Abstract
Marine bacterioplanktons are thought to play a vital role in Southern Ocean ecology and ecosystem function, as they do in other ocean systems. However, our understanding of phylogenetic diversity, genome-enabled capabilities and specific adaptations to this persistently cold environment is limited. Bacterioplankton community composition shifts significantly over the annual cycle as sea ice melts and phytoplankton bloom. Microbial diversity in sea ice is better known than that of the plankton, where culture collections do not appear to represent organisms detected with molecular surveys. Broad phylogenetic groupings of Antarctic bacterioplankton such as the marine group I Crenarchaeota, α-Proteobacteria (Roseobacter-related and SAR-11 clusters), γ-Proteobacteria (both cultivated and uncultivated groups) and Bacteriodetes-affiliated organisms in Southern Ocean waters are in common with other ocean systems. Antarctic SSU rRNA gene phylotypes are typically affiliated with other polar sequences. Some species such as Polaribacter irgensii and currently uncultivated γ-Proteobacteria (Ant4D3 and Ant10A4) may flourish in Antarctic waters, though further studies are needed to address diversity on a larger scale. Insights from initial genomics studies on both cultivated organisms and genomes accessed through shotgun cloning of environmental samples suggest that there are many unique features of these organisms that facilitate survival in high-latitude, persistently cold environments.
Keywords: Antarctic bacterioplankton, marine bacteria, marine microbial genomics, bacterioplankton diversity, Southern Ocean
1. Introduction
Polar marine environments are unique. They provide cold, saline water to fuel the deep-ocean conveyor belt (Baines & Condie 1998) and pump inorganic and organic carbon into the sequestered deep ocean (Sarmiento & Toggweiler 1984; Honjo 2004). Like the ocean below 200 m, the Southern Ocean is permanently cold with temperatures consistently below 4°C (Broecker & Peng 1982). In the majority of the Antarctic, seawater is at −1.8°C constantly. However, unlike the deep ocean, polar marine environments undergo extreme seasonal variations in sea-ice cover, light levels and day length. This greatly affects the biology and results in high rates of primary production and a strong pulse of biomass in summer (Holm-Hansen et al. 1977; Moline & Prezelin 1996) followed by a long winter of very low rates of photoautotrophy. Polar environments have been shown to be sensitive to global warming (Loeb et al. 1997; Smith et al. 2003; Moline et al. 2004), where minimal temperature differences can have significant effects on the extent and thickness of sea ice (Smetacek & Nicol 2005). Our knowledge of the concomitant dynamics and diversity of Antarctic marine bacterioplankton is still in its infancy. Only an adequate understanding of microbial diversity and genome-enabled capabilities in polar environments will enable us to complete the picture of polar ecosystem structure and function as well as establish the effects of climate change on the base of the polar food chain.
Observations of Antarctic bacterioplankton by light microscopy occurred at least as early as the Challenger expedition (1872–1876). These observations would have been limited to the larger bacterial species and predominantly phytoplankton. Cultivation-based studies of the domain bacteria appeared in journals almost 100 years later (Kriss et al. 1967) and have continued (Irgens et al. 1989; Delille 1996; Bowman et al. 1998; Bowman 2001) reporting cultivation of both cosmopolitan and endemic heterotrophic bacteria. However, it has been shown that cultivation-based studies overwhelmingly fail to describe either the microbial diversity of a given environment or the most active groups of organisms in that environment (Giovannoni 1991; Fuhrman et al. 1993; Amann et al. 1995). An exception may be the sea-ice bacterial community where cultivability well exceeds 1% of the viable population of bacteria (Brinkmeyer et al. 2003). And so, over the last decade, efforts to describe microbial diversity and activity in marine Antarctic environments have focused on molecular-based work.
This paper is an overview of molecular diversity and genomic research concerning Antarctic marine bacterioplankton. It focuses both on molecular applications making use of SSU rRNA gene diversity, fluorescence in situ hybridization (FISH) and real-time PCR, and on environmental genomics and genome sequencing of Antarctic micro-organisms. We have presented the results of recent research and integrated them with other studies that have focused on the diversity and genomics of micro-organisms in Antarctic surface waters. This environment should be differentiated from the cold, deep-water environments that make up 70% of the ocean by volume. While organisms in these two environments are at constant cold temperatures, their ecology is controlled by vastly different physical, chemical and biological variables. However, in discussing the advances in Antarctic microbiology through the use of modern molecular techniques, it is impossible to ignore some of the major milestones in microbial genomics that have occurred in Antarctic deep water (Moreira et al. 2004), lakes (Saunders et al. 2003), soils and other cold habitats on Earth (Rabus et al. 2004).
2. Material and methods
(a) PCR–DGGE
Seawater samples were collected on monthly intervals during both 1996–1997 and 2001–2002. DNA extraction and PCR–DGGE methods reported previously for the 1996–1997 samples (Murray et al. 1998) were used for the 2001–2002 samples except that samples (approx. 800 ng each) were run through 8% acrylamide, 1 mm thick, 30–60% denaturing gradient gels (62 V, 16 h) on a Bio-Rad D-Code system. Samples were stained for 15 min in SYBR-Green (Molecular Probes, Inc.) and then photographed (EDAS 290, Kodak). Here, we reanalysed the 1996–1997 data in comparison with the 2001–2002 dataset using Gel Compar II (Applied Maths, Inc.). Bands less than 1% of the total lane intensity were not considered. Pairwise comparisons using Sorenson's index (Magurran 1988), which was used originally to compare the Antarctic bacterioplankton DGGE profiles (Murray et al. 1998), were calculated for all months in comparison to the August (late winter) sample.
(b) Marine bacterial SSU rRNA gene diversity
Bacterioplankton diversity was surveyed using three approaches (bacterial PCR-amplified SSU rRNA gene clone library, shotgun fosmid clone library and culture collection) and included a SSU rRNA gene clone library from sea ice for comparison. The planktonic sample for the SSU rRNA gene library was collected in spring (designated AntCL1 and AntCL2; 17 October 2001, 20 m off LTER station A); the large-contig shotgun library was prepared from a sample collected in late winter 1996 (designated Ant; for details on library see Béjà et al. 2002a); and samples for the bacterial culture collection were collected in October 2001 from seawater and sea ice. Sea-ice cores were collected (6 October and 21 October 2001) using a SIPRE corer in the Arthur harbour region (300 m off Palmer Station: clones AntCL3A1–D12 and 100 m off Palmer Station: clones AntCL3E1–H12).
A standard approach was used here for SSU rRNA gene library creation and sequencing. Briefly, universal primers targeting bacteria (27F and 1492R) were used to amplify SSU rRNA genes from bacterioplankton DNA extracted from samples collected on 0.22 μm filters following prefiltration (GF/A filters, Whatman, Inc.). The amplification products were ligated into plasmid vectors and transformed into Escherichia coli cells following manufacturer's instructions (TOPO TA, Invitrogen, Inc.). Clones containing inserts were sequenced unidirectionally (T7 vector primer); then in most cases, clones with unique sequences were fully sequenced using the T3 vector primer and Bact533F. Sequences were run through CHECK_CHIMERA (Maidak et al. 1997) and Bellerophon (Huber et al. 2004) to identify potentially chimeric sequences.
Marine bacteria were cultivated using standard microbiological methods on several kinds of liquid and agar media at 4, 10 and 20°C. Media used were as follows (abbreviations were also used in strain IDs): SW media were prepared using coastal seawater and no other amendments; YE media included seawater amended with 5 mg l−1 yeast extract; SC media were used to enrich Actinobacteria (Adeleye et al. 2004); DMSP media were used to enrich DMSP-using micro-organisms (Gonzalez et al. 1999); and NZ media included seawater amended with 5 mg l−1 NZ-amine (Sigma–Aldrich, Inc.). Isolates were initially selected based on phenotypic characteristics and then screened using PCR–DGGE to eliminate redundancy as described above. Full-length SSU rRNA gene sequences were determined for isolates with unique melting points.
