandate, cell extracts were prepared and purified over a glutathione resin (Fig. 5B). If phosphorylation increases self-association, then one would expect to find HA-EWS/WT1 copurifying with GST-EWS/WT1 from pervanadate-treated cells. This was not the case because HA-EWS/WT1 copurified with GST-EWS/WT1 from untreated COS-7 cells, but not from prevandate-treated cells (Fig. 5B), indicating that phosphorylation prevents self-association of EWS/WT1.
Figure 5.
Phosphorylation of EWS/WT1 inhibits self-association. (A) Loss of EWS/WT1 self-association after phosphorylation. Cell lysate containing HA-tagged EWS/WT1 was incubated with either GST-EWS/WT1 or GST alone (indicated above panel). After affinity-selection, HA-tagged EWS/WT1 protein was quantitated by Western blot analysis using the anti-HA antibody, 12CA5. Treatment of cells with vehicle or with pervanadate is indicated at the top. The arrow indicates the position of migration of HA-EWS/WT1. Molecular mass markers are provided to the left and are derived from prestained NEB protein standards (broad range). (Lower) Western blotting shows that similar amounts of HA-EWS/WT1 are present in the input lysates. Ten percent of the input samples were separated on a 10% SDS/PAGE, transferred onto Immobilon PVDF membrane, and probed with anti-HA antibody, 12CA5. (B) EWS/WT1 self-association is not increased after phosphorylation. Ten micrograms of pcDNA3:HA-EWS/WT1 and pcDNA3:GST-EWS/WT1 was cotransfected into COS-7 cells and half of the cells were treated with pervanadate for 15 min before harvesting. After selection with glutathione-Sepharose beads, proteins were separated by 10% SDS/PAGE and interrogated by Western blotting using an anti-HA antibody (12CA5). Whether cells were pretreated with vehicle (−) or pervanadate for 15 min (+) before extracts were prepared is indicated at the top. (C) EWS/WT1 self-association and DNA binding domains are separable. A deletion mutant of EWS/WT1 containing six histidines, (His)6-EWS/WT1(245–362), was expressed in Escherichia coli, purified with Ni-nitrilotriacetic acid agarose resin, and probed with α-C-19 (Left). After transfer to nitrocellulose, Southwestern blot analysis was performed with WTE as probe (Right). The position of molecular mass markers are indicated on the left (NEB broad range markers) and the position of migration of (His)6-EWS/WT1(245–362) protein is indicated on the right.
Self-Association of EWS/WT1 Is Not Required for DNA Binding.
Because the DNA binding and self-association domains of EWS/WT1 overlap (Fig. 4C) and both are influenced by phosphorylation (Figs. 1C, 3C, and 5A), we addressed whether loss of DNA binding in response to this modification was an indirect consequence of loss of self-association (Fig. 5C). To this end, we generated a deletion mutant of EWS/WT1 containing six histidine residues, (His)6-EWS/WT1(245–362). This truncated protein contains the DNA binding domain of EWS/WT1, but cannot self-associate (Fig. 4C). Bacterially produced recombinant (His)6-EWS/WT1(245–362) was intact (Fig. 5C, Left) and was capable of binding WTE as assessed by Southwestern analysis (Fig. 5C, Right). These data indicate that although the EWS/WT1 DNA binding and self-association domains overlap, self-association is not required for DNA interaction.
