Abstract
FGFR3 is frequently activated by mutation in urothelial carcinoma (UC) and represents a potential target for therapy. In multiple myeloma, both over-expression and mutation of FGFR3 contribute to tumour development. To define the population of UC patients who may benefit from FGFR-targeted therapy, we assessed both mutation and receptor over-expression in primary UCs from a population of new patients. Manual or laser capture microdissection was used to isolate pure tumour cell populations. Where present, non-invasive and invasive components in the same section were microdissected. A screen of the region of highest tumour stage in each sample yielded a mutation frequency of 42%. Mutations comprised 61 single and 5 double mutations, all in hotspot codons previously identified in UC. There was a significant association of mutation with low tumour grade and stage. Subsequently, non-invasive areas from the 43 tumours with both non-invasive and invasive components were analysed separately. Eighteen of these had mutation in at least one region, including 9 with mutation in all regions examined, 8 with mutation in only the non-invasive component and one with different mutations in different regions. Of the 8 with mutation in only the non-invasive component, 6 were predicted to represent a single tumour and 2 showed morphological dissimilarity of fragments within the block, indicating possible presence of distinct tumour clones. Immunohistochemistry showed over-expression of FGFR3 protein in many tumours compared to normal bladder and ureteric controls. Increased expression was associated with mutation (85% of mutant tumours showed high-level expression). Overall, 42% of tumours with no detectable mutation showed over-expression including many muscle invasive tumours. This may represent a non-mutant subset of tumours in which FGFR3 signalling contributes to the transformed phenotype and which may benefit from FGFR-targeted therapies.
Keywords: FGFR3, bladder cancer, mutation, over-expression, therapeutic target
Introduction
Urothelial carcinomas (UC) comprise 2 major groups with distinct clinical and molecular features that are predicted to arise via different pathogenic pathways [1, 2]. At presentation 70-80% of UC are superficial, exophytic, papillary tumours that are well-differentiated (low grade) and do not penetrate the epithelial basement membrane. Such tumours are described as stage pTa. Tumours that have penetrated the basement membrane but not invaded muscle are stage pT1 and muscle invasive tumours, stage pT2 [3]. These invasive UC are believed to arise via a distinct pathogenetic pathway from low grade non-invasive UC [1]. An important recent finding is of activating mutations of FGF receptor 3 (FGFR3) in a significant proportion of UCs [4-6]. FGFR3 mutations are significantly associated with bladder tumours of low grade and stage [4, 7, 8], which have a more favourable disease course [9]. Mutation of FGFR3 is also found in some benign skin lesions in humans [10-12] and transgenic mice expressing mutant receptor from the cytokeratin 5 promoter show significant skin hyperplasia [12]. Thus FGFR3 mutation is associated with benign overgrowth of epithelial cells and tumours of low risk.
The only other tumour type in which significant involvement of FGFR3 has been reported is multiple myeloma in which 25% of cases have the translocation t(4;14)(p16;q32) that translocates FGFR3 on chromosome 4 into the IgH locus on chromosome 14 resulting in overexpression of wildtype FGFR3 from the derivative 14. FGFR3 mutations in the fusion protein are infrequent but have been associated with tumour progression [13]. This suggests that in this cell type both over-expression and mutational activation of the receptor can contribute to tumour development [13].
In bladder cancer as in multiple myeloma, increased expression of wildtype FGFR3 protein may be an alternative mechanism by which FGFR3 could contribute to tumour development. Although there is much information on the spectrum and frequency of FGFR3 mutation in bladder cancer, receptor expression has not yet been examined in detail. In the 3 studies of FGFR3 protein expression to date, conflicting results have been reported. Matsumoto et al., [14] reported expression of moderate or high levels of protein in 49% of tumours but found no relationship to tumour grade or stage. In 2 other studies a relationship between higher expression and lower tumour grade and stage was found [15, 16]. None of these studies were population based. Recently semi-quantitative PCR was used to examine RNA levels in a panel of tumours of known mutation status [17], demonstrating an association of higher levels of RNA expression and mutation status. To date the relationship between FGFR3 protein expression and mutation status has not been examined and it is not known whether over-expression of the wildtype receptor occurs in some cases.
Activated receptor tyrosine kinases represent potential therapeutic targets. Already several approaches to targeting FGFR3 have been examined in multiple myeloma and promising responses have been obtained with small molecule inhibitors and antibodies [18-22]. In these cancers, it has been demonstrated that several of the agents tested, directly affect the function of both over-expressed wildtype and mutant FGFR3. In bladder cancer, inhibition of the S375C or S249C mutant forms of the receptor inhibit bladder tumour cell proliferation in vitro, confirming that mutant FGFR3 is a good therapeutic target [17, 23]. As targeted therapies are developed, it will be important to define those patients most likely to benefit. This may include not only the large number of patients in which FGFR3 mutation is common but as in multiple myeloma this may extend to others in whose tumours wildtype receptor is overexpressed.
To determine the true frequency and distribution of mutation and protein overexpression, we have examined FGFR3 mutation status and protein expression levels and their relationship to a range of clinico-pathological parameters in bladder tumour samples from a population of new patients. We used both macro- and microdissection of tumour sections to examine mutation status of regions of the tissue samples with different stages. Our findings provide strong evidence for roles of both mutation and protein over-expression of FGFR3 in bladder cancer and provide novel insights into the genetic diversity of tumour tissues co-existing within the same patient.