SSU rRNA gene sequences were aligned with neighbouring sequences identified using Blast (Altschul et al. 1990) and Sequence Match (Cole et al. 2005). Sequences and alignment tools at the ribosomal database project were used for multiple sequence alignment (Cole et al. 2005). Phylogenetic relationships were determined with neighbour-joining method available in Mega v. 3.1 (Kumar et al. 2004). Sequences submitted to GenBank have the following accession numbers: DQ06709–DQ906772 and DQ92580–DQ925858.
Details describing the shotgun library bacterial composition, gene content and amino acid modification analysis of Antarctic fosmid sequences were recently described by Grzymski et al. (2006).
3. Ecology and diversity of Antarctic marine archaea
The first molecular ecological investigation in Antarctic waters targeting the bacterioplankton reported high abundance of archaeal rRNA picoplankton smaller than 1 μm in Antarctic surface waters (20–25% of total RNA) sampled in late winter/early spring (DeLong et al. 1994). The Antarctic SSU rRNA gene sequences corresponded to marine group I Crenarchaeota and marine group II Euryarchaeota that were just being reported at that time from temperate latitudes in the Pacific and Atlantic Oceans (DeLong 1992; Fuhrman et al. 1992; DeLong et al. 1994). This study helped to dispel the paradigm that marine group I Crenarchaeota were thermophiles by showing that these organisms exist in high abundance in coastal Antarctic surface waters. Subsequent studies investigating the temporal distribution of the marine group I Crenarchaeota showed that they appeared to vary substantially in per cent composition over the annual cycle in surface waters with higher abundance in winter as per cent of total RNA (Murray et al. 1998) or per cent of total cell counts (Church et al. 2003). Archaeal abundance also increased with depth, where cell concentrations were observed to vary between winter and summer (Church et al. 2003). Further studies demonstrated the circumpolar distribution of the planktonic archaea (Murray et al. 1999), as well as its variation over short time frames in coastal vertical profiles (Massana et al. 1998).
The diversity of both the Antarctic Crenarchaeota and Euryarchaeota appears to be quite limited in comparison to marine bacterial phylogenetic groups (Massana et al. 2001). Crenarchaeotal diversity has been reported to increase with depth and to be quite similar between assemblages sampled at both poles (Bano et al. 2004). The implications of this are not well understood, as it is possible that the SSU rRNA gene sequence variation under-represents genomic heterogeneity (Béjà et al. 2002a). The SSU rRNA gene is a very slowly evolving molecule with an estimated 1% sequence variation accumulating over 50 Myr (Ochman & Wilson 1987); interestingly, this coincides roughly with the timing of the formation of the Antarctic circumpolar current which allowed the cooling of the Antarctic continent. It is possible that the marine group I Crenarchaeota, which appear to be global residents of cold waters and are the most abundant marine prokaryote in the world oceans (Karner et al. 2001), have undergone adaptation and diversification at the genome level to facilitate their survival and persistence in cold waters of the Antarctic and deep sea.
Until recently, the biogeochemical role of the marine group I Crenarchaeota has only been hypothesized given the difficultly in detecting archaeal metabolic activity in situ (Massana et al. 1998). However, recently, a representative marine group I-affiliated crenarchaeon capable of chemoautotrophic growth using ammonia as an electron donor was isolated (Konneke et al. 2005). Subsequently, Francis et al. (2005) and Wuchter et al. (2006) reported on the diversity and distribution of archaeal ammonia monooxygenase genes in marine environments, supporting the hypothesis that ammonia-oxidizing Crenarchaeota are widespread. Another study has recently reported both 3H-leucine and 14C-bicarbonate uptake specifically by Crenarchaeota and Euryarchaeota in Antarctic circumpolar waters sampled between 200 and 3000 m (Herndl et al. 2005). The authors concluded that the Archaea may provide a significant CO2 sink in the ocean ecosystem, particularly deep waters, and they speculated that organic carbon (the proxy in their study was an amino acid) might be used as an energy source. Thus, the extent of metabolic capabilities of the marine group I Crenarchaeota and relatives in marine sediments are currently unknown. Future studies promise to open this door further, particularly in Antarctic waters, as another recent study has shown that Crenarchaeota in marine sediments appear to grow heterotrophically (Biddle et al. 2006).
4. Ecology and diversity of Antarctic marine bacteria
The bacterial lineage Cytophaga–Flavobacteria–Bacteroides (CFB) has been a strong focus of both culture dependent (Bowman et al. 1997; Staley & Gosink 1999; Junge et al. 2002) and independent (Brinkmeyer et al. 2003; Abell & Bowman 2005) studies in part owing to their apparent prevalence in Antarctic marine waters and sea ice (Staley & Gosink 1999). Molecular-based studies using PCR–DGGE approaches (Webster et al. 2004; Abell & Bowman 2005), clone libraries (Brinkmeyer et al. 2003), FISH (Brinkmeyer et al. 2003; Abell & Bowman 2005) and real-time PCR (Abell & Bowman 2005) have all shown CFB to be an important constituent of sea-ice microbial communities (SIMCO) and Antarctic marine waters. The Flavobacteria, in particular, are a group suspected to thrive in high concentration of dissolved organic matter in sea ice (Brinkmeyer et al. 2003) and in algae blooms where they constitute as much as 70% of the bacteria (Glöckner et al. 1999; Abell & Bowman 2005). The abundance of particle-associated Flavobacteria correlates well with chl a concentration (Abell & Bowman 2005) and during periods of high primary productivity. In contrast, marine group I Crenarchaeota were inversely correlated to chl a concentration (Murray et al. 1998).
Molecular ecological studies on the diversity and ecology of marine bacterioplankton are not limited to Archaea and the CFB, though most of the reported diversity from the plankton arises from cultivation-based studies (Maugeri et al. 1996; Bowman et al. 1997; Michaud et al. 2004) and a recent SSU rRNA and rDNA sequencing survey (Gentile et al. 2006). Psychrophilic sea-ice bacteria (γ-proteobacterial and CFB-related strains) were found to be quite distinct from those in underlying waters, whereas the psychrotrophic strains were largely in the same phylogenetic groups (Bowman et al. 1997). Planktonic surveys performed off the Antarctic Italian base at Terra Nova Bay reported greater diversity of rRNA-encoded gene sequences than for rDNA sequences (Gentile et al. 2006) with the greatest diversity in the γ-proteobacterial and CFB lineages. Comparisons of the results from two studies, one investigating diversity of bacterioplankton cultivars in Terra Nova Bay through comparisons of RFLP patterns (146 isolates were categorized into 52 distinct groups; Michaud et al. 2004) and another which compared RFLP patterns from a SSU rRNA gene clone library from McMurdo Sound seawater (160 clones fell into 103 groups; Webster et al. 2004), suggest that the diversity in the uncultivated fraction of the Antarctic bacterioplankton is relatively high and is comparable to other marine ecosystems.
