Abstract
Two competing effects at two vastly different scales may explain collagen’s current translation length. The necessity to have long molecules for maintaining mechanical integrity at the organism and supraorganism scales may be limited by the need to have small molecules capable of robust self-assembly at the nanoscale. The triglycine repeat regions of all 556 currently cataloged organisms with collagen-like genes were ranked by length. This revealed a sharp boundary in the GXY transcript number at 1032 amino acids (344 GXY repeats). An anomalous exception, however, is the intron-free Trichodesmium erythraeum collagen gene. Immunogold atomic force microscopy reveals, for the first time, the presence of a collagen-like protein in T. erythraeum. A phylogenetic protein sequence analysis which includes vertebrates, nonvertebrates, shrimp white spot syndrome virus, Streptococcus equi, and Bacillus cereus predicts that the collagen-like sequence may have emerged shortly after the divergence of fibrillar and nonfibrillar collagens. The presence of this anomalously long collagen gene within a prokaryote may represent an interdomain transfer from eukaryotes into prokaryotes that gives T. erythraeum the ability to form blooms that cover hundreds of square kilometers of ocean. We propose that the collagen gene entered the prokaryote intron-free only after it had been molded by years of mechanical selective pressure in larger organisms and only after large, dense food sources such as marine vertebrates became available. This anomalously long collagen-like sequence may explain T. erythraeum’s ability to aggregate and thus concentrate its toxin for food-source procurement.
Electronic supplementary material
The online version of this article (doi:10.1007/s00239-008-9111-7) contains supplementary material, which is available to authorized users.
Keywords: Collagen, Persistence length, Intron, Trichodesmium erythraeum, Interdomain transfer, Cyanobacteria
Introduction
Collagen is the most abundant protein as measured by the total mass present in humans (Di Lullo et al. 2002) and all organisms (Nimni et al. 1988). Fibrillar collagen is highly organized at critical mechanical stress locations such as bones, tendons, ligaments, nerve sheaths, skin, and cornea. In single-celled organisms, the necessity for a protein typically found in connective tissue is less clear. Exceptions are the collagen-like fragments found in Streptococcus equi (Liden et al. 2008) and Bacillus cereus (Daubenspeck et al. 2004). Nevertheless, collagen may have been one of the key emergent bridges from single-celled prokaryotes to multicellular eukaryotes. For a review of the importance of the triglycine repeat and how it is maintained in fibrillar collagens, see Kadler et al. (2007). Current consensus holds that the collagen gene began as a relatively short sequence that allowed for small groups of cells to organize into larger systems and perhaps eventually differentiate into complex colonies. Through the process of mutation and natural selection this gene eventually lengthened, diversified through gene duplication events, and was refined into its characteristic triple-helical, self-interacting structure, allowing for greater mechanical stresses to be sustained among cells (Boot-Handford and Tuckwell 2003; Buehler 2006b; Wada et al. 2006). This conferred the ability of the organism to maintain cellular diversity, accelerate quickly, absorb mechanical energy, and resist fracture (Buehler 2006b), allowing organisms that possessed it to rise to the “top” of the food chain.
The recently predicted, but yet undetected collagen-like gene found in Trichodesmium erythraeum by Orcutt et al. (2002) shares many characteristics with vertebrate collagen. It has a triglycine repeat region that is approximately 10% longer than that of most vertebrates, and it shares several identical or similar residues in its N and C termini. An intriguing possibility is that it acquired this gene through viral-mediated interdomain transfer from a larger marine organism.
T. erythraeum is a colonial marine cyanobacterium that can be seen with the naked eye. During periods of low wind stress and warm temperatures it forms blooms of surface aggregations that can be tens of thousands of kilometers wide (Capone et al. 1997). From NASA satellite images, blooms of this size have been seen (Negri 2006). Trichodesmium was given its name from its appearance: the Greek word “trichoma” for hair and “desmus” for bonded—”bonded-hair.” As the cells age, they become positively buoyant and rise to the surface (Walsby 1994). Once these segmented structures reach a critical length, they fracture, allowing new growth to occur at a new set of free ends as they enter their exponential growth phase (Bell et al. 2005). Its collagen-like gene may serve the purpose of maintaining colony contact even as individual cells lose direct contact with each other. These blooms or “rafts” consist of healthy and aged cells mixed with detritus in a “mucilaginous matrix” (Endean and Monks 1993). According to observations from the tropical and subtropical North Atlantic, Trichodesmium produces more nitrogen than any other macroscopic (0.5–4 mm) cyanobacteria and about half of the new nitrogen used for primary production (Capone et al. 1997). In fact, the biological productivity of large expanses of the ocean is often limited by the availability of nitrogen and Trichodesmium as an N2 fixer, thus making it of critical importance for supporting the metabolic requirements of a fast number of non-nitrogen-fixing organisms.
Genetic characterization of Trichodesmium species suggests that two distinct clades are present in the oceans: one including the closely related species T. thiebautii, T. tenue, T. hildebrandtii, and K. spiralis and the other containing only T. erythraeum as determined primarily by comparing introns and intein presence (Orcutt et al. 2002). The T. erythraeum ribonucleotide reductase (RIR) gene was found to encode four inteins and three group II introns, which is extremely unlikely to have occurred by chance, considering the rarity of inteins and introns (Liu et al. 2003) within this genus.
If early collagen genes were to survive as “the protein of choice” in large, multicellular organisms to maintain the mechanical integrity of tissues together under static loading conditions, and under extreme loading conditions such as avoiding predators or pursuing prey, then they must have been selected or “purified” by the two competing metrics: the ability to avoid rupture at high mechanical stress and the ability to self-assemble rapidly. This suggests the question: What is the optimal collagen translation length?
We present a theoretical framework for how the collagen translation length is balanced by the competing metrics of being long to support tensile loads while, at the same time, being short enough to robustly self-assembly. We also present experimental results demonstrating the presence of a collagen-like protein in T. erythraeum and a phylogenetic analysis. Experimental methods include immunogold atomic force microscopy, sequence alignment, and phylogeny tree construction.
Theory
The amount of mechanical stress, σ, a biological tissue can sustain is
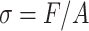 |
1 |
where F is the force applied to the tissue, and A is the tissue’s cross-sectional area. Organisms rely on mechanical integrity of their constituents insofar as these constituents do not have unjustifiable metabolic expenses. Recasting the stress equation as
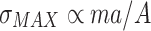 , where m is the organism mass, and a is the acceleration experienced by the organism, it is seen that organisms with greater mass and/or higher acceleration rates, must have tissues that can withstand greater stresses. Galileo (Galilei 1638, 1991) and later Huxley (1932, 1993) observed that the larger an organism becomes, the more of its mass must be structural. Their scaling laws take the form,
, where m is the organism mass, and a is the acceleration experienced by the organism, it is seen that organisms with greater mass and/or higher acceleration rates, must have tissues that can withstand greater stresses. Galileo (Galilei 1638, 1991) and later Huxley (1932, 1993) observed that the larger an organism becomes, the more of its mass must be structural. Their scaling laws take the form,
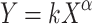 |
2 |
where Y represents the dimension of an organ or subsystem of an organism (measured in units of mass, length, area or volume), X represents the organism’s body size (measured in mass or volume), k is the “allometric coefficient,” and α is the “allometric exponent” that is a curve-fitting parameter. For example, it may be approximately cubic if X represents a linear dimension and Y represents a volumetric dimension. For the present application, we cast the allometric formula (Huxley 1932; Martin 1981) as 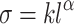 , where σ is the maximum mechanical stress an organism is likely to experience and l is the contour length of the collagen gene transcript.
, where σ is the maximum mechanical stress an organism is likely to experience and l is the contour length of the collagen gene transcript.