Materials and Methods
Patients and Tumour samples
Approval was obtained from the Leeds East Research Ethics Committee to search the computer records of the pathology department of the Leeds Teaching Hospitals NHS Trust for all new patients presenting with a bladder tumour in Leeds in 1998. To ensure that all patients were identified, search parameters were initially broad (all bladder tumours). The resulting list was examined manually to exclude referred patients and cross-referenced with data from previous years to exclude those with a prior history of bladder tumour. This was further confirmed by examination of the clinical records of the remaining patients. We thereby identified 172 patients presenting with a bladder tumour for the first time in 1998. Slides of the tumours were reviewed in all cases by a single consultant pathologist (PH) for consistent grading and staging [3, 24] and to select a representative formalin-fixed paraffin-embedded tumour block. Every patient diagnosed during this period with available tumour sample was included to avoid bias. Fourteen patients were excluded from the study because of lack of tumour sample (10 patients) and extracted tumour DNA of insufficient quality for PCR (4 patients). These 14 comprised 2 pTa, 2 pT1, 4 pT2, 5 unstaged (pTx), 1 case with carcinoma in situ (CIS), 5 grade 2, 8 grade 3 tumours and 1 tumour too poorly preserved to grade. The patient and tumour characteristics of the 158 tumours from which sufficient DNA could be extracted are shown in Table 1. Clinical records were reviewed during 2004 and 2005.
Table 1.
Patient and tumour characteristics
| Variables | Cases | FGFR3 | P χ2 | |
|---|---|---|---|---|
| WT, n (%) | Mutant, n (%) | |||
| Total | ||||
| 158 | 92 (58) | 66 (42) | ||
| Age (y) | ||||
| Mean | 71.7 | 71.3 | 72.3 | |
| Sex | NS | |||
| Male | 102 | 61 (59.8) | 41 (40.2) | |
| Female | 56 | 31 (55.4) | 25 (44.6) | |
| Tumour number | NS | |||
| Solitary | 112 | 68 (60.7) | 44 (39.3) | |
| Multiple | 29 | 14 (48.3) | 15 (51.7) | |
| Missing | 17 | 10 (58.8) | 7 (41.2) | |
| Tumour size | NS | |||
| < 3 cm | 70 | 38 (54.3) | 32 (45.7) | |
| ≥ 3 cm | 54 | 31 (57.4) | 23 (42.5) | |
| Missing | 34 | 23 (67.6) | 11 (32.4) | |
| Smoking | NS | |||
| Current | 49 | 30 (61.2) | 19 (38.8) | |
| Ex | 28 | 17 (60.7) | 11 (39.3) | |
| Never | 57 | 32 (56.1) | 25 (43.9) | |
| Missing | 24 | 13 (54.2) | 11 (45.8) | |
| Stage | ≤0.001 | |||
| Ta | 71 | 24 (33.8) | 47 (66.2) | |
| T1 | 29 | 18 (62.1) | 11 (37.9) | |
| T2 or higher | 47 | 40 (85.1) | 7 (14.9) | |
| Tx | 8 | 7 (87.5) | 1 (12.5) | |
| CIS | 3 | 3 (100) | 0 (0) | |
| Grade | ≤0.001 | |||
| 1 | 13 | 5 (38.5) | 8 (61.5) | |
| 2 | 60 | 20 (33.3) | 40 (66.7) | |
| 3 | 85 | 67 (78.8) | 18 (21.2) | |
| Recurrence (12 month) (Ta, T1 only) | NS | |||
| Yes | 26 | 10 (38.5) | 16 (61.5) | |
| No | 60 | 27 (45) | 33 (55) | |
| Missing | 15 | 6 (40) | 9 (60) | |
DNA extraction and FGFR3 mutation analysis
A reference haematoxylin and eosin stained section from every tumour sample was assessed to identify regions of tumour free from contaminating stroma or immune infiltrate, and to identify tumour regions of different stages (PH). Regions containing more than 80% tumour cells were removed from the section by scraping using a scalpel blade. Samples that contained mixed areas of normal and tumour tissue were dissected using laser capture microdissection (Arcturus, Mountain View, CA). DNA was extracted using the QIAamp DNA mini kit (Qiagen, Sussex, UK), according to the manufacturer’s instructions. In samples with mixed stage, DNA extracted from the highest stage part of the tumour was initially screened for mutation and information from these areas was used to generate the overall mutation score for the cohort. Regions of lower stage from the same tumour block were subsequently analysed separately. Direct sequencing was used to screen exons 7, 10 and 15. Primers used for PCR amplification have been described previously [25]. 2 μl samples of extracted DNA were amplified in 50 μl reactions using AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA). For sequencing, unincorporated primers and deoxynucleotides were removed using shrimp alkaline phosphatase and exonuclease I (Amersham Biosciences, Chalfont, UK). Sequencing reactions were carried out using both the forward and reverse PCR primers with the BigDye Terminator V1.1 Cycle Sequencing Kit (Applied Biosystems). Data analysis was carried out using Sequencing Analysis 3.0 software (Applied Biosystems) and by visual inspection of the electropherograms.
Immunohistochemistry
For detection of FGFR3 protein, 5 μm deparaffinized and rehydrated sections were treated with 3% hydrogen peroxide (Sigma, Poole, UK), microwaved for 20 minutes, and blocked with an Avidin Biotin blocking kit (Vector Laboratories, Peterborough, UK). Primary antibody (FGFR3 B9, Santa Cruz, CA, USA) was applied for 1 hour at room temperature and detected with a biotinylated secondary antibody and 3,3-diaminobenzidine (DAB). Slides were counterstained with hematoxylin, dehydrated, and mounted. Antibody specificity was confirmed using sections from tumours with known high or low RNA expression levels of FGFR3 measured previously by real-time RT-PCR [26]. Immunostaining was assessed by two independent observers (PH, MK), who were blinded to mutational status and clinical outcomes. Differences were settled by consensus following review of individual cases. A semi-quantitative scoring system was adopted: 0, all tumour cells negative; 1, faint but detectable positivity in some or all cells; 2, weak but extensive positivity; 3, strong positivity (regardless of extent). In addition to negative controls, sections of ureter and a tumour with known high-level expression were included in each run to represent normal findings (score 1) and strong positivity (score 3) respectively. These acted as reference sections for the scoring of tumours.
