Abstract
Background: The hepatitis B virus x gene (HBx) is a promiscuous transactivator implicated in the development of hepatocellular carcinoma (HCC). The present study was designed to investigate the molecular events regulated by HBx. Methods: Genomic and proteomic expression profiling was performed in Huh7 HCC cells transfected with HBx mutants with a C-terminal deletion. The gene and protein expression of wingless-type murine-mammary-tumour virus (MMTV) integration site family, member 5A (Wnt-5a) was validated by analyses of reverse transcription–polymerase chain reaction (RT–PCR), real-time RT–PCR, western blot and immunohistochemistry. Results: Differentially expressed genes and proteins were found in the transfected Huh7 HCC cells; most of them were involved in transcriptional regulation, although others including oncogenes or tumor suppressor genes, and molecules involved in cell junctions, signal transduction pathways, metabolism or the immune response were also observed. The expression of the Wnt-5a gene was elevated >10-fold in Huh7 cells transfected with the HBx3′-30 amino acid deletion mutant. However, the expression was downregulated by the transfection with the HBx3′-40 amino acid deletion mutant. The changes in Wnt-5a expression were also observed in human HCC tissues, compared with corresponding non-cancerous liver tissues. A negative correlation was found between the expression of Wnt-5a and HBx COOH mutations in HCC tissues. Conclusions: HBx mutants may participate in the development and progression of HCC, at least in part through the Wnt-5a pathway.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers and a leading cause of cancer death in many countries, mainly in Asia and Africa. Epidemiological evidence indicates that chronic hepatitis B virus (HBV) infection is one of the main causes of HCC (1,2). In addition to chronic inflammation and cirrhosis in the liver, HBV is suspected to have a direct role in liver carcinogenesis (1,2).
The hepatitis B X protein antigen participates in the pathogenesis of HCC (3). For example, high levels of intrahepatic hepatitis B X protein antigen directly correlate with the stage of liver lesions (4). Hepatitis B X protein antigen also binds to and inactivates or downregulates tumor suppressors and senescence-related factors (3). The protein inhibits cell proliferation by inducing late G1 arrest (5) and induces apoptosis in both a p53-dependent and -independent manner (6,7). Hepatitis B virus x gene (HBx) has been shown to induce HCC in several transgenic mouse models (8–10) and to have weak oncogenic potential in several immortalized murine cell lines in vitro (11–14). HBx can inhibit cellular DNA repair capacity in a p53-independent manner (15). HBx also increases the expression of p21cip-1/waf1/mda6 and p27kip-1 in primary mouse hepatocytes, and overexpression of p21 and p27 can inhibit Cdk2 and Cdk4 activity, leading to cell cycle arrest in the G1 phase (16), and activates Wnt/β-catenin signaling in hepatoma cells (17). The gene expression profiles observed by complementary DNA (cDNA) microarray showed that HBx can lead to aberrant expression of molecules involved in signal transduction, transcriptional regulation, protein degradation, cell cycle control, apoptosis, metastasis, immune response and metabolism in mouse hepatocyte cells (18). So, the role of HBx gene in the hepatocarcinogenesis is still not understood and controversial.
We have shown previously that HBx can specifically bind the p53 protein and stimulates the transactivational activity of p53 and HBV replication (6). Lin et al. (19) reported that the HBx-binding sites are located within the oligomerization and specific DNA-binding domains of p53, and that the p53-binding site is confined in a small region (102–136 aa) of the HBx C-terminal. The pro-transactivation and the p53-inhibitory functions of HBx are mutually exclusive, indicating that HBx can influence the growth of hepatocytes through various pathways.
Several different deletions in the HBx gene C-terminal region have been found in HBV-infected HCC patients (20–22). Murine and human cell lines transfected with HBx mutant plasmids show increases in colony formation, cell cycle progression and ras and myc transforming capacity (5). These results suggest that the HBx mutants with C-terminal deletions may be implicated in the development and procession of HCC and that the C-terminal is vital for the biological functions of the HBx protein. In our previous studies, we constructed 10 HBx mutants, including four N-terminal, two middle and four C-terminal mutants, and analyzed the biological activities of the hepatoma cells with those truncated mutants of the HBx gene. The results showed that HBx3′-40 at C-terminal mutant promoted cellular proliferation, focus formation, tumorigenicity, invasive growth and metastasis by promoting the cell cycle progression from G0/G1 to S phase; in contrast, HBx3′-30 at C-terminal mutant repressed cell proliferation by arresting cells in G1 phase (22). These suggesting different forms of HBx share with the different functions and it may be explained why the role of the HBx gene is contradiction in the hepatocarcinogenesis.