Combining the allometric equation with the stress equation yields
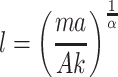 |
3 |
Restated, the contour length, l, of the collagen triglycine repeat should have a tendency to increase as the mass, m, of an organism, and the accelerations, a, it is subjected to increase. However, having a tissue with a large cross-sectional area, A will diminish the need for long fibril-forming molecules.
The relatively nonmotile organism T. erythraeum uses gas vesicles rather than flagella to stratify (Walsby 1994). Thus forces generated by its environment such as wind and ocean currents likely to rend its colonies apart, are represented by the ma term of Eq. 3. This force could also be expressed as being proportional to Stokes drag: −6πηrv, where η is the fluid viscosity, r is the cell radius, and v is the fluid velocity. Although T. erythraeum has not been observed to manufacture true collagen fibrils, this does not preclude the presence of single-, double-, or triple-helical collagen at the cell surfaces that provide a cohesive material for maintaining colony integrity.
At the molecular scale, a contour length that is significantly longer than the persistence length may become a liability to self-assembly. Restated, the contour length l competes with the persistence length l p. Once l surpasses l p, the probability of self-entanglement increases, thus diminishing the probability of a molecule such as collagen to aggregate radially and axially into fibrils. Recent work by Buehler (2006a) indicates qualitatively that longer molecules do have higher probabilities for their two ends to come into close proximity through the accumulation of molecular-scale bends. For a review on the mechanisms of fibrillogenesis see Hulmes (2002).
The persistence length is an important parameter in determining self-assembly mechanics of structural proteins at the molecular scale (Wilhelm and Frey 1996). For triple-helical type II collagen, l p = 11.2 nm (Sun et al. 2004), and for type I collagen l p = 14.4 nm for triple-helical (Sun et al. 2002). The persistence length for triple-helical collagen has been predicted by Buehler (2006a) to be 23.4 nm. Indeed, persistence length and bending stiffness are related through the relationship
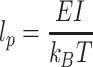 |
4 |
where E is the elastic modulus (with dimensions of energy per unit volume), I is the second moment of inertia (with dimensions of length to the fourth-power, L 4 [Gere 2004]), k B is Boltzmann’s constant, and T is temperature measured in kelvins. The persistence length thus represents the ratio between the order-preserving energy of the structure EI and its entropy-creating thermal energy k B T. By comparison, a microtubule’s persistence length is 100 μm to 5 mm (Pampaloni et al. 2006). The “critical question” facing a “freshly translated” tropocollagen triple helix is whether the contour length-to-persistence length ratio allows for the possibility of self-interaction and, thus, circular structure formation rather than fibril aggregation. Similar events have been observed in other filament-forming proteins such as actin (Tang et al. 2001). If this self-binding were to occur at an appreciable rate, fibrillogenesis would be impeded, thus reducing collagen’s efficacy at performing its structural role. Since the persistence length is the length at which the direction of one end becomes uncorrelated with that of the other (Doi and Edwards 1986), a molecular structure with a contour length of al p may be approximated as the length at which a looping self-interaction becomes likely, where a is a scalar. The distribution of entropically driven shapes is dependent on a variety of conditions such as solvent media, local molecular interactions, and microenvironmental constraints. To proceed with a formal analysis, the creation or simulation of tropocollagen molecules of increasing lengths with the N and C termini cleaved and lysines and hydroxylysines at appropriate locations to form cross-links is required. Experimental work or simulations at appropriate temperatures would produce probability curves of loop-structure formation. Preliminary work has been done by Buehler (2006a).
Materials and Methods
Triglycine repeat ranking
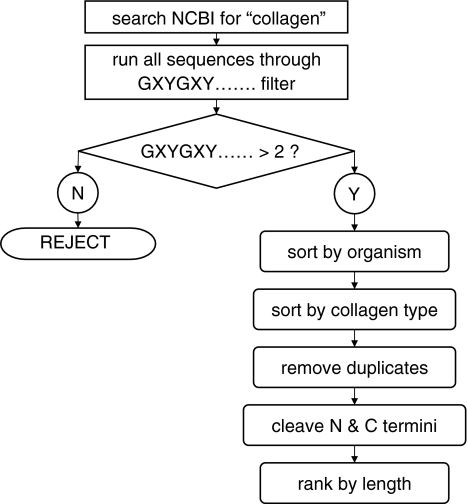
We performed searches in the NCBI database for the term “collagen.” All returned protein sequences were then “purified” by eliminating the N and C termini upstream and downstream of the GXY portion, using a custom-written Bourne shell script (Fig. 1) that searches for uninterrupted “G??” sequences, where “?” represents any character. The script counts the total number of characters in the triglycine repeat portion, then divides this number by three to obtain the triglycine repeat length. These lengths were then ranked by number and plotted.
Fig. 1.
Algorithm for producing data depicted in Fig. 2. All current sequences listed in the NCBI database with “collagen” in the description were downloaded, along with ascension number, species name, and collagen type. These were then filtered according to species and collagen type, their N and C termini were cleaved, and the triglycine repeat numbers ranked by length
Light microscopy
Samples of T. erythraeum were provided by John Waterhouse, Woods Hole Oceanographic Institute. Upon receipt, samples of T. erythraeum suspended in 50-ml vials of native ocean water were aliquoted into 1.5-ml vials and immediately stored at −70°C. Frozen samples were thawed at room temperature and centrifuged at 10,000 g for 5 min. Pellets were then pipetted onto standard microscopy slides (Fisher Scientific, USA), coverslipped, and imaged in phase-contrast mode at 200× on an Olympus IX81 inverted light microscope. Images were captured digitally with a SPOT-RT camera (Diagnostic Instruments, Sterling Heights, MI).
Environmental scanning electron microscopy
Samples from the same aliquot as used for light microscopy were also prepared for environmental scanning electron microscopy in a Phillips XL30 ESEM (FEI, Hillsboro, OR) with a gaseous secondary electron detector (GSED) at a chamber pressure of 4.0 Torr and 15 kV. Samples were placed directly on an aluminum stub, which was then placed on a sample holder inside the ESEM chamber in environmental (wet) mode. Excess water was evaporated from the sample to facilitate observation of the sample. By circulating water molecules and maintaining the pressure of the chamber within the range of 2–10 Torr, ESEM allows for observation of samples that are insulating without the need for a coating and keeps hydrated samples moist.