Results
FGFR3 mutation is associated with tumour grade and stage
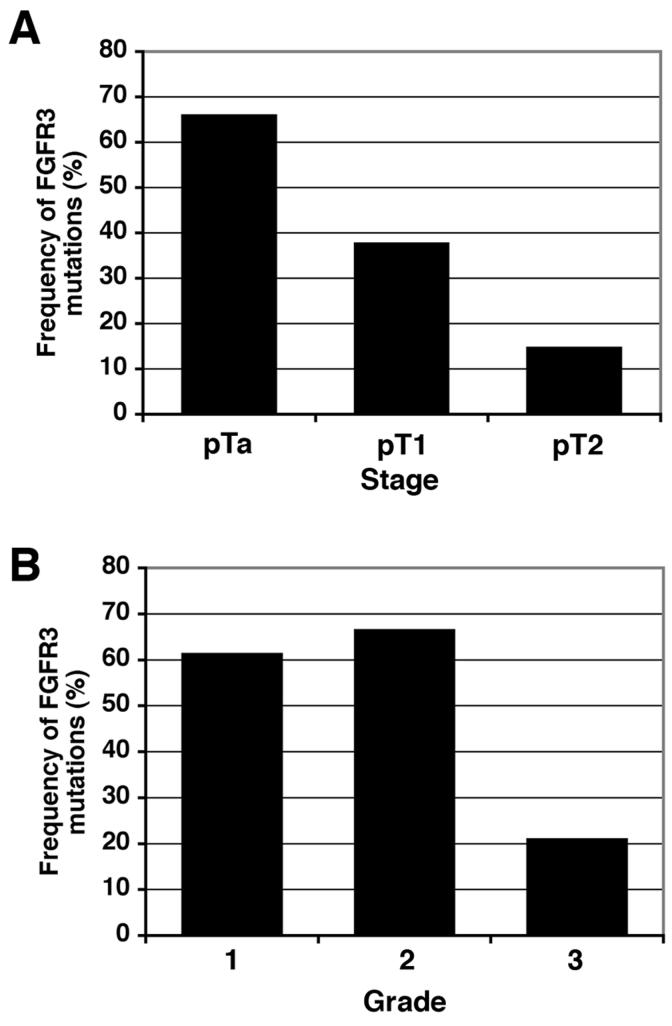
We used direct sequencing to screen 158 urothelial carcinoma (UC) samples for mutations in FGFR3 exons 7, 10 and 15 that contain all the mutations described in UC to date. Nine different mutations were found affecting eight codons 248, 249, 372, 373, 375, 382, 393, and 652 (Table 2). Seven of these mutations were in the extracellular domain (248 and 249) or the transmembrane domain (372, 373, 375, 382, and 393). The other two mutations were different base-pair substitutions of codon 652, located in the kinase domain. Initially, we examined the regions of highest stage in each specimen and detected 71 mutations in 66 tumours (42%) (Table 1). The most common mutations were: S249C (n=34; 47.9%), Y375C (n=16; 22.5%) and R248C (n=8; 11.3%). Five samples contained two mutations (1 × R248C+G382R; 1 × Y375C+K652E; 1 × G382R+K652Q; 2 × S249C+Y375C). The distribution of mutations with respect to diagnostic (highest) stage and grade is shown in Figure 1 and Table 1. FGFR3 mutation was significantly more prevalent in tumours of low tumour stage and grade (p<0.001, Pearson χ2 test). No significant association of mutation frequency was observed with sex, tumour multiplicity, tumour size, or smoking habit (Table 1).
Table 2.
FGFR3 mutations identified
| Exon | Nucleotide change | Amino acid change | Number of mutations | Frequency of mutation (%) |
|---|---|---|---|---|
| 7 | c.742C>T1 | R248C | 8 | 11.3 |
| 7 | c.746C>G | S249C | 34 | 47.9 |
| 10 | c.1114G>T | G372C | 5 | 7.1 |
| 10 | c.1117A>T | S373C | 1 | 1.4 |
| 10 | c.1124A>G | Y375C | 16 | 22.5 |
| 10 | c.1144G>A | G382R | 2 | 2.8 |
| 10 | c.1178C>A | A393E | 1 | 1.4 |
| 15 | c.1954A>C | K652E | 3 | 4.2 |
| 15 | c.1954A>G | K652Q | 1 | 1.4 |
nucleotide position according to the FGFR3b isoform numbering from the initiating ATG
Figure 1.
Distribution of FGFR3 mutations in UC tumours. A. FGFR3 mutation distribution according to tumour stage. B. FGFR3 mutation distribution according to histological grade.
Differences in mutation status within tumour tissues from the same patient
Forty-three of the 158 tumour samples contained tumour fragments or regions of more than one histological stage within the same tissue block. Only the mutation status of the tumour regions that represented the diagnostic stage (i.e. the highest stage present) was included in the data described above. To determine if regions of lower stage had the same mutation status as the highest stage, we screened all regions for mutation. Eighteen tumours (42%) had mutation in at least one part of the specimen. Nine had the same mutation in all regions of the tumour analysed, eight tumours had mutation only in the low stage region and one had different mutations in the low and high stage regions of the tumour. Laser capture microdissection of pure tumour cell populations confirmed that mutations in high stage regions were not masked by increased normal cell contamination. Three of these samples had two mutations. One (R248C+S249C) was present in a region of pT1 tumour in a sample where pTa contained S249C alone and pT2 contained S248C. The second (S249C+Y375C) was present in a pTa region of a sample where S249C alone was found in the pT1 region. The third showed S249C+S375C in pTa but only wildtype sequence in pT1 and pT2 regions in the same specimen.