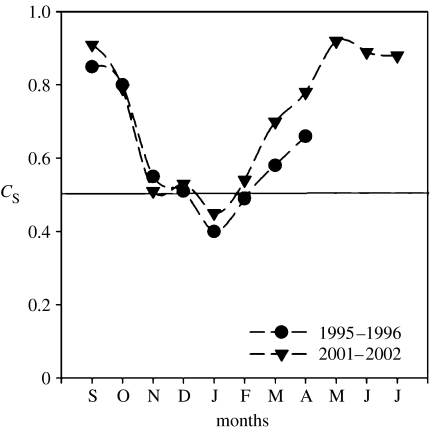
Using PCR–DGGE, Murray et al. (1998) reported significant shifts of bacterioplankton communities as the seasons changed. These shifts were accompanied by significant decreases of marine group I Crenarchaeota in the water during periods of high biomass. Here, we have analysed a second set of bacterioplankton samples over the annual cycle and found that bacterial SSU rRNA gene variation in DGGE profiles compared over the annual cycle for two different years, 6 years apart, indicates that the variation over the annual cycle is consistent and the timing of shifts in assemblage composition is surprisingly reproducible (figure 1). Similarity of PCR–DGGE profiles calculated using Sorenson's index suggests that the community is drastically different between winter and summer (figure 1; less than 45% similarity in both years).
Figure 1.
Seasonal variation in bacterial community structure over two annual cycles in nearshore waters off Palmer Station, Antarctica. Data are based on the presence/absence of bands in denaturing gradient electrophoresis. Pairwise similarity (Sorenson's index, CS) calculated for all months with respect to the August profile is plotted on the y-axis and the months are plotted on the x-axis. The black line is drawn at the 50% similarity level. Data from 1996–1997 were presented previously (Murray et al. 1998).
Though bacterial SSU rRNA gene clone library sequencing of Antarctic bacterioplankton is relatively under-explored, a few studies investigating SIMCO diversity (Brown & Bowman 2001; Brinkmeyer et al. 2003) have found that many environmental sequences are closely related to those found in SIMCO culture collections—this is unusual, and in contrast to nearly all reports from free-living plankton (Amann et al. 1995). Archaea have also been documented in Antarctic sea ice (Junge et al. 2004), where they represent a greater proportion of total cells (0–3.4%) at cold temperatures (−20°C). SIMCO bacterial diversity is dominated by sequences affiliated with the α-Proteobacteria, γ-Proteobacteria and the Bacteriodetes phylum and to a lesser degree, Actinobacteria, green sulphur bacteria (Chlorobiaceae), Chlamydiales and Verrucomicrobiales; the last three groups were not recovered in cultivation-based reports (Bowman et al. 1997; Brinkmeyer et al. 2003).
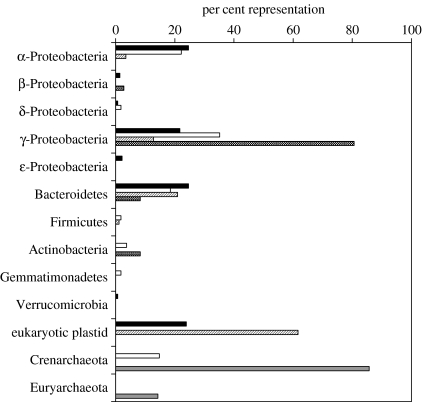
In order to supplement the findings summarized above, we report here the bacterial diversity found in waters and in sea ice offshore of Anvers Island, Antarctic Peninsula from (i) a SSU rRNA gene clone library (mid-October 2001), (ii) a culture collection from the same location and season, and (iii) a large-contig shotgun library (late August 1996; details of the library creation can be found in Béjà et al. 2002a). In late winter–spring surface waters off Palmer Station, three phylogenetic groups of bacteria dominated both a PCR-based bacterial SSU rRNA gene clone library and a large insert environmental clone library (figure 2). The γ-Proteobacteria comprised 20 and 35% of the bacterial clone library and large insert library, respectively. The α-Proteobacteria and the CFB were about equally represented at 20% in both libraries. In contrast, despite attempts to isolate bacteria on several types of low organic carbon media, a bacterial culture collection isolated at the same time of year was overwhelmingly dominated by γ-Proteobacteria with a small fraction (10% each) comprising CFB and Actinobacteria. In the sea ice, dominated by algae, the main phylogenetic groups of bacteria were the CFB and Actinobacteria (figure 2).
Figure 2.
Distribution of bacterial and archaeal phylogenetic groups based on SSU rDNA sequences in a PCR-based bacterial clone library prepared from seawater in mid-October 2001 (black bars), a PCR-based bacterial clone library prepared from sea ice in mid-October 2001 (stripped bars), in an environmental shotgun library prepared from picoplankton DNA collected in late August 1996 (open bars), in a bacterial culture collection from seawater and sea ice collected in October 2001 and 2002 (dotted bars) and a PCR-based archaeal clone library prepared with seawater in mid-October 2001 (grey bars).
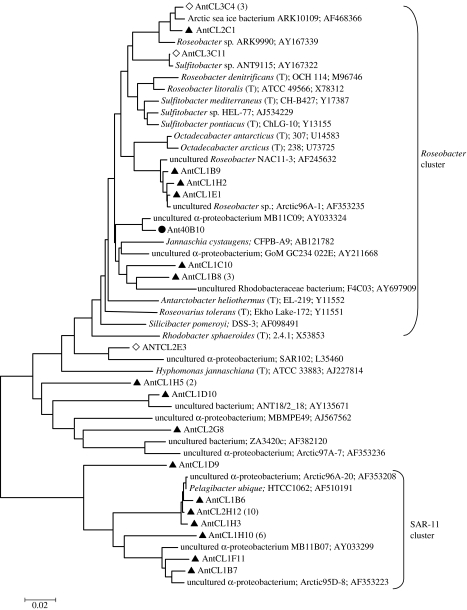
The phylogenetic relationships between the three most abundant classes of bacteria were determined to get a more detailed look at the planktonic diversity within these groups. The dominant α-proteobacterial sequences branched closely with Pelagibacter ubique (Rappé et al. 2002; Giovannoni et al. 2005), and other SAR-11-related bacteria from Arctic waters (figure 3). The α-proteobacterial sequences we detected were most closely related to sequences sampled in other polar studies (Bano & Hollibaugh 2002; Brinkmeyer et al. 2003; Arrieta et al. 2004). We detected several Roseobacter and Sulfitobacter-related sequences; however, we did not detect any sequences that were highly related to Octadecabacter antarcticus, a common genus in sea ice and seawater that contains organisms capable of producing gas vesicles. Several sequences (AntCL1B9, AntCL1H2 and AntCL1E1) branching off the Octadecabacter group (95% related) grouped tightly with the RCA cluster known only through environmental surveys recently described to be very abundant in the Southern Ocean (Selje et al. 2004).
Figure 3.
Neighbour-joining tree showing phylogenetic relationships between Antarctic marine plankton and sea-ice α-Proteobacteria and many of their nearest neighbours isolated or directly sequenced from marine environments. Sequences from SSU rRNA gene bacterioplankton libraries are indicated by filled triangles, sea-ice bacterial clones by open diamonds and shotgun clone library by filled circles.