Immunogold AFM
Bovine collagen was used as a control and E. coli were used as a negative control. Fresh samples of E. coli were supplied by MinJun Kim of Drexel University. This technique was chosen after initial efforts with immunohistochemistry revealed that T. erythraeum emits strong autofluorescence at each of the three standard fluorescence microscopy colors, FITC, TRITC, and rhodamine, making standard immunofluorescence techniques impractical. E. coli were cultured and stored as described elsewhere (Steager et al. 2007). Both of the bacteria samples were thawed at room temperature and lightly vortexed. Five hundred microliters of T. erythraeum and 200 μl of E. coli were each pipetted into separate microcentrifuge tubes. The two specimens were then placed in a microcentrifuge for 2 min at 10,000 g. After the supernatant was removed, 500 μl of 1.8% formaldehyde (diluted from Fisher Scientific BP531-500) was added. Specimens were then vortexed and kept at 5°C. Bovine collagen strands (Sigma C9879) (Einbinder and Schubert 1951) were teased apart into a single fragment of approximately 100 μm3 and placed in a 1.5-ml microcentrifuge tube. All three specimens were then spun for 1 min at 13,000 rpm and the supernatant was removed. Two hundred microliters of PBS (0.01 M, pH 7.4) was added to each while the samples were kept at 5°C. Fifty microliters of rabbit collagen (I + II + III + IV + V primary antibody ab24117; Abcam, Cambridge, MA) was diluted in 1.5 ml of PBS (0.01 M, pH 7.4). There is some contention as to what the specific binding sites are for collagen antibodies. It is generally assumed that the triglycine repeat portion is nonimmunogenic, and that the globular N and C termini are antigenic. Thus we chose a polyclonal antibody with a high likelihood of binding to a wide array of collagens, since the T. erythraeum collagen is yet to be classified. The mixture was vortexed and then centrifuged at 10,000 g for 5 min to remove any impurities. The specimens were centrifuged for 1 min at 10,000 g and the supernatant was removed. Two hundred fifty microliters of the supernatant of the diluted primary antibody was added to each specimen. Next the specimens were vortexed and kept at 5°C for 30 min. To prepare the anti-rabbit IgG (whole-molecule) gold colloid secondary antibody (Sigma G7402), 1400 μl of 0.5 M NaCl (diluted from Ricca Chemical 7215-16), 1.4 mg of BSA (Sigma A7030), 0.7 μl of Tween 20, and 70 μl of FBS (Fisher Scientific BW-14-503F) were added to a microcentrifuge tube and vortexed. Seventy-two microliters of this solution was discarded, 100 μl of secondary antibody was added, and then the mixture was vortexed. The three specimens were centrifuged for 2 min at 13,000 rpm and the supernatant was removed. Three hundred microliters of PBS was added to T. erythraeum and E. coli and the specimens were centrifuged for 2 min at 10,000 g. The supernatant was removed, and the wash was repeated. Six hundred microliters of PBS was added to the bovine collagen and the specimen was centrifuged for 2 min at 10,000 g. The supernatant was removed, and 250 μl of diluted 1000:1 secondary antibody was added to all three specimens. The specimens were then vortexed and kept in the dark at 5°C for 1 h. The specimens were centrifuged for 5 min at 10,000 g and the supernatant removed. Three hundred microliters of PBS was then added to each specimen and the samples were vortexed. The specimens were centrifuged for 5 min at 10,000 g and the supernatant was removed. This process was repeated two more times, for a total of three washes with PBS. Fifty microliters of PBS was added to each specimen, which were kept at −20°C in the dark until imaging. Samples were imaged on a Digital Instruments Series 3100 Nanoscope in air-tapping mode with DNP-S tips (Veeco Probes) at resonance frequency (∼300 kHz). Image sizes ranged from 153 to 500 nm2, and all were sampled at a resolution of 256 × 256 pixels. Images were postprocessed using the Digital Instruments 5.12r5 Nanoscope software. Individual features with diameters between 2 and 15 nm were selected visually and counted in four images from each of the three samples: positive control, bovine collagen (BC), negative control, E. coli (EC), and the experimental samples, T. erythraeum.
Homology scoring and phylogenetic tree construction
To determine the likely evolutionary path of T. erythraeum collagen, we employed a BLAST query (Altschul et al. 1990; Gish 2006; Karlin and Altschul 1990) to find homologous sequences throughout the NCBI database. This returned approximately 8000 sequences from 547 different organisms. In order to limit the size of our tree we selected 26 collagen expressing organisms for the phylogenic tree (Table 1). Sequences were chosen on the following basis: (1) their identity to the T. erythraeum sequence (these primarily included fibrillar collagen found in vertebrates), (2) their environment (for example, in the analysis we included collagen sequences from several nonvertebrate, marine-dwelling organisms such as the thermal tubeworm, Riftia pachyptila, Mueller’s freshwater sponge, Ephydatia muelleri, the hydrothermal worm, Alvinella pompejana, and the common mussel, Mytilus edulis), and (3) their physiology (or likely phylogenetic relationship to T. erythraeum). The number of sequences in this group is relatively small. For this reason we included Streptococcus equi, shrimp white spot syndrome virus, and Bacillus cereus. These three sequences, which are also intron-free, were included to examine likely divergence points of the respective sequences.
Table 1.
Collagen sequence ascession numbers and organisms used for T. erythraeum collagen phylogeny tree construction
| Entrez-ID | Species | Type | Chain | Seq. length |
|---|---|---|---|---|
| AAC35289 | Alvinella pompejana (hydrothermal worm) | Collagen | 890 | |
| 42783914 | Bacillus cereus ATCC 10987 | Collagen | 1321 | |
| 38649122 | Danio rerio (zebrafish) | Type I | α1 | 1447 |
| 34784485 | Danio rerio (zebrafish) | Type I | α3 | 1449 |
| 157030 | Drosophila melanogaster (fruit fly) | Type IV | α1 | 1775 |
| 1920343A | Ephydatia muelleri (Mueller’s freshwater sponge) | Collagen | 1095 | |
| 5921192 | Gallus gallus (chicken) | Type I | α2 | 1362 |
| 15546070 | Gallus gallus (chicken) | Type II | α1 | 1420 |
| 124056487 | Homo sapiens (human) | Type I | α1 | 1464 |
| 450394 | Homo sapiens (human) | Type II | α1 | 1487 |
| 124056490 | Homo sapiens (human) | Type III | α1 | 1466 |
| 83301500 | Mus musculus (house mouse) | Type I | α1 | 1453 |
| 200215 | Mus musculus (house mouse) | Type II | α1 | 1459 |
| AAB96638 | Mytilus edulis (common mussel) | Collagen | 922 | |
| 14164347 | Oncorhynchus mykiss (rainbow trout) | Type I | α1 | 1449 |
| 14164351 | Oncorhynchus mykiss (rainbow trout) | Type I | α2 | 1356 |
| 14164349 | Oncorhynchus mykiss (rainbow trout) | Type I | α3 | 1458 |
| 56565281 | Paralichthys olivaceus (bastard halibut) | Type I | α1 | 1447 |
| 56565283 | Paralichthys olivaceus (bastard halibut) | Type I | α2 | 1352 |
| 82123471 | Rana catesbeiana (bullfrog) | Type I | α1 | 1445 |
| 18202034 | Rana catesbeiana (bullfrog) | Type I | α2 | 1355 |
| AAB24972 | Riftia pachyptila (thermal tubeworm) | Collagen | 1027 | |
| 15021422 | Shrimp white spot syndrome virus | Collagen | 1684 | |
| 37498968 | Streptococcus equi | Collagen | 491 | |
| CAG02093 | Tetraodon nigroviridis (pufferfish) | Collagen | 1399 | |
| 71671489 | Trichodesmium erythraeum IMS101 | Collagen | 1340 |
These sequences were multiply aligned using the ClustalW program (Higgins et al. 1994) and then refined using the TreeRefiner program (Manohar and Batzoglou 2005). Formatting was performed using Boxshade 3.21 from EMBnet. The final tree was created using the Jukes-Cantor (1969) method of determining evolutionary distance and then using the sequence neighbor-joining (NJ) method (Gascuel 1997) as implemented in the Matlab R2007a bioinformatics toolbox (Cai et al. 2005). Gaps were treated as ‘missing values’ rather than as ‘differences’ so as not to overestimate resulting distance values. Bootstrap values were obtained from the ClustalW program using the default parameters of 1000 bootstrap repetitions and are reported as percentages (Higgins et al. 1994).
We used the NJ method since we are comparing multiple organisms from vastly different environmental niches. This is justified since it is likely that the selection criteria for the maintenance of the triglycine repeat region of marine cyanobacteria are as different from those for a fibrillar vertebrate collagen as are the selection criteria for afibrillar versus fibrillar collagens. We chose the NJ method rather than the unweighted pair group with arithmetic mean (UPGMA) method, since UPGMA only provides information about the relative order of the evolutionary path (e.g., Wiersma et al. 2005), whereas the NJ method provides an estimate of evolutionary distance.
Finally, to test whether the triglycine region artificially “forced alignment” between the T. erythraeum sequence and the other sequences included in the tree construction, we ran ClustalW with the glycine weight set to zero. This more rigorous approach was performed to evaluate the phylogeny purely on the nonglycine regions. With the glycines, which comprise nearly one-third of the sequence, absent from the analysis alignment and phylogeny, tree construction is more stringent, thus making the results more compelling.