Where possible we had selected separate tumour fragments within each block to represent tumour of different stages to facilitate clean dissection. Thus, the tumour regions of different stages analysed were not usually physically continuous. As all the resected tissues from a given patient were placed in the same container at cystoscopy regardless of the number of separate tumours seen, an individual tumour block could in theory contain fragments of different tumours. Of the eight samples with different mutation status in non-invasive and invasive components, 2 were reported as multifocal, 4 as single tumours and no information was recorded for 2 cases. We assessed whether there was continuity between the superficial and invasive components and whether tumour morphology was compatible with a single tumour entity or suggested the possibility of more than one tumour clone within the sample. Three tumours did not show continuity between stages anywhere within the block but all showed similar tumour morphology throughout, suggesting a single originating tumour. One of these was recorded as a single tumour at the time of surgery, one was multifocal and for one there was no information. An example (from single tumour) is shown in Figure 2 (A,B). The non-invasive component (A) contained a Y375C mutation but the invasive component (B) was wildtype. Two samples showed continuity of pTa and pT1/pT2 regions within the dissected fragment (one recorded as single tumour, one with no information) and one tumour (single tumour) showed continuity of different stages in part of the section, although not the dissected fragments. In all these cases, it is predicted that the individual dissected regions represent different regions of the same tumour or in the case of the multifocal tumour, possibly physically distinct tumours derived from the same tumour clone. Two samples showed morphological differences within the block. In the first, the pTa and pT2 regions analysed were in different tumour fragments, were dissimilar and no continuity of stages was observed anywhere in the section. This sample was derived from a patient with 2 distinct tumours resected. In the other tumour, the mutation-containing pTa region was physically continuous with pT1 within the section but there was morphological dissimilarity, raising the possibility that these were distinct, but physically merging tumours (Figure 2C). This sample contained S249C+Y375C in the pTa components, was wildtype in pT1 and pT2 and was recoded as a single tumour at the time of surgery.
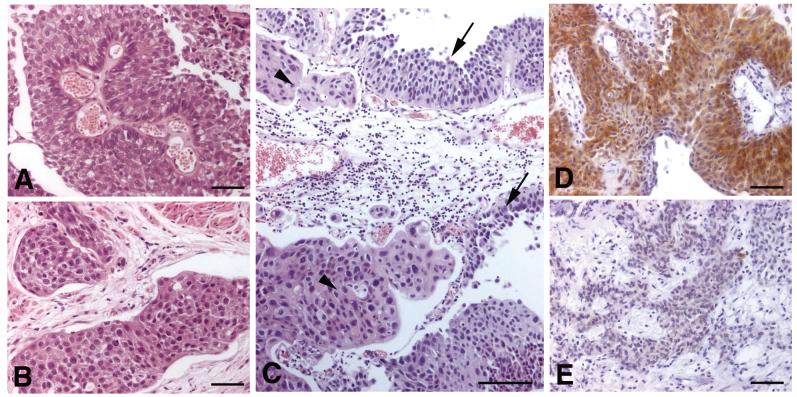
Figure 2.
Morphology and FGFR3 expression in tumour samples. A and B, Different, non-continuous regions from the same tumour section showing similar morphology. The non-invasive component (A) contained a Y375C mutation and the invasive component (B) was wildtype; C, Morphologically dissimilar regions of tumour that are physically merging. In this sample the low grade tumour (arrows) showed FGFR3 mutation (S249C+Y375C) and the high grade tumour (arrowheads) was wildtype; D and E, Distinct regions of the same sample showing different FGFR3 staining pattern. A, B, C, bars = 100μm; D, E, bars = 50μm.
FGFR3 protein expression is related to tumour stage, grade and mutation status
Next we examined FGFR3 protein expression by immunohistochemistry to determine if expression levels are related to tumour stage and grade. Immunohistochemistry was successfully carried out on 149 samples, including 8 which could not be staged (pTx). We were able to define 4 staining patterns (Figure 3A-D). Normal urothelial samples showed very faint but detectable staining in some or all cells (staining pattern 1)(Figure 3A). In contrast, some tumours were completely negative (staining pattern 0)(Figure 3B). Two further categories were defined based on weak but extensive positivity (pattern 2)(Figure 3C) or intense positivity in some or all cells (pattern 3)(Figure 3D). There was a high level of concordance between scorers using these 4 categories (80%). Of particular note, an increasingly greater proportion of tumours were negative (score 0) with advancing stage (9% of pTa, 19% of pT1 and 28% of pT2 tumours). This contrasted with the situation for normal (score 1) staining which was more evenly spread (21% of pTa, 23% of pT1 and 26% of pT2 tumours) and suggests that complete loss of expression has biological significance.
Figure 3.
Patterns of FGFR3 staining in normal urothelium and bladder tumours. Normal ureteric urothelium (A) and bladder tumour samples (B-D). A, staining pattern 1; B, staining pattern 0; C, staining pattern 2; D, staining pattern 3; bars = 100μm.