In the present study, we analyzed the genomic and proteomic expression of HCC cells transfected with HBx with C-terminus mutations, in order to better define the effects of the HBx mutants in the development and procession of HCC associated with HBV infection.
Materials and methods
Cell line and culture
The Huh7 cell line (characterized by p53 mutation with A:T → G:C at codon 220) was cultured in Dulbecco’s minimum essential medium (Invitrogen Life Technologies, Grand Island, NY) with 10% heat-inactivated fetal calf serum, 100 000 units/l penicillin and 100 mg/l streptomycin at 37°C in a 5% CO2 incubator.
Plasmid constructs
In order to evaluate the biological activities of HBx and to determine its transactivational effects, a series of C-terminal truncation mutants were made, since the HBx transactivational function has been mapped to residues within these regions (23). To express HBx mutants in the mammalian cells, the DNA fragment precisely encompassing the full-length HBx or HBx mutants with deletions of 30 or 40 amino acids at the C-terminus before the first stop codon were cloned into the pcDNA3.0 vector (Invitrogen Life Technologies) using HindIII/XhoI and KpnI/EcoRI sites. The mutants used in this study were generated by a polymerase chain reaction (PCR) cloning method using a PTKHH2 plasmid (a head-to-tail dimer of wild-type HBV, adw subtype) as the template. The sequences of primers for the full length and two HBx mutants are shown in supplementary material 1 (available at Carcinogenesis Online). The PCR products were cloned into a pcDNA3.0 expression vector. Positive clones were sequenced to confirm their identity. The negative control plasmid was the vector, pcDNA3.
Stable transfection experiments
In order to stably transfect clones expressing wild and mutant HBx polypeptides, cells were plated into six-well plates at a density of 1 × 105 cells per well and transferred into serum-free medium for 24 h before transfection experiments. We used 1 μg of the pcDNA3.0 vector to carry the full length and two mutant HBx plasmids according to the manufacturer’s instructions. Empty vectors were used as negative controls. Standard calcium phosphate DNA coprecipitation was used to obtain stable transfectants. To transfect the pcDNA3.0 vectors into cells, the Lipofectamine™ 2000 Mammalian Transfection Kit was used following the manufacturer’s protocol (Invitrogen Life Technologies). At 24 h after transfection, the medium was replaced with 10% fetal bovine serum (Gibco-BRL, Grand Island, NY, USA) and 200 μg/ml G418 containing 1% penicillin–streptomycin (Invitrogen Life Technologies), and the cells were grown for an additional 2 weeks.
Detection of stable transfection
To identify the stably transfected clones expressing wild and mutant HBx polypeptides, the Neo gene was amplified from the genomic DNA of the transfected cells by PCR. The DNA from the transfected cells was extracted from 106 cells using the standard phenol–chloroform extraction and ethanol precipitation method. The following primers were used for amplification: forward primer 5′-ACAAGATGGATTGCACGCAGG-3′ and reverse primer 5′-TTCTCGGCAGGAGCAAGGTGAG-3′, resulting in a 327 bp expected product. PCR was performed using a Thermal Cycler 9600 (PerkinElmer, Foster City, CA) under the following conditions: after denaturation at 94°C for 5 min, DNA amplification was performed for 38 cycles at 94°C for 45 s, at 56°C for 45 s and at 72°C for 60 s, with a final extension at 72°C for 10 min and then was kept at 4°C. The PCR products were analyzed by electrophoresis on a 1% agarose gel stained with ethidium bromide and sequenced as described previously (22). At least two independent DNA extractions and PCRs were performed for each sample, and four sequencing reactions (two for each strand) were carried out to confirm sequencing data for each sample. The plasmid pHBXB1, containing the full-length HBx gene cloned into the pcDNA3 vector, was used as positive controls. For western blot analysis, cells cultured in 35 mm culture dishes were scraped and lysed. Immunoblots were probed with the corresponding primary antibodies (1:400) and developed by using the EnVision system (K139211, Dako, Carpinteria, CA) and the western blot detection kit (ECL Plus, Santa Cruz Biotechnology, Delaware Avenue Santa Cruz, CA) (24).