Results
Triglycine repeat ranking
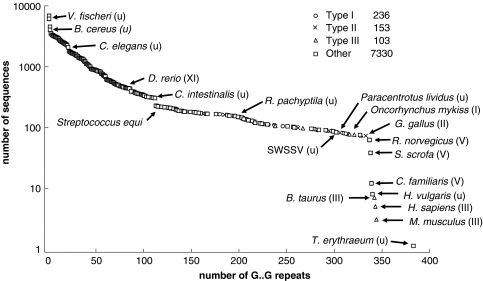
The longest uninterrupted GXY repeats returned came from fibrillar collagens. Afibrillar collagens such as type IV were returned by the script but were not among the longest because of their imperfect GXY repeats. We found that the predicted translation from the intron-free gene for the collagen-like gene found in T. erythraeum has a triglycine repeat length that is approximately 10% longer than that of any other sequenced organism (Fig. 2). The “shelf” present at approximately 454 repeats may represent the molecular contour length where loop assembly becomes more likely than fibril assembly, thus offering evidence as to why this length is not surpassed in nature. The apparent lack of fibrils, combined with the lack of posttranslational modification enzymes for collagen, suggests that T. erythraeum may be using its collagen-like protein in a manner similar to the nonfibrillar basement membrane collagens, where lattice-like structures are prevalent.
Fig. 2.
A log-linear plot of the triglycine repeats present in all of the presently sequenced collagen-gene containing organisms. The abrupt shelf at 344 triglycine repeats may represent an optimization whereby the necessity to have a long molecule for strong, compliant tissues competes with the necessity to have a short molecule for robust self-assembly. In the labels, roman numerals indicate collagen types and “u” indicates an unclassified collagen
The shelf in the graph in Fig. 2 indicates that this critical length for the collagen triple helix occurs at about 344 triglycine repeats. The obvious outlier, T. erythraeum, at 383 triglycine repeats suggests that this shelf is created by the necessity for the contour length, l, to be less than some critical multiple of the persistence length, l p. Indeed, Buehler (2006b) has predicted that past a contour length of ∼200 nm, extra length becomes a liability. This is explained through the following mechanics argument. A single collagen triple helix has a rupture force of approximately 22.5 nN (Buehler 2006a). By contrast, hydrogen bonds have a rupture forces three orders of magnitude lower (Gao et al. 2002), and individual cross-link rupture forces are approximately one order of magnitude lower (Sulchek et al. 2006). Thus, to break a single collagen triple helix, approximately 1000 hydrogen bonds, or approximately 10 cross-links, must be present. Therefore, a single collagen triple helix with a fixed cross-sectional area that accumulates more cross-links than the covalent bonds of its triple helix can support is more likely to rupture than a shorter triple helix with fewer cross-links. From this we conclude that there is no evolutionary advantage for individual triple helices to grow beyond a length that permits excessive accumulation of cross-links.
Light microscopy
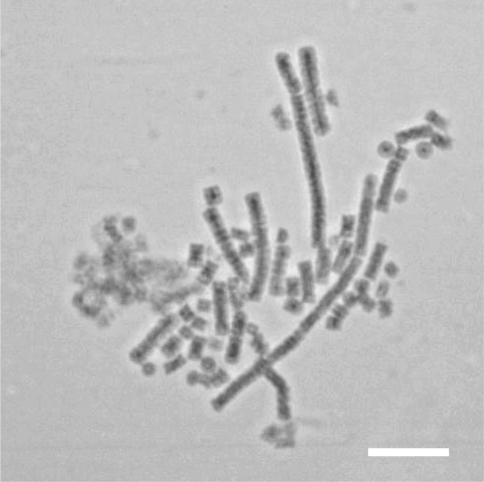
Under light microscopy, the rod-like appearance of T. erythraeum is apparent (Fig. 3). Its cell duplication modality is one of linear aggregation, with daughter cells being produced axially. Lateral aggregation is also apparent from this image. Although maintenance of laboratory cell culture is difficult, presumably all cells under natural and laboratory conditions are capable of division (Bell et al. 2005), thus a breakage along the length of the axially aggregating structure allows for accelerated growth of the colony at the newly formed ends.
Fig. 3.
Under light microscopy, the rod-like appearance of T erythraeum is apparent. Scale bar = 30 μm
Environmental scanning electron microscopy
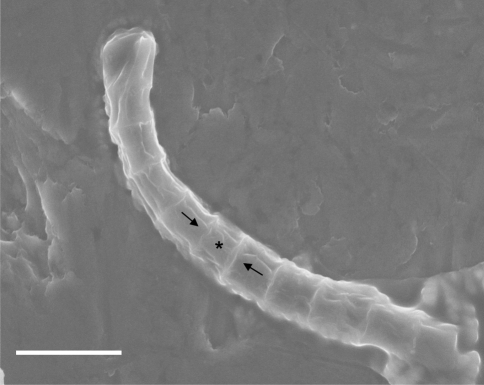
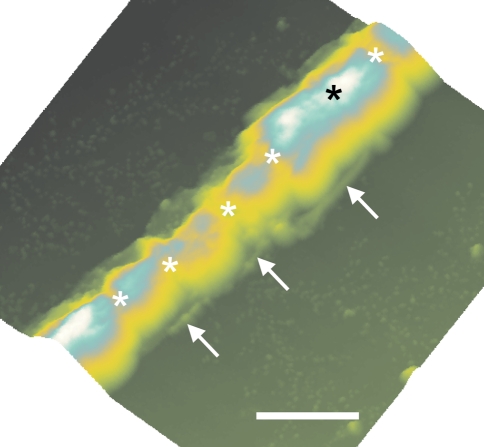
Under higher magnification using environmental scanning electron microscopy (Fig. 4), the axial aggregation of the cells is clearly present. A buttressed appearance is present, suggesting a polarity that may indicate the direction of growth. At this magnification, neither fibrils nor protofibrils appear to be present.
Fig. 4.
Environmental scanning electron microscopy results. Intracellular junctions are clearly present. The asterisk indicates a possible parental cell. The two arrows indicate the polarity of its two probable daughter cells. Scale bar = 10 μm
Atomic force microscopy
The cell junctions seen under both light microscopy and electron microscopy are also clearly seen under air-tapping-mode atomic force microscopy (Fig. 5). Also apparent in this view are traces of what may be protofibrils along the length of the cell structure. An alternative explanation for the presence of the fibrous structures seen along the lengths of the cells is that they are the result of precipitates formed during drying onto the mica surface.
Fig. 5.
Under atomic force microscopy, boundaries separating the individual cells are readily visible. Also noticeable in this sample preparation are fibrous-like structures that appear to bridge these cell junctions. Although banding was not observed, these fibrous structures may be collagen protofibrils. Scale bar = 5 μm
Immunogold atomic force microscopy
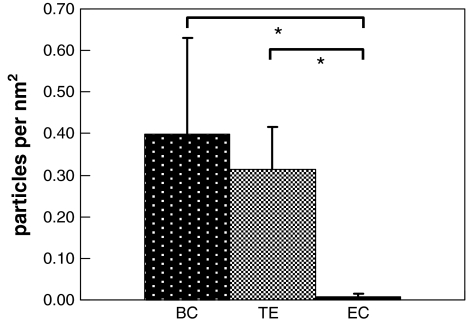
The expression of a protein in T. erythraeum with an affinity for polyclonal collagen antibodies has not previously been reported. Here, we present the first results indicating that such a protein is being expressed and is present at the cell surface. The density of immunolabeled gold nanospheres in T. erythraeum was approximately 0.3 particle/nm2. Bovine collagen, which was used as a positive control, showed a nanosphere density of approximately 0.4 particle/nm2. E. coli, a negative control, showed negligible labeling (Figs. 6 and 7). According to the antibody manufacturer, the specific location of reaction between the antibody and the collagens in this study is unknown. The antibody to collagen types I–V we selected is a cocktail of antibodies prepared by mixing semispecific antibodies to all these types. Rabbits were immunized with individual types but cross-response of antisera to several collagen types was typical. All the individual collagen types were generated as native molecules separated by differential salt precipitation after extraction into mild acidic media from pepsin-digested washed tissue (placenta, skin, cartilage). The antibody reacts predominantly to native collagen (some common determinants of five types) but somewhat less binding was observed to heat-denatured collagen types also. Finally, the manufacturer states that the reactivity to telopeptides cannot be excluded because pepsin digestion was minimal. It is commonly held that the triglycine region of collagen is nonimmunogenic: its telopeptide region is likely responsible for antibody binding. However, since Trichodesmium erythraeum does not appear to have the posttranslational enzymes necessary to cleave its N and C termini, these are likely still present and prevent fibrillogenesis. However, it may be that T. erythraeum’s collagen-like protein still maintains some ability to self-assemble and perhaps behaves more like the basement membrane collagens, e.g., types IV and VI.