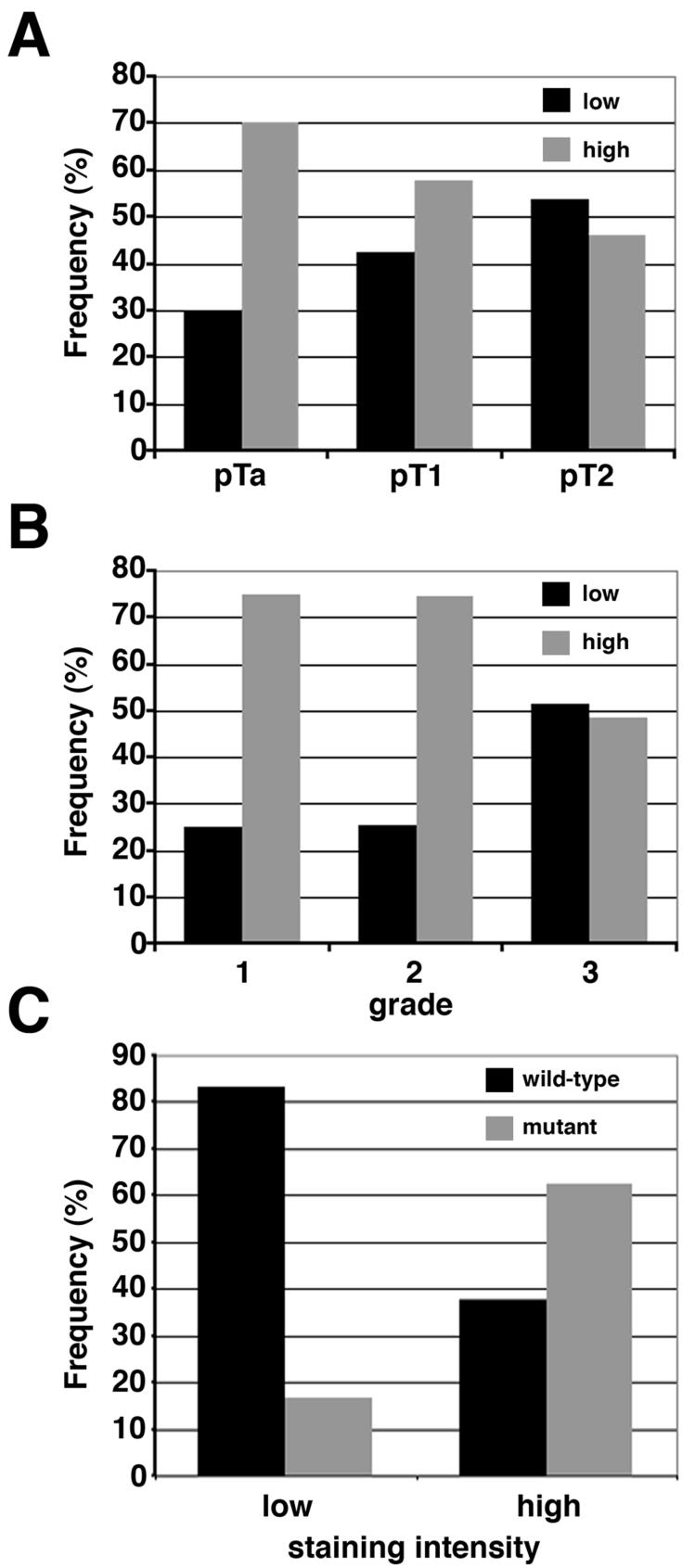
When samples were divided according to both staining intensity and mutation status there were small numbers of samples in some categories. Thus, for statistical analysis, reduced or normal expression patterns (0 and 1) were grouped into a “low” and increased expression patterns (2 and 3) into a “high” category. Concordance between scorers was then 90%. The distribution of FGFR3 expression with respect to stage is shown in Figure 4A and grade in Figure 4B. The expression of FGFR3 was significantly higher in non-invasive (pTa) compared to invasive (pT2) tumours (p<0.05; two sided, Fisher’s exact test) and was significantly associated with low grade (grades 1 and 2) compared with high grade (grade 3) (p<0.005).
Figure 4.
Distribution of FGFR3 staining intensity in UC tumours. The staining intensity of the FGFR3 antibody was classed as the same as or below normal (low) or above normal (high). A. FGFR3 staining intensity according to stage (black bars=low, grey bars=high). B. FGFR3 staining intensity according to grade (black bars=low, grey bars=high). C. Distribution of FGFR3 staining classification according to the presence (grey bar) or the absence (black bar) of FGFR3 mutation.
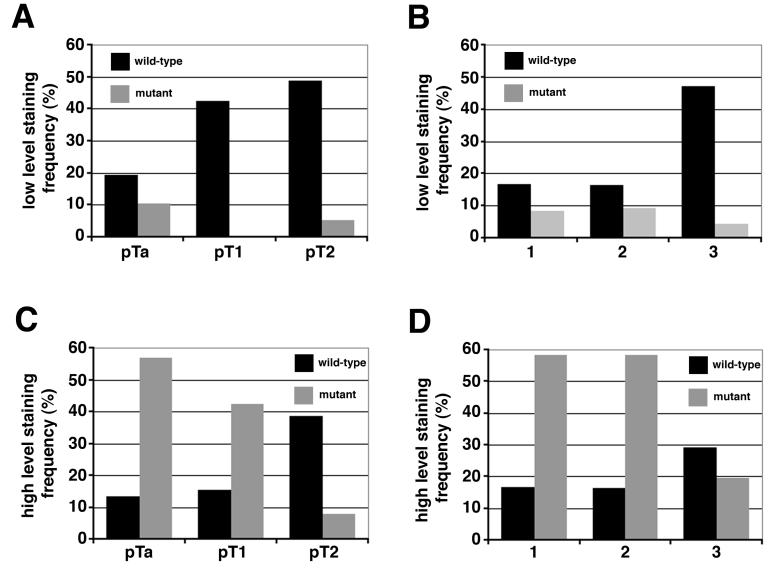
A highly significant association between expression level and mutation status was observed (p<0.0001), with high levels of FGFR3 predominantly expressed in tumours with FGFR3 mutation (Table 3, Figure 4C). Overall 85% of mutant tumours showed over-expression, most of which were low grade (74% of mutant high expressers were grade 1 or 2) or stage (73% non-invasive pTa)(Table 3, Figure 5). By contrast, of the 42% of wildtype tumours showing overexpression, 66% were high grade and 68% were invasive (pT1 or pT2). Thus, although only 25% of invasive tumours harboured a mutation, an additional 29% showed over-expression of wildtype receptor (Figure 5C and D). Overall therefore, 54% of these invasive tumours showed either mutation or over-expression of FGFR3 compared with 81% of non-invasive tumours, of which the majority (67%) harboured a mutation. No significant association was found between FGFR3 expression and age, sex, smoking habit, tumour multiplicity, size or recurrence.