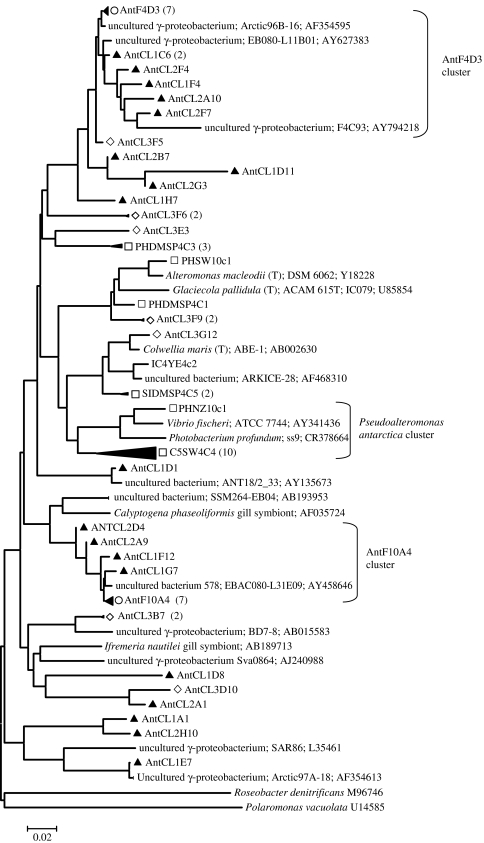
Organisms affiliated with γ-Proteobacteria, the most cosmopolitan marine planktonic bacteria, were well represented in both environmental surveys and our culture collections though there was no overlap in species detection. The most abundant SSU rRNA gene sequences in the environmental PCR and shotgun clone libraries grouped with two distinct clusters that contain no known cultivated representatives (figure 4). The Ant10A4-related sequences branch off a cluster of sulphur-oxidizing symbionts and the closest cultivated member of the Ant4D3 group, which appears to be common at both poles, is most closely affiliated with an isolate in the Oligotrophic Marine Group (HTTC2180) at 91% SSU rRNA gene identity (Cho & Giovannoni 2004). We fully sequenced environmental fosmid clones affiliated with each to get more information concerning these little known groups (Ant4D3, Grzymski et al. 2006, and Ant10A4).
Figure 4.
Neighbour-joining tree showing phylogenetic relationships between Antarctic marine plankton and sea-ice γ-Proteobacteria and many of their nearest neighbours isolated or directly sequenced from marine environments. Symbols are as described in figure 3 but shotgun clone libraries are indicated by open circles and in addition, open squares represent sequences for cultivated organisms and nodes representing both SSU rRNA gene libraries.
Our culture collection was dominated with a diverse group of γ-Proteobacteria mostly limited to a region of the tree with other well-studied organisms (e.g. Pseudoalteromonas antarctica, Vibrio fischeri, Alteromonas macleodii). Consistent with the findings of Brinkmeyer et al. (2003), several sea-ice environmental SSU rRNA sequences (AntCL3F9, AntCL3G12 and AntCL3E3) were most closely related to cultured organisms (e.g. Glaciecola pallidula, Colwellia maris).
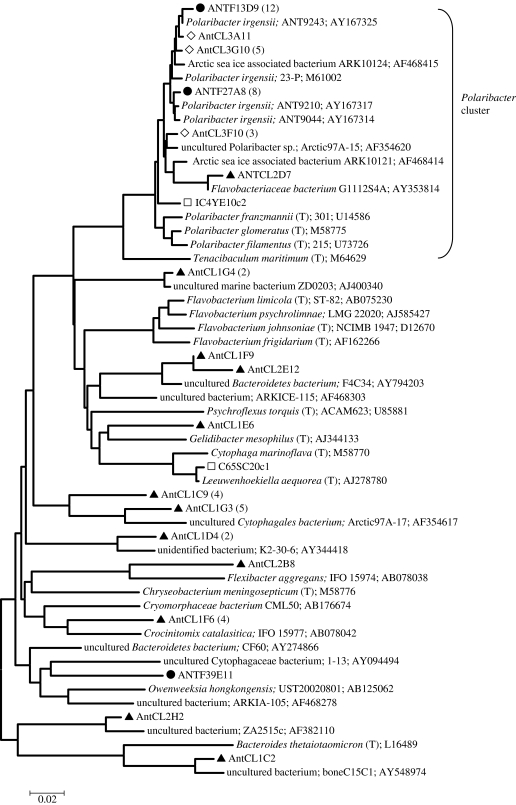
The majority of Bacteriodetes sequences we detected were affiliated with the Flavobacteriaceae family, one of the most common groups in marine (Kirchman 2002; Kirchman et al. 2003) and polar (Abell & Bowman 2005) ecosystems. In particular, we detected a high number of Polaribacter irgensii-related sequences (figure 5). Unlike the dominant γ-Proteobacteria groups which were only represented by uncultivated members, the Polaribacter genus is represented by both cultivated and environmentally detected sequences. Polaribacter irgensii-related sequences, though not confined to polar environments, are closest to organisms and environmental sequences from both Arctic and Antarctic environments, though they are thought to be more highly represented in the Antarctic (Brinkmeyer et al. 2003) and were one of the most common Antarctic zone groups detected in a latitudinal survey (Abell & Bowman 2005). We also detected a number of sequences grouping in clusters known only from environmental surveys, many in the Flavobacteriace (Abell & Bowman 2005). The most abundant of these grouped in the Agg58/Cryomorphaceae cluster and were most similar to uncultivated sequences from the Arctic (Arctic97A-17; Bano & Hollibaugh 2002). Overall, the Bacteriodetes diversity detected, particularly in the free-living planktonic library, was high, consistent with the suspected importance of this group in polar ocean systems where they may be particularly associated with algal blooms (Simon et al. 1999) and particles (Abell & Bowman 2005).
Figure 5.
Neighbour-joining tree showing phylogenetic relationships between Antarctic marine plankton and sea-ice Bacteriodetes-related sequences and many of their nearest neighbours isolated or directly sequenced from marine environments. Symbols are the same as in figures 3 and 4.
5. Antarctic environmental genomics
The environmental genomics approach, which has recently become popular in part due to the lower cost of sequencing (Tyson et al. 2004; Venter et al. 2004), has well-established roots in Antarctic bacterioplankton studies albeit on a smaller scale (Béjà et al. 2002; de la Torre et al. 2003; Lopez-Garcia et al. 2004; Moreira et al. 2004, 2006; Grzymski et al. 2006). In fact, Antarctic environmental genomics has perhaps outpaced complete microbial genome sequencing until very recently. Large DNA inserts (30–45 kb) provide more robust information than ribosomal RNA gene phylogenetic diversity, including gene content and biological potential, which is particularly valuable for organisms that largely remain uncultivable (Béjà et al. 2002a).
These larger genome fragment datasets provided information about genomic divergence that was otherwise lost when comparing SSU rRNA gene sequences from highly similar phylotypes. In a study comparing marine group I crenarchaeotal genome fragments from Antarctica and Pacific temperate waters, strain variability was shown to be considerably different in four Antarctic Crenarchaetoa despite almost identical SSU rRNA gene sequences (greater than 99.8% identical; Béjà et al. 2002a). Furthermore, these data established chromosomal organization in marine group I Crenarchaeota despite there being no cultivated representatives at that time. While protein prediction only suggests functional diversity, this greatly expands our understanding of microbial ecology and the metabolic potential of uncultivable organisms (Béjà et al. 2002a; de la Torre et al. 2003; Moreira et al. 2004; Grzymski et al. 2006). So far this has best been shown in the pervasiveness of proteorhodopsin genes in bacteria including the SAR-86, γ-Proteobacteria and ubiquitous SAR-11 cluster of α-Proteobacteria (Béjà et al. 2001, 2002b; de la Torre et al. 2003; Venter et al. 2004; Giovannoni et al. 2005), all of which have been detected in environmental genomic surveys. These studies extended the use of environmental genomics to describe functional gene diversity.
Since studies of functional gene diversity (i.e. nirK, RuBisCO, AmoA) often preclude phylogenetic classification of the sequences, either the inclusion of highly conserved genes in the environmental fragment or the preponderance of evidence from Blast results can be used to identify the affiliation of the genome fragment. de la Torre et al. (2003) used 13 ribosomal proteins and Blast results of the remaining predicted proteins to conclude that a proteorhodopsin-containing BAC from the central North Pacific was most probably affiliated with an α-proteobacterium. Less definitively, the PR-containing Antarctic fragment and another highly syntenous fragment (96% nucleotide identity over 20 kB) could only be identified as Proteobacteria affiliates (36% of predicted proteins most similar to γ-Proteobacteria). Similarly, the Antarctic fosmid clone Ant24C4 was identified as an α-Proteobacteria from Blast results of its 42 ORFs and another fosmid clone Ant29B7 fragment was affiliated with the Bacteroidetes phylum from phylogenetic analysis of groEL (Grzymski et al. 2006).