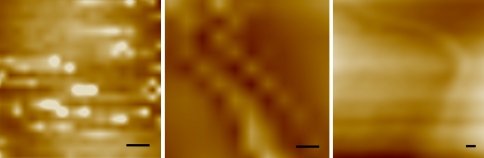
Fig. 6.
Examples of immunogold atomic force microscopy labeling results for (A) bovine collagen (BC; positive control), (B) T. erythraeum (TE; experimental), and (C) E. coli flagellum (EC; negative control). Scale bar = 15 nm
Fig. 7.
Immunogold atomic force microscopy labeling results for bovine collagen (BC; positive control), T. erythraeum (TE; experimental), and E. coli (EC; negative control). *p < 0.05, Student’s t-test
Alignment
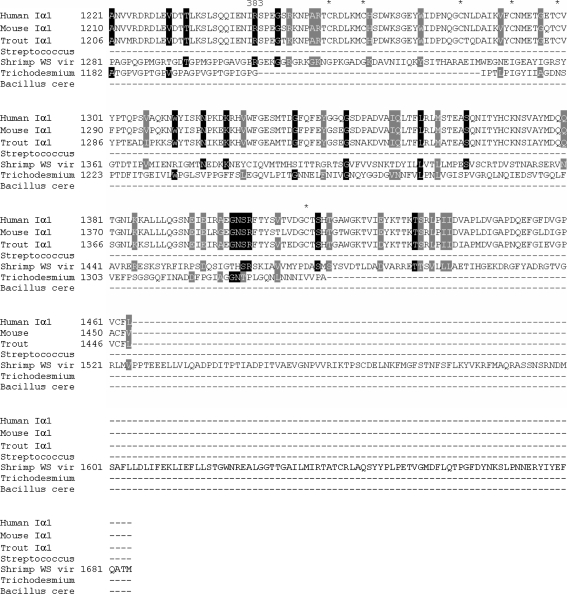
The protein alignment of Homo sapiens (human) Iα1, Mus musculus (mouse) Iα1, Oncorhynchus mykiss (trout) type I α1, shrimp white spot syndrome virus collagen of an unclassified type, Streptococcus equi collagen-like sequence, and Bacillus cereus collagen-like sequence, with the predicted sequence from T. erythraeum, indicates that the identity shared with shrimp white spot virus, human, bovine, and trout is the greatest (Fig. 8). Entrez IDs used are given in Table 1. The long, uninterrupted triglycine repeat regions similar among humans, mouse, shrimp white spot syndrome virus, and T. erythraeum suggest the intriguing hypothesis that T. erythraeum inherited its collagen-like gene via a viral-mediated interdomain transfer. The majority of genes found in WSSV share homologues with eukaryotes rather than prokaryotes (Yang et al. 2001), indicating that WSSV may indeed be a predominant vector for gene transfer from eukaryotes. Beginning with residue 91 for both human and bovine, residue 170 for shrimp white spot virus, and residue 2 for T. erythraeum, the triglycine repeat region runs uninterrupted for 383 GXYs. The two other prokaryotes included in the analysis, Streptococcus equi and Bacullis cereus, have 102 and 100 triglycine repeats, respectively. Alignment results wherein glycines were excluded were nearly identical to those in Fig. 8. This was primarily due to the shared identity at the N and C termini as well as the result that many of the differences between the T. erythraeum sequence and the others analyzed involved substitutions of residues with similar physiochemical properties.
Fig. 8.
Alignment of the predicted T. erythraeum collagen sequence with three vertebrates with high identity scores: Homo sapiens (human) type I α1, Mus musculus (mouse) type I α1, and Oncorhynchus mykiss (trout) type I α1. Also included are the collagenous sequences found in shrimp white spot syndrome virus, Streptococcus equi, and Bacillus cereus. Black highlighting indicates >50% identity. Gray highlighting represents conservative substitutions. Asterisks indicate the six cystines present at the C terminus of the vertebrates. The collagenous and noncollagenous regions are indicated. Numbers appearing above the sequences indicate the number of triglycine repeats
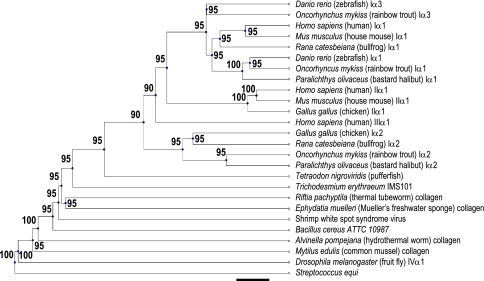
Phylogeny
The phylogeny tree for T. erythraeum’s collagen gene, in which we included several of its most similar fibrillar collagens, indicates that the acquisition of T. erythraeum’s collagen-like gene occurred at a time when fibrillar and nonfibrillar collagens were diverging (Fig. 9). It sits at a very interesting intersection in the phylogeny tree, namely, at the junction between several invertebrates (below) and several vertebrates with Clade A fibrillar collagens (above). It also sits very close to shrimp WSSV. Also notable is that the collagen-like sequences from the prokaryotes Streptococcus equi and Bacillus cereus appear on more distant branches. These sequences were included primarily just to examine where these degenerate collagen-like sequences might fall within a phylogenetic tree. Their remoteness from the fibrillar collagens included indicates an ancient divergence. Near the bottom of Fig. 9 is a type IV collagen, which is separated by a relatively large distance from the fibrillar collagens at the top of the figure, and is consistent with results from others who indicate that the divergence of fibrillar and afibrillar collagens is more ancient that divergences of individual fibrillar collagens (e.g., Aouacheria et al. 2004; Morvan-Dubois et al. 2003; van der Rest and Garrone 1991). For a discussion of afibrillar collagen evolution see Aouacheria et al. (2006). (Aouacheria et al. 2006) Since the tree structure was not affected by gaps, and tree-branch lengths represent approximations for divergence times, the placement of the divergence time of T. erythraeum’s collagen-like gene from that of its closest neighbors such as thermal worms and SWSSV, indicates that the collagen-like gene of T. erythraeum had a common ancestor that predates modern vertebrates, but that it may have inherited its collagen-like gene from a now extinct vertebrate. The tree that resulted from the unweighted glycine analysis showed no difference in divergence order, and only slight differences in divergence times.
Fig. 9.