Discussion
The results reported herein begin to explore the functional regulation of the EWS/WT1 oncoprotein. Whereas WT1 is generally a repressor of transcription, EWS/WT1 is a potent activator. Many potential WT1 target genes have been identified on the basis of the presence of potential binding sites within their promoters. These include those encoding for early growth response 1 (EGR-1), epidermal growth factor receptor, insulin-like growth factor 2, insulin-like growth factor 1 receptor (IGF1R), platelet-derived growth factor A (PDGFA), PAX2, c-MYC, BCL2, the transforming growth factor-β molecules 1, 2, and 3, retinoic acid receptor α, colony-stimulating factor 1, c-MYB, p21, and WT1 itself, among many others (16, 17). EWS/WT1 has been demonstrated to activate transcription of the IGF1R gene (7) and EGR-1 (8) in transient transfections and of the endogenous PDGFA promoter (15). Our results demonstrate that EWS/WT1 is a tyrosine- and serine-phosphoryated protein and that specific interaction with DNA is abolished when EWS/WT1 is phosphorylated (Figs. 1 and 3). There are reported differences regarding the ability of EWS/WT1 to activate specific downstream targets of WT1 (e.g., see ref. 15) and a possible explanation for this could be that cell-type specific differences in the phosphorylation status of EWS/WT1 may be responsible. Regulation of DNA binding by phosphorylation of sequence-specific transcription factors is well documented and can be positively (18, 19) or negatively (20, 21) influenced. In addition, the EWS product can be phosphorylated by protein kinase C through an IQ domain and this phosphorylation inhibits its ability to bind RNA (22). The EWS/WT1 chimeric product does not retain the IQ domain and therefore is not expected to be a substrate for protein kinase C. However, we have found that it is a substrate for purified protein kinase A, and therefore it is likely that some of the observed serine phosphorylation (Fig. 1B) is caused by modification by this kinase (data not shown).
Our finding of tyrosine phosphorylation of EWS/WT1 in vivo (Fig. 1) and that c-Abl can associate with, and phosphorylate, EWS/WT1 in vitro (Figs. 2 and 3), suggests that c-Abl may reside upstream of EWS/WT1. Although the large number of serine residues (40 serines in the NTD and eight serines in WT1 zinc fingers 2, 3, and 4) in the vicinity of tyrosine residues within the EWS/WT1 NTD does not allow us to compare phospho-peptide maps of EWS/WT1 isolated from transfected cells treated with pervanadate (Fig. 1B, phosphorylated on serines and tyrosines) to EWS/WT1 phosphorylated in response to c-Abl (Fig. 3D, phosphorylated on tyrosines), there are several suggestive lines of evidence that EWS/WT1 lies downstream of the c-Abl kinase. EWS/WT1 has a high content of tyrosine, glutamine, alanine, serine, threonine, glycine, and proline residues, some of which are organized in a repeated and degenerated polypeptide motif with a frequently occurring serine-tyrosine dipeptide (NSYGQQS) (6). A database search revealed that the EWS/WT1 NTD shares homology with the CTD of the large subunit of eukaryotic RNA polymerase II (6). Interestingly, this subunit is efficiently phosphorylated on tyrosines by c-Abl (14) and the site of phosphorylation is within the CTD, with c-Abl capable of phosphorylating most, if not all, of the 52 tyrosine residues in the CTD (14). The consequences of this phosphorylation event on RNA polymerase II is to convert transcriptionally paused complexes into elongation competent molecules (23). It is doubtful that EWS/WT1 competes with RNA polymerase II for the c-Abl kinase, or for similar accessory factors, because this competition would predict interference with RNA polymerase II function and inhibition of transcription. Rather, we prefer a model where EWS/WT1 may recruit basal transcription factors necessary for initiation or elongation. Consistent with this prediction, Petermann et al. (24) recently have demonstrated that EWS/Fli-1 associates with HsRPB7 (a subunit of RNA polymerase II). The NTD of EWS/WT1 contains two putative c-Abl SH3 binding sites although it remains to be established whether c-Abl is interacting via these motifs (Fig. 2A). Our results do not preclude EWS/WT1 from interacting with (and being a target of) other kinases, and recent experiments have demonstrated that EWS can interact with the SH3 domain of Bruton’s tyrosine kinase (25). The EWS/WT1 fusion protein also contains several potential tyrosine phosphorylation sites and SH2 domain interacting motifs, such as YXXP, which have been shown to interact with the SH2 domain of Abl, rasGAP, or Crk and YXXV, which have been shown to interact with Src family members or SHPTP2 (26).