Comparative analyses within highly similar populations and between distant ecosystems are necessary to understand the ecology and evolution of marine bacterioplankton including the effects of gene loss, rearrangement and horizontal gene transfer. The comparative analysis of Antarctic marine crenarchaeotal genome fragments has revealed multiple horizontal gene transfers (Lopez-Garcia et al. 2004). These horizontal gene transfers are hypothesized to play a role in the low-temperature adaptation of marine group I Crenarchaeota. The complex evolutionary history of some of these genes was demonstrated by the phylogenetic analysis of ORFs surrounding the conserved 16S-23S-ITS-GSAT region of Crenarchaeota. The same large insert library also resulted in the recovery of the first group II Euryarchaeota genome fragment to contain the 16S rRNA gene (Moreira et al. 2004). Incidentally, environmental genomic surveys in the North Pacific subtropical gyre recently found group II Euryarchaeota-encoded proteorhodopsin genes in surface water samples (Frigaard et al. 2006), consistent with the prevalence of these organisms in the photic zone (DeLong 1992; Massana et al. 1997; DeLong et al. 2006). The genome fragment analysis of ORFs perhaps revealed a novel succinate dehydrogenase (Moreira et al. 2004). Again, the work emphasized the use of genome fragments to infer metabolic potential in the uncultivable organisms. Metabolic potential must be emphasized until more data are available (e.g. expression studies, metabolic assays).
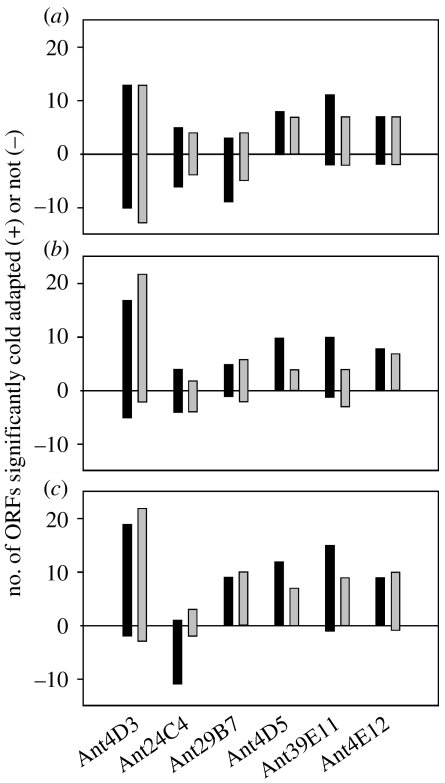
Recently, six environmental genome fragments from nearshore Antarctic coastal waters were described (Grzymski et al. 2006). These bacterial fragments from a late winter library (Béjà et al. 2002a) were chosen for complete sequencing based on their hypothesized ecological relevance (one γ-Proteobacteria, one putative α-Proteobacteria, two Bacteriodetes) and uniqueness (one Gemmatimonadetes, one Actinobacteria). The genome fragments that contained SSU rRNA genes were only 85–91% similar to 16S rRNA genes from cultivated relatives. Apart from describing the content of the six fragments, the authors also analysed the predicted proteins against the SwissProt database and found amino acid usage patterns indicative of cold adaptation. While this analysis has been performed on various psychrophile enzymes such as α-amylase and β-glucosidase (Feller & Gerday 1997; Huston et al. 2004) and whole genomes (Saunders et al. 2003; Methé et al. 2005), this was the first analysis conducted across diverse bacterial taxa. The most significant amino acid changes in the predicted proteins were reductions in salt-bridge-forming residues (arginine), reduced proline content and reduced hydrophobic clusters (figure 6; Grzymski et al. 2006). In addition, predictions of protein disordered regions were significantly longer in Antarctic sequences than in mesophile homologues. The lengthened disordered regions (regions without a defined three-dimensional structure) are hypothesized to aid in processes that are retarded at cold temperature such as various association and disassociation reactions (Dunker et al. 2002; Grzymski et al. 2006).
Figure 6.
Three indices of amino acid modification in the predicted coding regions of Antarctic proteins: (a) reduced proline content, (b) reduction in the arginine ratio (arg/arg+lys), and (c) reduction in acidic residues (Asn+Gln/Asn+Gln+Glu+Asp). Proteins compared here had at least two homologues with Blast e-scores less than 10−15. Black bars represent number of Antarctic open reading frames in comparison to mesophilic homologues with G+C content ±2.5% that of the Antarctic sequence. Grey bars represent data compared to the UniSwiss database (all characterized proteins). All data are significant based on a one-sample Student's t-test (p<0.05).
6. Genome sequencing of polar micro-organisms
As of spring 2006, there are nine bacterial and one archaeal polar microbial genomes in GenBank. Several are accompanied by publications: two Euryachaeota (Methanogenium frigidum and Methanococcoides burtonii; Saunders et al. 2003), two γ-Proteobacteria (Colwellia psychrerythraea 34H Methé et al. 2005; Pseudoalteromonas haloplanktis , Medigue et al. 2005) and a δ-proteobacterium (Desulfotalea psychrophila; Rabus et al. 2004). At least nine other polar marine strains are in various stages of completion (table 1) in addition to at least ten other psychrophilic strains including fish pathogens and deep-sea strains. Seven out of the 16 polar bacteria are from Antarctic surface waters or sea ice: two CFBs, Psychroflexus torquis (isolated from a sea-ice algal assemblage), P. irgensii (isolated from nearshore waters off the Antarctic Peninsula), a Marine Actinobacterium PHSC20C1 (also from Antarctic Peninsula waters), O. antarcticus 307, an α-proteobacterium strain isolated from McMurdo Sound, and γ-Proteobacteria strains, P. haloplanktis, Oleispira antarctica and Shewanella frigidimarina, which were isolated from either seawater or sea ice.
Table 1.
Polar bacterial and archaeal genome projects from both marine and terrestrial environments. (The projects without accession numbers have not been submitted to GenBank and for these cases, the URLs for genome projects are listed if available.)