Evolutionary distance between the collagen gene(s) found in T. erythraeum and the collagen genes in several disparate organisms. Numbers next to divergence nodes represent bootstrap values. Scale bar = 0.1 substitution per site
Discussion
We have provided the first evidence that the collagen-like gene in the marine cyanobacterium T. erythraeum is expressed and present on the cell surface. We have also provided theoretical evidence that the collagen translation length may be determined by competing metrics of strength and self-assembly. Phylogenetic analysis indicates that the collagen-like gene appeared in this organism after the divergence of fibrillar and afibrillar collagens, but before the divergence of the fibrillar collagens. Finally, we argue that the maintenance of this gene within the genome of T. erythraeum provides it with a selective advantage in that it allows aggregations that enable it to “prey” on larger organisms through concentration of its neurotoxin and through mechanical gill-clogging mechanisms. Four discussion sections—theoretical, experimental, bioinformatic, and ecological—follow.
Theoretical
The triglycine portion of the collagen gene transcript appears to have reached an evolutionary “ledge” at approximately 340 GXY. This ledge likely appears at this specific length for two reasons: (1) if it were to become longer, this would represent a liability for self-assembly; and (2) a longer triple helix would not necessarily increase fibril strength, as additional length would allow the possibility of additional cross-links and hydrogen bonds to accumulate along its length in excess of what the covalent bonds of the triple helix itself can support.
Long collagen transcripts are driven by the need for a sufficient number of cross-links to develop between a single triple helix and its approximately 24 neighboring triple helices through hydrogen bonding and hydroxyproline-lysine cross-links. However, the need to self-assemble keeps the collagen molecule from becoming too long so that self-interaction prior to fibrillogenesis is less likely. The persistence length/contour length ratio has recently been discussed, but not systematically studied to determine the likelihood of self-interaction (Buehler 2006a).
In rope building, long subfibers are clearly an asset for developing great tensile strength. Shear lag theory (Weitsman and Beltzer 1992) states that the tensile force within a subcomponent of a tension member is proportional to the amount of shear stress developed within it. This theory remains valid for the molecular scale within and among individual collagen triple helices: the ratio between the sum of the strength of collagen’s cross-links and that of the triple helix itself determines its success or failure as an effective molecular rope. Adding additional binding sites along a triple helix might overwhelm the bond strength along a single triple helix (Buehler 2006b).
Experimental
We have provided the first evidence that the abnormally long triglycine repeat within the collagen-like gene of T. erythraeum is being expressed. The collagen of T. erythraeum, which shares a great deal of identity with the Clade A fibrillar collagens of large vertebrates, apparently does not form fibrils. Other discoveries of “superlong” collagen molecules from the cuticle of marine tube worms and annelids (Gaill et al. 1991), interpreted from histograms taken from rotary-shadowed TEM images, indicate that there may be collagen molecules up to 2.4 μm in length. However, since this publication, the full collagen sequence data for the two organisms Riftia pachyptila and Alvinella pompejana, from which these samples were taken, indicate that their collagen protein sequences are substantially shorter: 1027 and 890, respectively. Gel electrophoresis data from Gaill et al. (1991) indicated that these two marine organisms do have massive collagens and that banded fibrils are present in the cuticle and interstitial tissues of both. However, the possibility exists that the molecular lengths measured by Gaill et al. were fibril fragments rather than individual triple helices.
The lack of fibrils present in our samples indicates that although the collagen-like sequence of T. erythraeum shares a great degree of identity with the Clade A fibrillar collagens, the lack of posttranslational enzymes required for N- and C-terminal cleavage prevents fibrillogenesis. Clearly further work is required to determine if the N termini of the T. erythraeum collagens are capable of performing the trimerizing required to initiate tropocollagen formation.
Bioinformatic
There are multiple alternatives for the origin of the collagen-like gene of T. erythraeum. One possibility is that it acquired its collagen-like gene intron-free through a horizontal transfer at a time when large vertebrates were prevalent, during the Devonian or Silurian epochs. The second alternative is that it evolved from a shorter prokaryotic version of the protein via repeat expansion. The relative likelihood of these two alternatives may be evaluated through more exhaustive phylogenetic analyses once additional sequence data become available from a greater number of organisms. Nakamura et al. (2004) (Nakamura et al. 2004) found, in analyzing whole genomes of 116 prokaryotes, that 14% of open reading frames were subjected to recent horizontal gene transfer. The most frequently transferred genes were those related to cell surface function, DNA binding, and pathogenicity.
Support for the former alternative is that shrimp white spot syndrome virus also carries a long, intron-free collagen sequence, with a length of 5054 bp (van Hulten et al. 2001). Lateral gene transfer from within a given species, from organelles to the nucleus, is a commonly observed occurrence (e.g., Adams et al. 1999) and has been used to estimate the amount of time organelles such as mitochondria have inhabited eukaryotic cells (Parkinson et al. 2005). Recently, lateral gene transfer has also been observed and discussed among prokaryotes, P → P, among eukaryotes, E → E, from prokaryotes to eukaryotes, P → E, and, recently, from eukaryotes to prokaryotes, E → P (Jenkins et al. 2002). An E → P event, however, might help explain why the gene for an extracellular matrix protein typically associated with vertebrates or multicellular metazoans might have found its way into contemporary prokaryotes through an event such as viral infection or naked DNA transfer. If this occurred in T. Erythraeum, it may have happend via a spliced mRNA intermediate as discussed by Andersson (2005). The lability of exposed RNA makes this less likely, but the abundance of introns in eukaryotic collagen DNA (50 for human type I αI) and the lack thereof in T. erythraeum make it likely that the infectious agent path is the only way for interdomain transfer to have occurred (e.g., Davis 2002). Another alternative is that this event occurred via a viral transfection event as mentioned by Gogarten (2003). The ecological relationships among viral phages and prokaryotes is vast and complex and may offer a viable explanation as to how this particular collagen-like gene entered the prokaryotic genome (Weinbauer 2004).
Regarding the second alternative, namely, that the origin of Trichodesmium erythraeum’s collagen-like gene was from a repeat expansion mechanism, the probability of this happening compared to the probability of transfection is difficult to estimate, but is presumably less involved and more probable in an organism with a plasmid-based DNA instruction set. Similar hypotheses have been put forth as an explanation for the large variety of collagen genes currently observed naturally (Boot-Handford and Tuckwell 2003).
The primary argument against our contention that the collagen-like gene found in T. erythraeum was inherited through horizontal gene transfer is that two vertebrate species such as human vs. fish (trout) sequences are nearly identical, whereas less identity is shared between T. erythraeum and any of the three vertebrates in Fig. 8. More specifically, 400 million years of evolution between two vertebrates such as trout and human has not greatly changed the amino acid sequence of the repeat region. However, with a few notable exceptions, such as the approximately 15% identity in the C terminus, and a few tenuous identities in the N terminus, the nonglycine residues of T. erythraeum within the triglycine region appear to share little identity with those of the vertebrates. Why, then, should T. erythraeum maintain such a long, interrupted, intron-free collagen-like sequence in its genome? This may be partially explained as follows: the fibrillar and afibrillar collagens have diverged to a much greater extent within individual species than, for example, human type I collagen and trout type I collagen have. Thus it is not merely the presence of a perfectly uninterrupted triglycine repeat region, but also the preservation of other critical residues such as prolines and lysines that make triple-helix formation and fibrillogenesis possible. This is reasonable since greater selective pressure is placed on organisms that rely on perfect triglycine repeats and their associated cross-bridge-forming lysines and rotationally stiff prolines to maintain fibril-forming collagens. Indeed, fibril formation confers a selective advantage: osteogenesis imperfecta. In an organism such as T. erythraeum that apparently does not form fibrils, but does apparently rely on its collagen-like protein for survival, the maintenance of the identity of its second and third residues is likely not critical, whereas the maintenance of its triglycine repeat appears to be especially valued.