RNA isolation
Total RNA from the Huh7 cells transfected with full-length HBx or HBx mutants was isolated using the Trizol reagent, according to the manufacturer’s instructions (Invitrogen Life Technologies). The concentration of RNA was quantified by measuring the ultraviolet absorbence at 260 nm using a Pharmacia GeneQuant RNA/DNA calculator. The RNA was further purified on an affinity resin column (RNeasy; Qiagen, Chatsworth, CA) and converted to cDNA using the Superscript cDNA synthesis kit (Gibco-BRL, Gaithersburg, MD). Double-stranded cDNA was then purified by a phase lock gel (Eppendorf, Westbury, NY) with phenol–chloroform extraction.
cDNA microarray expression analysis
To determine the relative gene expression profiles in Huh7 cells transfected with HBx mutants, we analyzed the relative abundance of messenger RNA (mRNA) using high-density oligonucleotide microarrays containing probes for 21 074 transcripts (including 20 228 known human genes and 846 ESTs, human oligo array; Agilent, G4110B Agilent technologies Co. Ltd., shanghai, China) according to the protocols described previously (25,26). In brief, 2 μg of sample mRNA and 2 μg of reference mRNA were labeled with Cy3-NTP and Cy5-NTP (PerkinElmer), respectively, using reverse transcriptase (Invitrogen Life Technologies) for 2 h at 42°C. The two labeled cDNA probes were separated from unincorporated nucleotides by filtration, mixed and hybridized to the microarray at 60°C overnight. After hybridization, arrays were scanned with an Agilent microarray scanner. The image files were analyzed with the Agilent 2100 Bioanalyzer program (Agilent) to quantitate the relative amount of mRNA in the HBx mutant and full-length HBx-transfected Huh7 cells samples. All non-flagged array elements for which the fluorescent intensity in each channel was >1.5 times than the local background were considered well measured (27). Genes for which <75% of measurements across all the samples met this standard were excluded from further analysis. The Cy3:Cy5 normalized intensity ratio for each expressed sequence tag was averaged from duplicate spots in replicate experiments. Genes with expression levels that differed by at least 2-fold from the mean in at least one sample were selected for further evaluation.
Preparation of protein samples
Soluble protein fractions were extracted from the Huh7 cells transfected with full-length HBx or HBx mutants. The samples were lyophilized prior to the extraction, and the extraction was accomplished using the Klose method as described previously (28,29).
Two-dimensional electrophoresis
Two milligrams of the total protein was used for each electrophoresis experiment. Aliquots of proteins in sample buffer were applied onto immobilized pH 3–10 non-linear gradient strips (immobilized pH gradient strips; Bio-Cyte, Hercules, CA and Bio-Rad, Hercules, CA). Isoelectrofocusing was performed at 70 000 voltage hours. The second dimension was performed on 10–16% linear gradient polyacrylamide gels (17 cm × 17 cm × 1.5 mm) at 30 mA per gel constant current for ∼5 h, until the dye front reached the bottom of the gel. After protein fixation with a solution containing 40% methanol and 5% phosphoric acid for 1 h, the gels were stained with Coomassie blue G250 for 2 h. The gels were then destained with a solution containing 0.1% ethanol and 10% acetic acid, scanned in a Bio-Rad G710 densitometer and converted into electronic files, which were then analyzed with PDQUEST computer software (Bio-Rad, Hercules, CA).
Protein identification
For identification, several protein samples per spot of interest were excised from the gels stained with Coomassie brilliant blue. Peptide mass spectrometry fingerprinting after tryptic in-gel digestion was used as described previously (29). Identification of proteins was accomplished by searching the National Center for Biotechnology Information and/or Mascot databases with MS-Fit or PeptIdent, respectively. A mass deviation of 0.1 Da and two incomplete cleavages were allowed. Identifications were accepted when they covered at least 30% of the whole sequence and when comparable molecular mass/P values were obtained from the databases.