| domain | phylogenetic group | micro-organism | origin of strain | status of genome/accession number or URL | reference |
|---|---|---|---|---|---|
| Archaea | Euryarchaeota | Methanogenium frigidum | Ace Lake, Antarctica | draft/http://psychro.bioinformatics.unsw.edu.au/blast/mf_blast.php | Saunders et al. (2003) |
| Archaea | Euryarchaeota | Methanococcoides burtonii DSM6242 | Ace Lake, Antarctica | draft/CP000300 | Saunders et al. (2003) |
| Bacteria | γ-Proteobacteria | Colwellia psychrerythraea 34H | Arctic Marine Sediments | complete/CP000083 | Methé et al. (2005) |
| Bacteria | γ-Proteobacteria | Shewanella frigidimarina NCMB400 | Sea Ice, Antarctica | draft/http://genome.jgipsf.org/draft_microbes/shefr/shefr.home.html | |
| Bacteria | γ-Proteobacteria | Psychrobacter arcticus 273-4 | Siberian permafrost | complete/CP000082 | |
| Bacteria | γ-Proteobacteria | Psychrobacter cryohalolentis K5 | Siberian permafrost (40k yr old) | complete/CP000323, CP000324 | |
| Bacteria | γ-Proteobacteria | Oleispira antarctica RB-8 | Rod Bay, Ross Sea, crude oil-enriched surface water | in progress | |
| Bacteria | γ-Proteobacteria | Pseudoalteromonas haloplanktis TAC 125 | Coastal Antarctic seawater, Terra Adelie | complete/CR954246, CR954247 | Medigue et al. (2005) |
| Bacteria | δ-Proteobacteria | Desulfotalea psychrophila LSv54 | Arctic marine sediments, Svalbard | complete/CR522870, CR522871, CR522872 | Rabus et al. (2004) |
| Bacteria | Firmicutes | Exiguobacterium 255-15 | Siberian permafrost | draft/AADW00000000 | |
| Bacteria | Bacteriodetes | Psychroflexus torquis ATCC 700755 | sea-ice algal assemblage, Prydz Bay, Antarctica | draft/AAPR00000000 | |
| Bacteria | Bacteriodetes | Polaribacter filamentous 215 | surface seawater, 350 km north of Deadhorse, Alaska | in progress | |
| Bacteria | Bacteriodetes | Polaribacter irgensii 23-P | nearshore marine waters off Antarctic Peninsula | draft/AAOG00000000 | |
| Bacteria | Actinobacteria | Marine Actinobacterium PHSC20C1 | nearshore marine waters of Antarctic Peninsula | draft/AAOB00000000 |
The CFB genomes represent two of the three sequences available from polar marine habitats and promise to offer much in the way of understanding features that facilitate their dominance in these ecosystems. For example, initial investigations have revealed ice-binding and cold-shock proteins suggesting specialization to cold environments (table 2). The bipolar distribution of P. irgensii (Staley & Gosink 1999), its prevalence in sea ice and seawater, and cellular features such as a proteorhodopsin homologue and gas vesicle formation suggest that this organism will be a model for polar marine microbiology and cold adaptation studies in the future.
Table 2.
Examples of proteins involved in cold resistance in several polar microbial genomes (based on function in the environment, adaptation to cold and natural product synthesis). (All these proteins are present in the P. irgensii 23-P genome.)
| cold-resistant proteins |
|---|
| cold-shock (acclimation) protein (EAR12042, EAR12901 and EAR11908) |
| soluble acyl-ACP desaturase (EAR13799) |
| ATP-dependent RNA helicase, DEAD/DEAH box family protein (EAR12918 and EAR11919) |
| β-d-galactosidase, putative (EAR13130) |
| putative ribonuclease H1 (EAR13453) |
A study of the draft genome sequences of two Ace Lake methanogens, M. frigidum and M. burtonii, was the first comparative analysis of archaeal genomes that spanned psychrophile to hyperthermophile lifestyles (Saunders et al. 2003). The analysis of the two Antarctic methanogens detailed genotype characteristics of cold adaptation such as increased glutamine and threonine usage and decreased leucine usage in the predicted proteins of these genomes. Five predicted proteins were unique to the two organisms and have putative roles in adaptation to the cold. Three of these had cold-shock domain folds. The comparative analysis of nine methanogen genomes spanning an optimal growth temperature (OGT) of 83°C also showed that G+C content is a major factor influencing tRNA stability in organisms with an OGT greater than 60°C. G+C content was not significantly different below that temperature (Saunders et al. 2003).
Similar comparative analyses were done on the Arctic psychrophile bacterium C. psychrerythraea 34H and 24 other organisms with wide-ranging OGTs (Methé et al. 2005). These data suggested a decrease in charged amino acids in the psychrophile when compared with mesophiles and thermophiles. It also appeared that serine usage increased as OGT decreased. In the P. haloplanktis genome, which appears to be well equipped for growth in the cold, a significant bias towards asparagine residues was found (Medigue et al. 2005).
7. Conclusions
In conclusion, the state of knowledge concerning polar marine micro-organisms from ecological and genomic perspectives is in the early phase of exponential growth. Initial studies on diversity of bacterioplankton suggest that the diversity appears to rival that found in other ocean systems, though many polar phylotypes harbour a distinct biogeographic signal (Pommier et al. 2005). This information is essential if we are to be able to understand the specific adaptations of organisms to life in the cold, describe the evolutionary history of psychrophiles, and, importantly, predict the impacts of climate change, such as reduction in sea ice and its impact on the carbon cycle in polar oceans. Understanding the regulation and roles of ice-binding proteins (in P. irgensii), dioxygenases (in P. haloplanktis; Medigue et al. 2005), and cold-shock proteins in marine group I Crenarchaeota (Béjà et al. 2002a) are key first steps in understanding adaptations facilitating survival and leading to development of diverse polar marine bacterioplankton assemblages.
Acknowledgments
This work was the result of efforts by B. J. Carter, who is graciously thanked for his assistance in this research. A. E. Kelley played a key role in field research support. Both Carter and Kelley developed and maintained the culture collection. We also thank the Ribosomal Database Project for computational support. Sequencing collaborations at Joint Genome Institute were instrumental in this research. RPSC is acknowledged for their assistance in sample collection in 1997 and 2002. NSF-OPP 0085435 grant to A.E.M. supported this work in part.
Footnotes
One contribution of 10 to a Theme Issue ‘Antarctic ecology: from genes to ecosystems. Part 2: evolution, diversity and function’.
References
- Abell G.C.J, Bowman J.P. Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol. Ecol. 2005;51:265–277. doi: 10.1016/j.femsec.2004.09.001. doi:10.1016/j.femsec.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Adeleye I.A, Eruba S, Ezeani C.J. Isolation and characterisation of antibiotic producing microorganisms in composted Nigerian soil. J. Environ. Biol. 2004;25:313–316. [PubMed] [Google Scholar]
- Altschul S.F, Gish W, Miller W, Myers E.W, Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amann R, Ludwig W, Schleifer K. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta J.M, Weinbauer M.G, Lute C, Herndl G.J. Response of bacterioplankton to iron fertilization in the Southern Ocean. Limnol. Oceanogr. 2004;49:799–808. [Google Scholar]
- Baines P.G, Condie S. Observations and modelling of Antarctic downslope flows: a review. In: Jacobs S.S, Weiss R, editors. Ocean, ice and atmosphere: interactions at the Antarctic continental margin. vol. 75. 1998. pp.. 29–49 Washington, DC: American Geophysical Union. [Google Scholar]
- Bano N, Hollibaugh J.T. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 2002;68:505–518. doi: 10.1128/AEM.68.2.505-518.2002. doi:10.1128/AEM.68.2.505-518.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano N, Ruffin S, Ransom B, Hollibaugh J.T. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl. Environ. Microbiol. 2004;68:781–789. doi: 10.1128/AEM.70.2.781-789.2004. doi:10.1128/AEM.70.2.781-789.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjà O, Spudich E.N, Spudich J.L, Leclerc M, DeLong E.F. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. doi:10.1038/35081051 [DOI] [PubMed] [Google Scholar]
- Béjà O, Suzuki M.T, Heidelberg J.F, Nelson W.C, Preston C.M, Hamada T, Eisen J.A, Fraser C.M, Delong E.F. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol. 2002a;68:335–345. doi: 10.1128/AEM.68.1.335-345.2002. doi:10.1128/AEM.68.1.335-345.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjà O, Suzuki M.T, Heidelberg J.F, Nelson W.C, Preston C.M, Hamada T, Eisen J.A, Fraser C.M, Delong E.F. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002b;415:630–633. doi: 10.1038/415630a. doi:10.1038/415630a [DOI] [PubMed] [Google Scholar]
- Biddle J.F, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl Acad. Sci. USA. 2006;103:3846–3851. doi: 10.1073/pnas.0600035103. doi:10.1073/pnas.0600035103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J. P. 2001 Methods for psychrophilic bacteria. In Methods in microbiology, vol. 30, pp. 591–614.