Ecological
Based on prior satellite imagery and our own evidence of filamentous structures found in unlabeled atomic-force microscopy preparations, we have presented evidence suggesting that this filamentous type of collagen might be a mat-forming matrix similar to other afibrillar collagens. A gene does not long remain within a genome unless it is serving the purpose of increasing organism (and gene) survivability (Dawkins 1989). Any “excessive” genes that do not contribute to fitness are quickly eliminated from the genome (Pal et al. 2003). We suggest that the remarkably long collagen-like protein found in T. erythraeum confers a selective advantage to its host organism by enabling it to maintain large colonies that give it the selective advantage of concentrating its secreted toxin (Cox et al. 2005; Wolk 1973), thus potentially enriching its available food supply.
Early in life’s history, when there were no multicellular organisms, organic energy supplies were likely scarce and diffuse. In this environment, it would likely have been to a single-celled organism’s advantage to diffuse or actively move away from its neighbors to reduce competition for resources. However, if a large, swimming energy source were present, the ability to colonize might prove to be advantageous (Burchard 1981; Martin 2002). A single bacterium attempting to intoxicate and kill an organism in the ocean would have little chance of success. But if colonization were to be made possible by the inclusion of an extracellular matrix protein, and large volumes and high concentrations of toxin could be produced, a larger food source might be killed and used as a food source. This purported selective advantage to colonize in a community that lacks signaling and motility necessitates the need for a glue to hold the bacterial colony together. This extraordinarily long collagen-like triglycine sequence may have provided this early glue, effectively creating the “earth’s first fishing net.”
While other collagen fragments appear in bacteria such as Bacillus cereus, these are likely used to attach to host extracellular matrix rather than for colonization purposes. Indeed, no other marine species of prokaryote has been shown to colonize to the extensive degree that T. erythraeum does. This cooperative nature of a group of primitive cells may even provide clues as to the origins of multicellular primitive organisms such as the hydra and sponges.
Interestingly, the triglycine repeat motif of collagen shares similar characteristics with two diseases: fragile X syndrome (Fu et al. 1991) and Huntington’s disease (Andrew et al. 1993). Genes responsible for both of these diseases cause the repeat of either a single amino acid or a triad of amino acids. While the molecular machinery that enables the “gene stuttering” necessary for producing collagen, the glue of multicellular life, it may have the detrimental effect of allowing some disease states to persist.
Electronic supplementary material
Acknowledgments
We would like to thank Pia Rossi for her help with environmental scanning electron microscopy, The State of Pennsylvania Department of Health award “Nanotechnology Meets Neuroscience” 4100026196-240418, the Nanotechnology Institute of Philadelphia for provision of the AFM, the Keck Foundation, and the National Science Foundation (Grant BES-0216343) for provision of the ESEM. Equally beneficial were fruitful discussions with David Hulmes, Fredrick Silver, and Donald McEachron.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Bradley E. Layton, Email: bradley.e.layton@drexel.edu
Adam J. D’Souza, Email: ajd45@drexel.edu
William Dampier, Email: William.dampier@drexel.edu.
Adam Zeiger, Email: zig@drexel.edu.
Alia Sabur, Email: alia@drexel.edu.
Jesula Jean-Charles, Email: tua34589@temple.edu.
References
- Adams KL, Song K, Roessler PG, Nugent JM, Doyle JL, Doyle JJ, Palmer JD. Intracellular gene transfer in action: dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc Natl Acad Sci USA. 1999;96:13863–13868. doi: 10.1073/pnas.96.24.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nature Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Aouacheria A, Cluzel C, Lethias C, Gouy M, Garrone R, Exposito JY. Invertebrate data predict an early emergence of vertebrate fibrillar collagen clades and an anti-incest model. J Biol Chem. 2004;279:47711–47719. doi: 10.1074/jbc.M408950200. [DOI] [PubMed] [Google Scholar]
- Aouacheria A, Geourjon C, Aghajari N, Navratil V, Deleage G, Lethias C, Exposito JY. Insights into early extracellular matrix evolution: spongin short chain collagen-related proteins are homologous to basement membrane type IV collagens and form a novel family widely distributed in invertebrates. Mol Biol Evol. 2006;23:2288–2302. doi: 10.1093/molbev/msl100. [DOI] [PubMed] [Google Scholar]
- Bell PRF, Uwins PJR, Elmetri I, Phillips JA, Fu FX, Yago AJE. Laboratory culture studies of Trichodesmium isolated from the great Barrier Reef Lagoon, Australia. Hydrobiologia. 2005;532:9–21. doi: 10.1007/s10750-004-8768-1. [DOI] [Google Scholar]
- Boot-Handford RP, Tuckwell DS. Fibrillar collagen: the key to vertebrate evolution? A tale of molecular incest. Bioessays. 2003;25:142–151. doi: 10.1002/bies.10230. [DOI] [PubMed] [Google Scholar]
- Buehler MJ. Atomistic and continuum modeling of mechanical properties of collagen: elasticity, fracture, and self-assembly. J Mater Res. 2006;21:1947–1961. doi: 10.1557/jmr.2006.0236. [DOI] [Google Scholar]
- Buehler MJ. Nature designs tough collagen: explaining the nanostructure of collagen fibrils. Proc Natl Acad Sci USA. 2006;103:12285–12290. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard RP. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- Cai JJ, Smith DK, Xia X, Yuen K-y. MBEToolbox: a Matlab toolbox for sequence data analysis in molecular biology and evolution. BMC Bioinform. 2005;6:6. doi: 10.1186/1471-2105-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone DG, Zehr JP, Paerl HW, Bergman B, Carpenter EJ. Trichodesmium, a globally significant marine cyanobacterium. Science. 1997;276:1221–1229. doi: 10.1126/science.276.5316.1221. [DOI] [Google Scholar]
- Cox PA, Banack SA, Murch SJ, Rasmussen U, Tien G, Bidigare RR, Metcalf JS, Morrison LF, Codd GA, Bergman B. Diverse taxa of cyanobacteria produce beta-N-methylamino-L-alanine, a neurotoxic amino acid. Proc Natl Acad Sci USA. 2005;102:5074–5078. doi: 10.1073/pnas.0501526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubenspeck JM, Zeng HD, Chen P, Dong SL, Steichen CT, Krishna NR, Pritchard DG, Turnbough CL. Novel oligosaccharide side chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J Biol Chem. 2004;279:30945–30953. doi: 10.1074/jbc.M401613200. [DOI] [PubMed] [Google Scholar]
- Davis BK. Molecular evolution before the origin of species. Prog Biophys Mol Biol. 2002;79:77–133. doi: 10.1016/S0079-6107(02)00012-3. [DOI] [PubMed] [Google Scholar]
- Dawkins R. The selfish gene. Oxford, New York: Oxford University Press; 1989. [Google Scholar]
- Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J Biol Chem. 2002;277:4223–4231. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- Doi M, Edwards SF. The theory of polymer dynamics. Oxford: Oxford Science; 1986. [Google Scholar]
- Einbinder J, Schubert M. Binding of Mucopolysaccharides and dyes by collagen. J Biol Chem. 1951;188:335–341. [PubMed] [Google Scholar]
- EMBnet (2007) Available at: http://www.ch.embnet.org/software/BOX_form.html. Accessed September 7, 2007
- Endean R, Monks SA GJ, Llewellyn LE. Apparent relationships between toxins elaborated by the cyanobacterium Trichodesmium erythraeum and those present in the flesh of the narrow-barred Spanish mackerel Scomberomorus commersoni. Toxicon. 1993;31(9):1155–1165. doi: 10.1016/0041-0101(93)90131-2. [DOI] [PubMed] [Google Scholar]