Validation of microarray data
To validate the microarray results and to further clarify a difference in the expression pattern for wingless-type murine-mammary-tumour virus (MMTV) integration site family, member 5A (Wnt-5a) in HBV-associated HCC, we carried out semiquantitative reverse transcription–polymerase chain reaction (RT–PCR), western blotting, immunohistochemistry and HBx sequencing. Independent tissue samples from 41 HCC patients and corresponding non-cancerous liver tissues were obtained during surgeries at the Changhai hospital between August 2003 and October 2004. All patients had HBsAg-positive serum. Histopathological diagnosis was made according to the World Health Organization histological classification of tumors of the liver and intrahepatic bile ducts (2000) (30).
RT–PCR and real-time RT–PCR
Total RNA was isolated, and cDNA was synthesized from frozen HCC tissues and corresponding non-cancerous liver tissues, and the RT–PCR and relative quantitation by real-time RT–PCR was performed using SYBR-green detection of PCR products in real-time with a Light Cycler (Roche Diagnostics, Mannheim, Germany) as described previously (31). The primer sequences for the gene were as follows: Wnt-5a, 5′-ACCACATGCAGTACATCGGAG-3′ and 5′-GAGGTGTTATCCACAGTGCTG-3′ (103 bp) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene used as an internal quantitative control: 5′-TCCACCACCCTGTTGCTGTA-3′ and 5′-ACCACAGTCCATGCCATCAC-3′ (452 bp). The cycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 amplification cycles of 95°C for 30s, 60°C for 30s and 72°C for 1.5 min. For quantification, the relative standard curves were created for each gene product. GAPDH was used for normalization of the concentration. Relative expression was calculated as a ratio of the expression in the tumor compared with that in the corresponding non-cancerous liver tissue.
Western blot analysis
Total protein was extracted from frozen tumor tissues and their corresponding non-cancerous liver tissues as described above. Protein samples were quantified by the Bradford assay and denatured at 100°C for 5 min in a protein sample buffer containing 1% sodium dodecyl sulfate and 1% dithiothreitol. Twenty microgram of total protein extracts were loaded in each lane and subjected to 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to Bio-Rad membranes. Specific antisera used to probe the membranes included mouse anti-Wnt-5a (AF645, 1: 500, R&D Systems Co. Ltd., Shanghai, China) and rabbit polyclonal anti-β-actin (Santa Cruz Biotechnology) antibodies. Subsequently, membranes were probed with the corresponding secondary antibodies and developed using the EnVision system (K139211, Dako) and visualized with enhanced chemiluminescence (Santa Cruz Biotechnology).
Immunohistochemistry
To evaluate and localize Wnt-5a in HCC tissues, immunohistochemical staining was performed on carcinomas and the corresponding non-cancerous liver tissue sections by using anti-Wnt-5a (1:50) with the EnVision Plus System (Dako). Negative controls were included by omitting the primary antibody, and a known positive control was included with each batch.
PCR and sequence analysis of HBx
To demonstrate the status of HBx mutations and their correlation with Wnt-5a expression in HCC tissues, genomic DNA was extracted from frozen HCC tissues and corresponding non-cancerous liver tissues, using the standard phenol–chloroform extraction and ethanol precipitation method. HBx-Alu PCR and sequencing were performed as described previously (22). At least two independent DNA extractions and PCRs were performed for each sample, and four sequencing reactions (two for each strand) were carried out to confirm the results for each sample. The plasmid pHBXB1, containing the full-length HBx gene cloned into the pcDNA3 vector, was used as positive control.
Statistical analysis
All data and statistical analyses were carried out by using a commercially available statistical software package (SPSS 11.0; SPSS, Chicago, IL). The differences between Wnt-5a expression in tumor and the corresponding non-tumorous liver tissues were statistically analyzed by the paired Student’s t-test for single comparisons and one-way analysis of variance for multiple comparisons. The relationship between Wnt-5a protein expression and clinicopathological features of HCCs was statistically analyzed by using the Pearson χ2 test. Spearman’s bivariate correlation was used to determine whether there is a positive or negative correlation between the HBx mutation, Wnt-5a RNA and protein expression levels. P < 0.05 was considered statistically significant.