- Bowman J.P, Brown M.V, Nichols D.S. Biodiversity and ecophysiology of bacteria associated with Antarctic sea ice. Antarct. Sci. 1997;9:134–142. [Google Scholar]
- Bowman J.P, McCammon S.A, Lewis T, Skerratt J.H, Brown J.L, Nichols D.S, McMeekin T.A. Psychroflexus torquis gen. nov., sp. nov., a psychrophilic species from Antarctic sea ice, and reclassification of Flavobacterium gondwanense (Dobson .1993) as Psychroflexus gondwanense gen. nov., comb. nov. Microbiology-UK. 1998;144:1601–1609. doi: 10.1099/00221287-144-6-1601. [DOI] [PubMed] [Google Scholar]
- Brinkmeyer R, Knittel K, Jurgens J, Weyland H, Amann R, Helmke E. Diversity and structure of bacterial communities in arctic versus Antarctic pack ice. Appl. Environ. Microbiol. 2003;69:6610–6619. doi: 10.1128/AEM.69.11.6610-6619.2003. doi:10.1128/AEM.69.11.6610-6619.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broecker W, Peng T.-H. Eldiago Press; New York, NY: 1982. Tracers in the Sea. [Google Scholar]
- Brown M.V, Bowman J.P. A molecular phylogenetic survey of sea-ice microbial communities (SIMCO) FEMS Microbiol. Ecol. 2001;35:267–275. doi: 10.1111/j.1574-6941.2001.tb00812.x. doi:10.1111/j.1574-6941.2001.tb00812.x [DOI] [PubMed] [Google Scholar]
- Cho J.C, Giovannoni S.J. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ. Microbiol. 2004;70:432–440. doi: 10.1128/AEM.70.1.432-440.2004. doi:10.1128/AEM.70.1.432-440.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church M.J, DeLong E.F, Ducklow H.W, Karner M.B, Preston C.M, Karl D.M. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol. Oceanogr. 2003;48:1893–1902. [Google Scholar]
- Cole J, Chai B, Farris R, Wang Q, Kulam S, McGarrell D, Garrity G, Tiedje J. The ribosomal database project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. doi:10.1093/nar/gki038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre J.R, Christianson L.M, Béjà O, Suzuki M.T, Karl D.M, Heidelberg J, DeLong E.F. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc. Natl Acad. Sci. USA. 2003;100:12 830–12 835. doi: 10.1073/pnas.2133554100. doi:10.1073/pnas.2133554100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delille D. Biodiversity and function of bacteria in the Southern Ocean. Biodivers. Conserv. 1996;5:1505–1523. doi:10.1007/BF00051989 [Google Scholar]
- DeLong E.F. Archaea in coastal marine environments. Proc. Natl Acad. Sci. USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. doi:10.1073/pnas.89.12.5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E.F, Wu K.Y, Prezelin B.B, Jovine R.V.M. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. doi:10.1038/371695a0 [DOI] [PubMed] [Google Scholar]
- DeLong E.F, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. doi:10.1126/science.1120250 [DOI] [PubMed] [Google Scholar]
- Dunker A.K, Brown C.J, Lawson J.D, Iakoucheva L.M, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. doi:10.1021/bi012159+ [DOI] [PubMed] [Google Scholar]
- Feller G, Gerday C. Psychrophilic enzymes: molecular basis of cold adaptation. Cell Mol. Life Sci. 1997;53:830–841. doi: 10.1007/s000180050103. doi:10.1007/s000180050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis C.A, Roberts K.J, Beman J.M, Santoro A.E, Oakley B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl Acad. Sci. USA. 2005;102:14 683–14 688. doi: 10.1073/pnas.0506625102. doi:10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N.U, Martinez A, Mincer T.J, DeLong E.F. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. doi:10.1038/nature04435 [DOI] [PubMed] [Google Scholar]
- Fuhrman J.A, McCallum K, Davis A.A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. doi:10.1038/356148a0 [DOI] [PubMed] [Google Scholar]
- Fuhrman J.A, McCallum K, Davis A.A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile G, Guiliano L, D'Auria G, Smedile F, Azzaro M, Domenico M.D, Yakimov M.M. Study of bacterial communities in Antarctic coastal waters by a combination of 16S rRNA and 16S rDNA sequencing. Environ. Microbiol. 2006;8:2150–2161. doi: 10.1111/j.1462-2920.2006.01097.x. doi:10.1111/j.1462-2920.2006.01097.x [DOI] [PubMed] [Google Scholar]
- Giovannoni S. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Wiley; New York, NY: 1991. pp. 177–203. [Google Scholar]
- Giovannoni S.J, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. doi:10.1126/science.1114057 [DOI] [PubMed] [Google Scholar]
- Glöckner F.O, Fuchs B.M, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.M, Kiene R.P, Moran M.A. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzymski J.J, Carter B.J, DeLong E.F, Feldman R.A, Ghadiri A, Murray A.E. Comparative genomics of DNA fragments from six Antarctic marine planktonic bacteria. Appl. Environ. Microbiol. 2006;72:1532–1541. doi: 10.1128/AEM.72.2.1532-1541.2006. doi:10.1128/AEM.72.2.1532-1541.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndl G.J, Reinthaler T, Teira E, Aken H.M.V, Veth C, Pernthaler A, Pernthaler J. Contribution of Archaea to total prokaryotic production in the deep Atlantic ocean. Appl. Environ. Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. doi:10.1128/AEM.71.5.2303-2309.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm-Hansen O, El-Sayed S.Z, Franceschini G.A, Cuhel R.L. Primary productivity and factors controlling phytoplankton growth in the Southern Ocean. In: Llana G.A, editor. Adaptations within Antarctic ecosystems. Gulf; Houston, TX: 1977. pp. 11–50. [Google Scholar]
- Honjo S. Particle export and the biological pump in the Southern Ocean. Antarct. Sci. 2004;16:501–516. doi:10.1017/S0954102004002287 [Google Scholar]
- Huber T, Faulkner F, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Huston A.L, Methé B, Deming J.W. Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl. Environ. Microbiol. 2004;70:3321–3328. doi: 10.1128/AEM.70.6.3321-3328.2004. doi:10.1128/AEM.70.6.3321-3328.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irgens R.L, Suzuki I, Staley J.T. Gas vacuolate bacteria obtained from marine waters of Antarctica. Curr. Microbiol. 1989;18:261–265. doi:10.1007/BF01570303 [Google Scholar]
- Junge K, Imhoff F, Staley T, Deming J.W. Phylogenetic diversity of numerically important arctic sea-ice bacteria cultured at subzero temperature. Microb. Ecol. 2002;43:315–328. doi: 10.1007/s00248-001-1026-4. doi:10.1007/s00248-001-1026-4 [DOI] [PubMed] [Google Scholar]
- Junge K, Eicken H, Deming J.W. Bacterial activity at −2 to −20 degrees C in Arctic wintertime sea ice. Appl. Environ. Microbiol. 2004;70:550–557. doi: 10.1128/AEM.70.1.550-557.2004. doi:10.1128/AEM.70.1.550-557.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner M.B, DeLong E.F, Karl D.M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. doi:10.1038/35054051 [DOI] [PubMed] [Google Scholar]
- Kirchman D.L. The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microb. Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- Kirchman D.L, Yu L.Y, Cottrell M.T. Diversity and abundance of uncultured Cytophaga-like bacteria in the Delaware Estuary. Appl. Environ. Microbiol. 2003;69:6587–6596. doi: 10.1128/AEM.69.11.6587-6596.2003. doi:10.1128/AEM.69.11.6587-6596.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konneke M, Bernhard A.E, de la Torre J.R, Walker C.B, Waterbury J.B, Stahl D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. doi:10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- Kriss A.E, Mishustina I.E, Mitskevich N, Zemtsova E.V. St Martins Press; New York, NY: 1967. Microbial population of oceans and seas. [Google Scholar]