Several mechanisms could account for the effects of tyrosine phosphorylation of EWS/WT1 by c-Abl on DNA binding. In many cases where phosphorylation inhibits DNA binding, the phosphorylation sites are located either within, or near, the DNA binding domain (28). In such cases, it is likely that phosphorylation interferes with DNA binding by electrostatic repulsion between phosphate groups on the protein and phosphates on the DNA (20, 21). Although we have not yet determined the mechanism for inhibition of EWS/WT1 DNA binding by tyrosine phosphorylation (Fig. 3), it is probably not caused by direct tyrosine phosphorylation of the DNA binding domain because this domain contains only one tyrosine residue not present in a known consensus sequence for tyrosine phosphorylation. In those cases in which the phosphorylation sites are separate from the DNA binding domain, it is most likely that phosphorylation alters the conformation of the protein in such a way that its DNA binding activity is altered (29). In vivo phosphorylation of EWS/WT1 is likely to be more complex. Sakamoto et al. (12) have shown that the WT1 can be phosphorylated on Ser-365 and Ser-393 of zinc fingers 2 and 3, respectively in response to forskalin treatment. The experiments herein do not rule out that similar phosphorylation events also can influence EWS/WT1 DNA binding ability but rather demonstrate a very specific effect of tyrosine phosphorylation.
We also have shown a unique property of EWS/WT1, self-association. The responsible domain maps to the fusion junction moiety of the chimeric protein and is negatively influenced by phosphorylation (Figs. 4 and 5). We show that although the DNA binding and self-association domains overlap, DNA binding does not depend on self-association (Fig. 5C). Although the functional significance self-association still remains undefined, it is possible that binding of several EWS/WT1 molecules to sites on a given gene leads to homotypic associations that translate into transcriptional effects, as has been proposed for the WT1 tumor suppressor gene (30). The self-association of EWS/WT1 represents an excellent target for drug design given that it resides within the junction portion of EWS/WT1 and therefore is unique to this chimeria.
Acknowledgments
We thank Dr. Michel L. Tremblay and members of his lab for their generous help, especially Jean-François Côté and John Wagner for thoughtful discussions. J.K. is supported by a fellowship from the McGill Faculty of Medicine. J.P. is a Medical Research Council of Canada Scientist. This work was supported by a grant from the Medical Research Council of Canada to J.P.
Abbreviations
- NTD
amino-terminal domain
- GST
glutathione S-transferase
- HA
hemagglutinin
- CTD
carboxyl-terminal domain
- SH3
Src homology 3
- PVDF
poly(vinylidene difluoride)
References
- 1.Gerald W L, Rosai J, Ladanyi M. Proc Natl Acad Sci USA. 1995;92:1028–1032. doi: 10.1073/pnas.92.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ladanyi M, Gerald W L. Cancer Res. 1994;54:2837–2840. [PubMed] [Google Scholar]
- 3.Ladanyi M. Diag Mol Pathol. 1995;4:162–173. doi: 10.1097/00019606-199509000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Lee K, Pelletier J. Oncogene. 1998a;16:1021–1030. doi: 10.1038/sj.onc.1201616. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Lee K, Pelletier J. Oncogene. 1998b;16:1973–1979. doi: 10.1038/sj.onc.1201716. [DOI] [PubMed] [Google Scholar]
- 6.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, Jong P-d, Rouleau G, et al. Nature (London) 1992;356:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 7.Karnieli E, Werner H, Rauscher F J, III, Benjamin L E, LeRoith D. J Biol Chem. 1996;271:19304–19309. doi: 10.1074/jbc.271.32.19304. [DOI] [PubMed] [Google Scholar]