Results
Detection of HBx deletion mutant plasmid
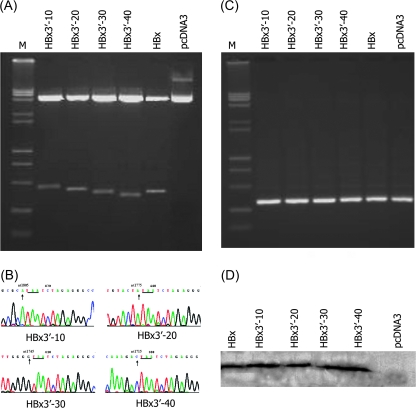
To investigate the effects on the gene expressions of different HBx mutants, the PCR products were cloned into a mammalian expression vector, we constructed four C-terminal mutants of hepatoma cells with truncated mutants of the HBx gene. The fragment size matched the predicted fragment by restriction digest (Figure 1a), suggesting that the correct plasmid construct was present. This was further confirmed by sequencing (Figure 1b), and the Neo gene was amplified (Figure 1c). The levels of HBx expression, as measured by western blotting, were similar for full-length HBx and the two truncated HBx (HBx3′-30 and HBx3′-40) expression vectors. The purified different bands had a molecular size of ∼17 kDa, corresponding with the size of the HBx protein (Figure 1d). The results of the biological activities of hepatoma cells with truncated mutants of the HBx gene showed that HBx3′-40 at C-terminal mutant promoted cellular proliferation, the focus formation and elevated tumorigenicity and invasive growth and metastasis by promote the cell cycle progression from G0/G1 to S phase; in contrast, HBx3′-30 at C-terminal mutant repressed cell proliferation by block in G1 phase, when compared with full-length HBx group (date not shown).
Fig. 1.
Detection of HBx deletion mutant plasmid. (A) The fragment size was matched the predicted fragment by restriction digest, suggesting that the correct plasmid construct was present. (B) The sequencing results have been confirmed theirs success constructed. (C) The Neo gene amplified by PCR. (D) The full-length HBx and the four truncated HBx expression vectors was expressed the hepatitis B X protein antigen by western blot, the purified bands had a molecular size of ∼17 kDa, corresponding with the size of the HBx protein.
Gene expression patterns of Huh7 cells transfected with HBx mutants
We further analyzed the effects of HBx mutants on the expression pattern of the transfected Huh7 cells using a cDNA microarray assay (as shown in supplementary material 2 available at Carcinogenesis Online). All differentially expressed genes are listed in supplementary material 3 (available at Carcinogenesis Online). The obvious differentially expressed (>5-fold increase) genes altered by HBx3′-30 and HBx3′-40 are partially summarized in Table I.
Table I.