- Fu YH, Kuhl DP, Pizzuti A, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Gaill F, Wiedemann H, Mann K, Kuhn K, Timpl R, Engel J. Molecular characterization of cuticle and interstitial collagens from worms collected at deep sea hydrothermal vents. J Mol Biol. 1991;221:209–223. doi: 10.1016/0022-2836(91)80215-G. [DOI] [PubMed] [Google Scholar]
- Galilei G (1638) Discorsi e dimostrazioni matematiche, intorno à due nuoue scienze, attenenti alla mecanica & i movimenti locali... Con una appendice del centro di grauità d’alcuni solidi. Appresso gli Elsevirii, Leida
- Galilei G. Dialogues concerning two new sciences. Buffalo, NY: Prometheus Books; 1991. [Google Scholar]
- Gao M, Wilmanns M, Schulten K. Steered molecular dynamics studies of titin I1 domain unfolding. Biophys J. 2002;83:3435–3445. doi: 10.1016/S0006-3495(02)75343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gere JM. Mechanics of materials. Belmont, CA: Thomson/Brooks/Cole; 2004. [Google Scholar]
- Gish W (2006) Available at: http://www.blast.wustl.edu. Accesed January 2006
- Gogarten JP. Gene transfer: gene swapping craze reaches eukaryotes. Curr Biol. 2003;13:R53–R54. doi: 10.1016/S0960-9822(02)01426-4. [DOI] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulmes DJ. Building collagen molecules, fibrils, and suprafibrillar structures. J Struct Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- Huxley J. Problems of relative growth. New York: L. MacVeagh. Dial Press; 1932. [Google Scholar]
- Huxley J. Problems of relative growth. Baltimore, MD: Johns Hopkins University Press; 1993. [Google Scholar]
- Jenkins C, Samudrala R, Anderson I, Hedlund BP, Petroni G, Michailova N, Pinel N, Overbeek R, Rosati G, Staley JT. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci USA. 2002;99:17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T, Cantor C. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–123. [Google Scholar]
- Kadler KE, Baldock C, Bella J, Boot-Handford RP. Collagens at a glance. J Cell Sci. 2007;120:1955–1958. doi: 10.1242/jcs.03453. [DOI] [PubMed] [Google Scholar]
- Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Scie USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liden A, van Wieringen T, Lannergard J, Kassner A, Heinegard D, Reed RK, Guss B, Rubin K. A secreted collagen- and fibronectin-binding streptococcal protein modulates cell-mediated collagen gel contraction and interstitial fluid pressure. J Biol Chem. 2008;283:1234–1242. doi: 10.1074/jbc.M704827200. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Yang J, Meng Q. Four inteins and three group II introns encoded in a bacterial ribonucleotide reductase gene. J Biol Chem. 2003;278:46826–46831. doi: 10.1074/jbc.M309575200. [DOI] [PubMed] [Google Scholar]
- Manohar A, Batzoglou S (2005) TreeRefiner: a tool for refining a multiple alignment on a phylogenetic tree. Proc IEEE Comput Syst Bioinform Conf, pp 111–119 [DOI] [PubMed]
- Martin MO. Predatory prokaryotes: an emerging research opportunity. J Mol Microbiol Biotechnol. 2002;4:467–477. [PubMed] [Google Scholar]
- Martin RD. Relative brain size and basal metabolic rate in terrestrial vertebrates. Nature. 1981;293:57–60. doi: 10.1038/293057a0. [DOI] [PubMed] [Google Scholar]
- Morvan-Dubois G, Le Guellec D, Garrone R, Zylberberg L, Bonnaud L. Phylogenetic analysis of vertebrate fibrillar collagen locates the position of zebrafish alpha 3(I) and suggests an evolutionary link between collagen alpha chains and hox clusters. J Mol Evol. 2003;57:501–514. doi: 10.1007/s00239-003-2502-x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nature Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- Negri A (2006) Available at: http://www.aims.gov.au/pages/research/trichodesmium/tricho-01.html
- Nimni ME, Olsen BrR, Kang AH. Collagen. Boca Raton, FL: CRC Press; 1988. [Google Scholar]
- Orcutt KM, Rasmussen U, Webb EA, Waterbury JB, Gundersen K, Bergman B. Characterization of Trichodesmium spp. by genetic techniques. Appl Environ Microbiol. 2002;68:2236–2245. doi: 10.1128/AEM.68.5.2236-2245.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD. Genomic function: rate of evolution and gene dispensability. Nature. 2003;421:496–497. doi: 10.1038/421496b. [DOI] [PubMed] [Google Scholar]
- Pampaloni F, Lattanzi G, Jonas A, Surrey T, Frey E, Florin EL. Thermal fluctuations of grafted microtubules provide evidence of a length-dependent persistence length. Proc Natl Acad Sci USA. 2006;103:10248–10253. doi: 10.1073/pnas.0603931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson CL, Mower JP, Qiu YL, Shirk AJ, Song K, Young ND, DePamphilis CW, Palmer JD. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol. 2005;5:73. doi: 10.1186/1471-2148-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steager EB, Patel JA, Kim C-B, Yi DK, Lee W, Kim MJ (2007) A novel method of microfabrication and manipulation of precise bacterial teamsters in low Reynolds number fluidic environments. Microfluid Nanofluid (published online)
- Sulchek T, Friddle RW, Noy A. Strength of multiple parallel biological bonds. Biophys J. 2006;90:4686–4691. doi: 10.1529/biophysj.105.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YL, Luo ZP, Fertala A, An KN. Direct quantification of the flexibility of type I collagen monomer. Biochem Biophys Res Commun. 2002;295:382–386. doi: 10.1016/S0006-291X(02)00685-X. [DOI] [PubMed] [Google Scholar]
- Sun YL, Luo ZP, Fertala A, An KN. Stretching type II collagen with optical tweezers. J Biomech. 2004;37:1665–1669. doi: 10.1016/j.jbiomech.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Tang JX, Kas JA, Shah JV, Janmey PA. Counterion-induced actin ring formation. Eur Biophys J. 2001;30:477–484. doi: 10.1007/s002490100178. [DOI] [PubMed] [Google Scholar]
- van der Rest M, Garrone R. Collagen family of proteins. FASEB J. 1991;5:2814–2823. [PubMed] [Google Scholar]
- van Hulten MC, Witteveldt J, Peters S, Kloosterboer N, Tarchini R, Fiers M, Sandbrink H, Lankhorst RK, Vlak JM. The white spot syndrome virus DNA genome sequence. Virology. 2001;286:7–22. doi: 10.1006/viro.2001.1002. [DOI] [PubMed] [Google Scholar]
- Wada H, Okuyama M, Satoh N, Zhang S. Molecular evolution of fibrillar collagen in chordates, with implications for the evolution of vertebrate skeletons and chordate phylogeny. Evol Dev. 2006;8:370–377. doi: 10.1111/j.1525-142X.2006.00109.x. [DOI] [PubMed] [Google Scholar]
- Walsby AE. Gas vesicles. Microbiol Rev. 1994;58:94–144. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG. Ecology of prokaryotic viruses. Fems Microbiol Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Weitsman Y, Beltzer AI. An eccentric shear-lag model and implications on the strength of fibrous composites. Int J Solids Struct. 1992;29:1417–1431. doi: 10.1016/0020-7683(92)90087-A. [DOI] [Google Scholar]
- Wiersma AC, Millon LV, Hestand MS, Van Oost BA, Bannasch DL. Canine COL4A3 and COL4A4: sequencing, mapping and genomic organization. DNA Seq. 2005;16:241–251. doi: 10.1080/10425170500136822. [DOI] [PubMed] [Google Scholar]
- Wilhelm J, Frey E. Radial distribution function of semiflexible polymers. Phys Rev Lett. 1996;77:2581–2584. doi: 10.1103/PhysRevLett.77.2581. [DOI] [PubMed] [Google Scholar]
- Wolk CP. Physiology and cytological chemistry blue-green algae. Bacteriol Rev. 1973;37:32–101. doi: 10.1128/br.37.1.32-101.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, He J, Lin X, Li Q, Pan D, Zhang X, Xu X. Complete genome sequence of the shrimp white spot bacilliform virus. J Virol. 2001;75:11811–11820. doi: 10.1128/JVI.75.23.11811-11820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.