Table 3.
Association of FGFR3 expression levels and presence of mutation
| GRADE/STAGE1 | Low FGFR3 expression | High FGFR3 expression | Total | P value (Fisher’s exact test) | ||
|---|---|---|---|---|---|---|
| wildtype | mutant | wildtype | mutant | |||
| 1 | 2 (17%) | 1 (8.3%) | 2 (16.7%) | 7 (58.3%) | 12 | 0.2364 |
| 2 | 9 (16%) | 5 (9.0%) | 9 (29.3%) | 32 (58.2%) | 55 | 0.007 |
| 3 | 34 (47.4%) | 3 (4.2%) | 21 (29.2%) | 14 (19.4%) | 72 | 0.0019 |
| Ta | 13 (19.4%) | 7 (10.5%) | 9 (13.4%) | 38 (56.7%) | 67 | <0.0001 |
| T1 | 11 (42.3%) | 0 | 4 (15.4%) | 11 (42.3%) | 26 | <0.0001 |
| T2 | 19(48.7%) | 2 (5.1%) | 15 (38.5%) | 3 (7.7%) | 39 | 0.0012 |
Tumours in which staining was scored differently in different regions of the section (n=10 including one pTx sample) are not included
Figure 5.
Comparison of FGFR3 staining intensity with mutation status. Tumours with a low (A and B) or high (C and D) level of FGFR3 staining were compared with stage (A and C) and grade (B and D). Frequency represents the percentage of tumours in a particular stage or grade. Black bars represent FGFR3 wildtype tumours and the grey bars represent FGFR3 mutant tumours.
Interestingly, 10 tumours showed regions of both high and low FGFR3 expression. The regions of different staining intensity were continuous in 8 of the 10 tumours. A decrease in FGFR3 staining intensity between the low and high stage regions was shown in seven tumours. Two of these were tumours with different mutation status in regions of different stage (Figure 2 D, E). In each case lower staining was recorded in the higher stage region, which lacked mutation.
Discussion
We have provided data for the first time on the frequency and type of FGFR3 mutations in an unselected population of patients presenting with bladder cancer. In addition, we used macro- and micro-dissected regions of individual tumour samples, thereby providing information on the relationship of FGFR3 mutation status to different stages of tumour development and within complex samples from individual patients. This is also the first study to examine the relationship between FGFR3 mutation status and FGFR3 protein expression.
Our results confirm that FGFR3 mutations are found predominantly in urothelial tumours of low stage and grade. We detected a frequency of FGFR3 mutations in low stage UC that was intermediate (58%) to those reported in studies performed in the Netherlands (67%) [27] and in Spain (50%) [9]. A difference between our study and previous reports is the number of Y375C mutations detected. This difference may be attributable to the method used to detect mutations. Our study and two previous studies [9, 28] used direct sequencing to identify FGFR3 mutations and showed a higher proportion of Y375C mutations (23%, 24% and 27%) compared to studies using initial mutation scanning by single strand conformation polymorphism (SSCP) analysis followed by sequencing [4-6, 8, 27, 29-31]. In total these studies using SSCP identified 455 mutations of which only 52 (11%) were Y375C. A recent study also raised the issue of the differential ability of SSCP and SnaPshot analysis to detect FGFR3 mutations [32]. SnaPshot proved more sensitive than SSCP, detecting almost double the number of Y375C mutations. We conclude that SSCP is an inadequate method for detection of Y375C mutations.
A strength of the present study is that we macro-dissected every tissue section that contained more than one pathological stage. We assigned each sample an overall diagnostic stage and grade that reflected the highest stage and grade present in the tissue block and designated overall mutation status according to what was found in this component of the sample. This avoided potential over-estimation of mutation frequency in patients of high diagnostic stage if mutation was present only in the low stage component of the sample. Overall, 43 tumour blocks contained more than one pathological stage and we subsequently determined the mutation status of each component. Eight tumours showed mutation only in a low stage region but not in higher stage regions of the sample and we confirmed these results by microdissection of invasive regions to ensure minimal normal tissue contamination. Had we pooled all tumour regions for analysis, the number of mutations in patients with a high stage diagnosis could have effectively doubled. As it can be predicted that subsequent clinical outcome is likely to depend on the behaviour of the most aggressive lesion, such skewing of results could lead to erroneous conclusions regarding the predictive or prognostic influence of FGFR3 mutation in high stage disease. Six of the eight tumours that contained mutations in the low but not the high stage regions were predicted to represent single tumours based on contiguity of regions of different stage and/or morphological similarity of tumour throughout the sample. This suggests either that the allele containing the FGFR3 mutation was lost in the high stage region or that a small subset of FGFR3 wildtype cells present in the low stage tumour had progressed. Two samples that may have contained more than one tumour clone were identified. In one of these, mixing of morphologically dissimilar elements was observed, the invasive component lacking FGFR3 mutation. A further interesting finding was that in one tumour the non-invasive region was homozygous for S249C while the muscle invasive (pT2) region was homozygous for the R248C mutation and the early invasive (pT1) region contained both mutations. Again this may suggest that there were distinct tumour clones present. Unfortunately insufficient DNA remained for studies of LOH or clonality in these samples but future studies should include these.