- 8.Rauscher F J, III, Benjamin L E, Fredericks W J, Morris J F. Cold Spring Harbor Symp Quant Biol. 1994;59:137–146. doi: 10.1101/sqb.1994.059.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinson C R, LaMarco K L, Johnson P F, Landschulz W H, McKnight S L. Genes Dev. 1988;2:801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- 11.Dahmus M E, Dynan W S. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 109–128. [Google Scholar]
- 12.Sakamoto Y, Yoshida M, Semba K, Hunter T. Oncology. 1997;15:2001–2012. doi: 10.1038/sj.onc.1201391. [DOI] [PubMed] [Google Scholar]
- 13.Ye Y, Raychaudhuri B, Gurney A, Campbell C E, Williams B R G. EMBO J. 1996;15:5606–5615. [PMC free article] [PubMed] [Google Scholar]
- 14.Baskaran R, Dahmus M E, Wang J Y J. Proc Natl Acad Sci USA. 1993;90:11167–11171. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S B, Kolquist K A, Nichols K, Englert C, Maheswaran S, Ladanyi M, Gerald W L, Haber D A. Nat Genet. 1997;17:309–313. doi: 10.1038/ng1197-309. [DOI] [PubMed] [Google Scholar]
- 16.Reddy J C, Licht J D. Biochem Biophys Acta. 1996;1287:1–28. doi: 10.1016/0304-419x(95)00014-7. [DOI] [PubMed] [Google Scholar]
- 17.Englert C, Maheswaran S, Garvin A J, Kreidberg J, Haber D A. Cancer Res. 1997;57:1429–1434. [PubMed] [Google Scholar]
- 18.Janknecht R, Hipskind R A, Houthaeve T, Nordheim A, Stunnenberg H G. EMBO J. 1992;11:1045–1054. doi: 10.1002/j.1460-2075.1992.tb05143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klimczak L J, Schindler U, Cashmore A R. Plant Cell. 1992;4:87–89. doi: 10.1105/tpc.4.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lüscher B, Christenson E, Litchfield D W, Krebs E G, Eisenman R N. Nature (London) 1990;344:517–522. doi: 10.1038/344517a0. [DOI] [PubMed] [Google Scholar]
- 21.Boyle W J, Smeal T, Defize L H K, Angel O, Woodgett J R, Karin M, Hunter T. Cell. 1991;64:573–584. doi: 10.1016/0092-8674(91)90241-p. [DOI] [PubMed] [Google Scholar]
- 22.Deloulme J C, Prichard L, Delattre O, Storm D R. J Biol Chem. 1997;272:27369–27377. doi: 10.1074/jbc.272.43.27369. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien T, Hardin S, Greenleaf A, Lis J T. Nature (London) 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 24.Petermann R, Mossier B M, Aryee D N, Khazak V, Golemis E A, Kovan H. Oncogene. 1998;17:603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 25.Guinamard R, Fougereau M, Seckinger P. Scand J Immunol. 1997;45:587–595. doi: 10.1046/j.1365-3083.1997.d01-447.x. [DOI] [PubMed] [Google Scholar]
- 26.Yamanashi Y, Baltimore D. Cell. 1997;88:205–211. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 27.Dupraz P, Rebai N, Klein S J, Beaulieu N, Jolicoeur P. J Virol. 1997;71:2615–2620. doi: 10.1128/jvi.71.4.2615-2620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurley J R, Dean A M, Sohl J L, Koshland D E J, Stroud R M. Science. 1990;249:1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- 29.Sprang S R, Acharya K R, Goldsmith E J, Stuart D I, Varvill K, Fletterick R J, Madsen N B, Johnson L N. Nature (London) 1988;336:215–221. doi: 10.1038/336215a0. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z Y, Qiu Q-Q, Enger K T, Deuel T F. Proc Natl Acad Sci USA. 1993;90:8896–8900. doi: 10.1073/pnas.90.19.8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexandropoulos K, Cheng G, Baltimore D. Proc Natl Acad Sci USA. 1995;92:3110–3114. doi: 10.1073/pnas.92.8.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]