- Kumar S, Tamura K, Nei M. Mega3:integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. doi:10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Loeb V, Siegel V, Holm-Hansen O, Hewitt R, Fraser W, Trivelpiece W, Trivelpiece S. Effects of sea-ice extent and krill or salp dominance on the Antarctic food web. Nature. 1997;387:897–900. doi:10.1038/43174 [Google Scholar]
- Lopez-Garcia P, Brochier C, Moreira D, Rodriguez-Valera F. Comparative analysis of a genome fragment of an uncultivated mesopelagic crenarchaeote reveals multiple horizontal gene transfers. Environ. Microbiol. 2004;6:19–34. doi: 10.1046/j.1462-2920.2003.00533.x. doi:10.1046/j.1462-2920.2003.00533.x [DOI] [PubMed] [Google Scholar]
- Magurran A.E. Princeton University Press; Princeton, NJ: 1988. Ecological diversity and its measurement. [Google Scholar]
- Maidak B.L, Olsen G.J, Larsen N, Overbeek R, McCaughey M.J, Woese C.R. The RDP (Ribosomal database project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. doi:10.1093/nar/25.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Murray A.E, Preston C.M, DeLong E.F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Taylor L.T, Murray A.E, Wu K.Y, Jeffrey W.H, DeLong E.F. Distribution of marine planktonic archaea in the Gerlache strait, Antarctic Peninsula, during early spring. Limnol. Oceanogr. 1998;43:607–617. [Google Scholar]
- Massana R, Pedros-Alio C, Casamayor E.O, Gasol J.M. Changes in marine bacterioplankton phylogenetic composition during incubations designed to measure biogeochemically significant parameters. Limnol. Oceanogr. 2001;46:1181–1188. [Google Scholar]
- Maugeri T.L, Gugliandolo C, Bruni V. Heterotrophic bacteria in the Ross Sea (Terra Nova Bay, Antarctica) Microbiologica. 1996;19:67–76. [PubMed] [Google Scholar]
- Medigue C, et al. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. doi:10.1101/gr.4126905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methé B.A, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl Acad. Sci. USA. 2005;102:10 913–10 918. doi: 10.1073/pnas.0504766102. doi:10.1073/pnas.0504766102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud L, Cello F.D, Brilli M, Fani R, Guidice A.L, Bruni V. Biodiversity of cultivable psychrotrophic marine bacteria isolated from Terra Nova Bay (Ross Sea, Antarctica) FEMS Microbiol. Lett. 2004;230:63–71. doi: 10.1016/S0378-1097(03)00857-7. doi:10.1016/S0378-1097(03)00857-7 [DOI] [PubMed] [Google Scholar]
- Moline M.A, Prezelin B.B. Long-term monitoring and analyses of physical factors regulating variability in coastal Antarctic phytoplankton biomass, in situ productivity and taxonomic composition over subseasonal, seasonal and interannual time scales. Mar. Ecol. Prog. Ser. 1996;145:143–160. [Google Scholar]
- Moline M.A, Claustre H, Frazer T.K, Schofield O, Vernet M. Alteration of the food web along the Antarctic Peninsula in response to a regional warming trend. Glob. Change Biol. 2004;10:1973–1980. doi:10.1111/j.1365-2486.2004.00825.x [Google Scholar]
- Moreira D, Rodríguez-Valera F, López-García P. Analysis of a genome fragment of a deep-sea uncultivated Group II euryarchaeote containing 16S rDNA, a spectinomycin-like operon and several energy metabolism genes. Environ. Microbiol. 2004;6:959–969. doi: 10.1111/j.1462-2920.2004.00644.x. doi:10.1111/j.1462-2920.2004.00644.x [DOI] [PubMed] [Google Scholar]
- Moreira D, Rodriguez-Valera F, Lopez-Garcia P. Metagenomic analysis of mesopelagic Antarctic plankton reveals a novel deltaproteobacterial group. Microbiology. 2006;152:505–517. doi: 10.1099/mic.0.28254-0. doi:10.1099/mic.0.28254-0 [DOI] [PubMed] [Google Scholar]
- Murray A.E, Preston C.M, Massana R, Taylor L.T, Blakis A, Wu K, DeLong E.F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters off Anvers Island, Antarctica. Appl. Environ. Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.E, Wu K.Y, Moyer C.L, Karl D.M, DeLong E.F. Evidence for circumpolar distribution of planktonic Archaea in the Southern Ocean. Aquat. Microb. Ecol. 1999;18:263–273. [Google Scholar]
- Ochman H, Wilson A.C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 1987;26:74–86. doi: 10.1007/BF02111283. doi:10.1007/BF02111283 [DOI] [PubMed] [Google Scholar]
- Pommier T, Pinhassi J, Hagstrom A. Biogeographic analysis of ribosomal RNA clusters from marine bacterioplankton. Aquat. Microb. Ecol. 2005;41:79–89. [Google Scholar]
- Rabus R, et al. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 2004;6:887–902. doi: 10.1111/j.1462-2920.2004.00665.x. doi:10.1111/j.1462-2920.2004.00665.x [DOI] [PubMed] [Google Scholar]
- Rappé M.S, Connon S.A, Vergin K.L, Giovannoni S.J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Sarmiento J.L, Toggweiler J.R. A new model for the role of the oceans in determining atmospheric pCO2. Nature. 1984;308:621–624. doi:10.1038/308621a0 [Google Scholar]
- Saunders N.F.W, et al. Mechanisms of thermal adaptation revealed from the genomes of the Antarctic Archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 2003;13:1580–1588. doi: 10.1101/gr.1180903. doi:10.1101/gr.1180903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selje N, Simon M, Brinkhoff T. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature. 2004;427:445–448. doi: 10.1038/nature02272. doi:10.1038/nature02272 [DOI] [PubMed] [Google Scholar]
- Simon M, Glöckner F.O, Amann R. Different community structure and temperatures optima of heterotrophic picoplankton in various regions of the Southern Ocean. Aquat. Microb. Ecol. 1999;18:275–824. [Google Scholar]
- Smetacek V, Nicol S. Polar ocean ecosystems in a changing world. Nature. 2005;437:362–368. doi: 10.1038/nature04161. doi:10.1038/nature04161 [DOI] [PubMed] [Google Scholar]
- Smith R.C, Fraser W.R, Stammerjohn S.E. Climate variability and ecological response of the marine ecosystem in the western Antarctic Peninsula (WAP) region. In: Greenland D, Goodin D.G, Smith R.C, editors. Climate variability and ecosystem response at long-term ecological research sites. Oxford University Press; New York, NY: 2003. pp. 158–173. [Google Scholar]
- Staley J.T, Gosink J.J. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 1999;53:189–215. doi: 10.1146/annurev.micro.53.1.189. doi:10.1146/annurev.micro.53.1.189 [DOI] [PubMed] [Google Scholar]
- Tyson G.W, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. doi:10.1038/nature02340 [DOI] [PubMed] [Google Scholar]
- Venter J.C, Remington K, Heidelberg J.F. Environmental genome shotgun sequencing of the Sargasso sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. doi:10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- Webster N.S, Negri A.P, Munro M, Battershill C.N. Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 2004;6:288–300. doi: 10.1111/j.1462-2920.2004.00570.x. doi:10.1111/j.1462-2920.2004.00570.x [DOI] [PubMed] [Google Scholar]
- Wuchter C, et al. Archaeal nitrification in the ocean. Proc. Natl Acad. Sci. USA. 2006;103:12 317–12 322. doi: 10.1073/pnas.0600756103. doi:10.1073/pnas.0600756103 [DOI] [PMC free article] [PubMed] [Google Scholar]