We have demonstrated that overexpression of FGFR3 protein is common in UC and that increased expression is more frequent in tumours of low stage. Three previous studies have examined FGFR3 protein expression in UC by immunohistochemistry [14-16]. Although all three studies have observed increased expression, there was some inconsistency in the percentage of tumours of different stages and grades that overexpressed FGFR3. Gomez-Roman et al reported similar results to our study with increased FGFR3 protein expression in 69.5% pTa, 72 % pT1 and 49 % of pT2 tumours [15]. Mhawech-Faucegli et al only examined superficial UC and showed increased protein expression in 76.6% pTa and 46.9% pT1 tumours [16]. Matsumoto et al found no relationship between FGFR3 expression and tumour stage and grade [14]. One study previously used semi-quantitative RT-PCR and identified a correlation between FGFR3 mRNA expression and mutation but the relationship to tumour stage and grade was not reported [17]. Our study now shows a strong correlation between protein expression and mutation of FGFR3 and its relationship to tumour grade and stage. The mechanism responsible for this upregulation of expression is not known. However it is unlikely to result from DNA amplification of the FGFR3 locus as 4p16.3 amplification has not been found in bladder tumours.
Interestingly, some tumours were found to express high levels of wildtype FGFR3 protein, suggesting that as in multiple myeloma, both mutant and wildtype FGFR3 can contribute to tumorigenesis. Activation of wildtype FGFR3 may occur via ligand independent dimerisation of the over-expressed protein, increased expression of ligand or via differential splicing that generates a splice variant such as FGFR3c with altered ligand specificity. Increased expression of FGFs is common in bladder cancer (reviewed in [33]) and we have shown that alternative splicing of FGFR3 that results in decreased specificity of ligand binding occurs in bladder cancer cell lines [34].
Many of the tumours we identified that overexpressed wildtype FGFR3 were high stage and grade tumours. As these tumours may develop via a different pathway from non-invasive tumours [2, 35], it is possible that the different mechanisms of activation of FGFR3 reflect either different downstream signalling consequences generated by mutant and wildtype receptor or different molecular mechanisms by which alterations occur in these two tumour groups.
If both mutation and over-expression of this receptor contribute to disease pathogenesis, our current findings implicate FGFR3 in 81% of pTa and 51% of pT2 tumours. It has been shown that in bladder cancer cell lines expressing high levels of wildtype FGFR3, targeted inhibition by antibodies results in reduced proliferation [15, 36] and small molecule inhibitors have been shown to inhibit myeloma cell lines with FGFR3 mutation [21]. We conclude that both wildtype and mutant FGFR3 may provide a valid therapeutic target in both non-invasive and invasive bladder cancers.
Acknowledgements
We are grateful to Filomena Esteves and Wendy Kennedy for carrying out immunohistochemistry and to Liz Sheldon for help with obtaining clinical information.
Grant support: This work was supported by a Programme grant from Cancer Research UK
Footnotes
All authors declare that they have no conflicts of interests.
References
- 1.Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27:361–73. doi: 10.1093/carcin/bgi310. [DOI] [PubMed] [Google Scholar]
- 2.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5:713–25. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 3.WHO Histological typing of urinary bladder tumours. International Histological Classification of Tumours. 1973:10. [Google Scholar]
- 4.Billerey C, Chopin D, Aubriot-Lorton MH, et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am J Pathol. 2001;158:1955–9. doi: 10.1016/S0002-9440(10)64665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cappellen D, De Oliveira C, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 6.Sibley K, Cuthbert-Heavens D, Knowles MA. Loss of heterozygosity at 4p16.3 and mutation of FGFR3 in transitional cell carcinoma. Oncogene. 2001;20:686–91. doi: 10.1038/sj.onc.1204110. [DOI] [PubMed] [Google Scholar]
- 7.Kimura T, Suzuki H, Ohashi T, Asano K, Kiyota H, Eto Y. The incidence of thanatophoric dysplasia mutations in FGFR3 gene is higher in low-grade or superficial bladder carcinomas. Cancer. 2001;92:2555–61. doi: 10.1002/1097-0142(20011115)92:10<2555::aid-cncr1607>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 8.van Rhijn BW, Lurkin I, Radvanyi F, Kirkels WJ, van der Kwast TH, Zwarthoff EC. The fibroblast growth factor receptor 3 (FGFR3) mutation is a strong indicator of superficial bladder cancer with low recurrence rate. Cancer Res. 2001;61:1265–8. [PubMed] [Google Scholar]
- 9.Hernandez S, Lopez-Knowles E, Lloreta J, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006;24:3664–71. doi: 10.1200/JCO.2005.05.1771. [DOI] [PubMed] [Google Scholar]
- 10.Hafner C, van Oers JM, Hartmann A, et al. High Frequency of FGFR3 Mutations in Adenoid Seborrheic Keratoses. J Invest Dermatol. 2006;126:2404–7. doi: 10.1038/sj.jid.5700422. [DOI] [PubMed] [Google Scholar]
- 11.Hafner C, van Oers JM, Vogt T, et al. Mosaicism of activating FGFR3 mutations in human skin causes epidermal nevi. J Clin Invest. 2006;116:2201–7. doi: 10.1172/JCI28163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logie A, Dunois-Larde C, Rosty C, et al. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum Mol Genet. 2005;14:1153. doi: 10.1093/hmg/ddi127. [DOI] [PubMed] [Google Scholar]
- 13.Chesi M, Brents LA, Ely SA, et al. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–36. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto M, Ohtsuki Y, Ochii K, et al. Fibroblast growth factor receptor 3 protein expression in urothelial carcinoma of the urinary bladder, exhibiting no association with low-grade and/or non-invasive lesions. Oncol Rep. 2004;12:967. [PubMed] [Google Scholar]
- 15.Gomez-Roman JJ, Saenz P, Molina M, et al. Fibroblast growth factor receptor 3 is overexpressed in urinary tract carcinomas and modulates the neoplastic cell growth. Clin Cancer Res. 2005;11:459. [PubMed] [Google Scholar]
- 16.Mhawech-Fauceglia P, Cheney RT, Fischer G, Beck A, Herrmann FR. FGFR3 and p53 protein expressions in patients with pTa and pT1 urothelial bladder cancer. Eur J Surg Oncol. 2006;32:231–7. doi: 10.1016/j.ejso.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Bernard-Pierrot I, Brams A, Dunois-Larde C, et al. Oncogenic properties of the mutated forms of fibroblast growth factor receptor 3b. Carcinogenesis. 2006;27:740–7. doi: 10.1093/carcin/bgi290. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Williams IR, Lee BH, et al. Constitutively activated FGFR3 mutants signal through PLC{gamma}-dependent and -independent pathways for hematopoietic transformation. Blood. 2005;106:328–37. doi: 10.1182/blood-2004-09-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grand EK, Chase AJ, Heath C, Rahemtulla A, Cross NC. Targeting FGFR3 in multiple myeloma: inhibition of t(4;14)-positive cells by SU5402 and PD173074. Leukemia. 2004;18:962–6. doi: 10.1038/sj.leu.2403347. [DOI] [PubMed] [Google Scholar]
- 20.Trudel S, Ely S, Farooqi Y, et al. Inhibition of fibroblast growth factor receptor 3 induces differentiation and apoptosis in t(4;14) myeloma. Blood. 2004;103:3521–8. doi: 10.1182/blood-2003-10-3650. [DOI] [PubMed] [Google Scholar]
- 21.Trudel S, Li ZH, Wei E, et al. CHIR-258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t(4;14) multiple myeloma. Blood. 2005;105:2941. doi: 10.1182/blood-2004-10-3913. [DOI] [PubMed] [Google Scholar]
- 22.Trudel S, Stewart AK, Rom E, et al. The inhibitory anti-FGFR3 antibody, PRO-001, is cytotoxic to t(4;14) multiple myeloma cells. Blood. 2006;107:4039–46. doi: 10.1182/blood-2005-10-4179. [DOI] [PubMed] [Google Scholar]
- 23.Tomlinson DC, Hurst CD, Knowles MA. Knockdown by shRNA indeitifies S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210399. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.UICC . TNM classification of malignant tumors, bladder. 3 ed Geneva: Union Internationale Contre le Cancer; 1978. pp. 113–7. [Google Scholar]
- 25.Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218–25. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 26.Tomlinson DC, Hurst CD, Knowles MA. Knockdown by shRNA identifies S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Rhijn BW, Vis AN, van der Kwast TH, et al. Molecular grading of urothelial cell carcinoma with fibroblast growth factor receptor 3 and MIB-1 is superior to pathologic grade for the prediction of clinical outcome. J Clin Oncol. 2003;21:1912–21. doi: 10.1200/JCO.2003.05.073. [DOI] [PubMed] [Google Scholar]
- 28.Zieger K, Dyrskjot L, Wiuf C, et al. Role of activating fibroblast growth factor receptor 3 mutations in the development of bladder tumors. Clin Cancer Res. 2005;11:7709–19. doi: 10.1158/1078-0432.CCR-05-1130. [DOI] [PubMed] [Google Scholar]
- 29.Rieger-Christ KM, Mourtzinos A, Lee PJ, et al. Identification of fibroblast growth factor receptor 3 mutations in urine sediment DNA samples complements cytology in bladder tumor detection. Cancer. 2003;98:737–44. doi: 10.1002/cncr.11536. [DOI] [PubMed] [Google Scholar]
- 30.van Rhijn BW, Montironi R, Zwarthoff EC, Jobsis AC, van der Kwast TH. Frequent FGFR3 mutations in urothelial papilloma. J Pathol. 2002;198:245–51. doi: 10.1002/path.1202. [DOI] [PubMed] [Google Scholar]
- 31.van Rhijn BW, Lurkin I, Chopin DK, et al. Combined microsatellite and FGFR3 mutation analysis enables a highly sensitive detection of urothelial cell carcinoma in voided urine. Clin Cancer Res. 2003;9:257–63. [PubMed] [Google Scholar]
- 32.van Oers JM, Lurkin I, van Exsel AJ, et al. A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clin Cancer Res. 2005;11:7743–8. doi: 10.1158/1078-0432.CCR-05-1045. [DOI] [PubMed] [Google Scholar]
- 33.Munro NP, Knowles MA. Fibroblast growth factors and their receptors in transitional cell carcinoma. J Urol. 2003;169:675–82. doi: 10.1097/01.ju.0000042721.63319.1d. [DOI] [PubMed] [Google Scholar]
- 34.Tomlinson DC, L’Hote CG, Kennedy W, Pitt E, Knowles MA. Alternative splicing of fibroblast growth factor receptor 3 produces a secreted isoform that inhibits fibroblast growth factor-induced proliferation and is repressed in urothelial carcinoma cell lines. Cancer Res. 2005;65:10441–9. doi: 10.1158/0008-5472.CAN-05-1718. [DOI] [PubMed] [Google Scholar]
- 35.Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27:361–73. doi: 10.1093/carcin/bgi310. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Torrecuadrada J, Cifuentes G, Lopez-Serra P, Saenz P, Martinez A, Casal JI. Targeting the extracellular domain of fibroblast growth factor receptor 3 with human single-chain fv antibodies inhibits bladder carcinoma cell line proliferation. Clin Cancer Res. 2005;11:6280. doi: 10.1158/1078-0432.CCR-05-0282. [DOI] [PubMed] [Google Scholar]