Positive and negative transcriptional regulation by HBx mutants in hepatoma cells (>5-fold increase)
| Genes title | Genebank ID | Abbreviation | Cytoband | Ratio (Cy3:Cy5) | Description | |
| HBx3′-30 | HBx3′-40 | |||||
| Wingless-type MMTV integration site family, member 5A | L20861.1 | Wnt-5a | 3p21-p14 | 14.94301 | 0.565567 | Signal transduction |
| Chaperone, ABC1 activity of bc1 complex like (Schizosaccharomyces Pombe) | BC005171.1 | CABC1 | 1q42.13 | 9.977776 | 6.510925 | Metabolism |
| 1-acylglycerol-3-phosphate O-acyltransferase 3 | Q9BTD5 | AGPAT3 | 21q22.3 | 9.188781 | 0.944973 | Unknown function |
| Atonal homolog 1 (Drosophila) | NM_005172.1 | ATOH1 | 4q22 | 6.838085 | 8.93694 | Transcriptional regulation |
| Transcribed sequence with strong similarity to protein prf:810024G (Homo sapiens) 810024G URF 2 | NM_173709.1 | MTND2 | 21 | 5.528639 | 6.818047 | Metabolism |
| Histone 1, H2bn | BC011372.1 | HISTIH2BN | 6p22-p21.3 | 1.119792 | 9.715353 | Chromosome structure |
| Similar to hypothetical protein | AL834319.1 | LOC221091 | 11q12.3 | 1 | 10.87552 | Cytoskeletal structure and adhesion |
| Carboxypeptidase D | NM_001304.3 | 17p11.1-q11.2 | 1 | 10.61998 | Transcriptional regulation | |
| DnaJ (Hsp40) homolog, subfamily A, member 3 | NM_005147.1 | Hsp40 | 16p13.3 | 0.92649 | 4.567822 | Apoptosis |
| Netrin-G1 ligand | AB046800.1 | NGC-1 | 11p12 | 0.856526 | 4.541515 | Unknown function |
| Ets variant gene 5 (Ets-related molecule) | BC007333.1 | ETV5 | 3q28 | 0.853321 | 4.376849 | Oncogene |
| Phosphatidylinositol transfer protein, cytoplasmic 1 | AAF06148.1 | PITPN1 | 17q24.3 | 0.849904 | 9.248197 | Signal transduction |
| Fructosamine-3-kinase-related protein | BC007611.1 | FN3KRP | 17q25.3 | 0.652204 | 5.909704 | Metabolism |
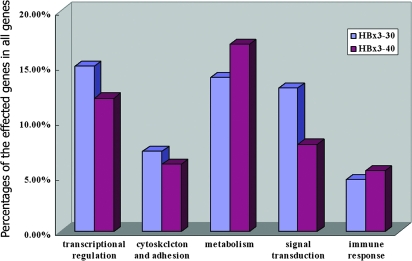
In HBx3′-30-transfected Huh7 cells, 193 of 20 228 genes (0.95%) showed altered expression compared with the same cells transfected with full-length HBx. Of the 154 elevated transcripts, 140 were increased 2- to 5-fold, 13 were increased 5- to 10-fold and the expression of Wnt-5a gene was elevated >10-fold. Of the 39 genes repressed by HBx3′-30, 38 were decreased 2- to 5-fold and 1 was decreased >5-fold. In HBx3′-40-transfected Huh7 cells, 165 of 20 228 genes (0.82%) showed altered expression compared with the same cells transfected with HBx. All differentially expressed genes are listed in supplementary material 3 (available at Carcinogenesis Online). Of the 143 elevated transcripts, 127 were increased 2- to 5-fold, 15 were increased 5- to 10-fold and the expression of 2 genes (carboxypeptidase D and one similar to a hypothetical protein) was elevated >10-fold. Of the 21 genes repressed by HBx3′-40, 18 were decreased 2- to 4-fold and 3 were decreased >4-fold. Of these differentially expressed genes, most are involved in transcriptional regulation, cytoskeletal structure and/or adhesion, signal transduction, metabolism, apoptosis or the immune response (Figure 2).
Fig. 2.
Bar graphs showing the profile of the differentially expressed genes in cells transfected with the HBx3′-30 and HBx3′-40 mutants. The differentially expressed genes are involved in transcriptional regulation, cytoskeletal integrity and adhesion, signal transduction, metabolism, apoptosis and the immune response.
Effects of HBx and its mutants on proteomic expression patterns of the transfected Huh7 cells
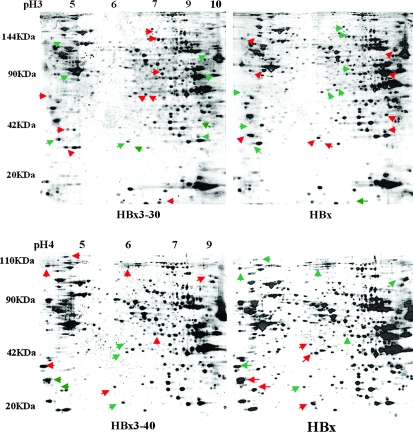
The silver-stained 2-D gels of the proteomes expressed in Huh7 cells transfected with HBx3′-30, HBx3′-40 and full-length HBx are presented in Figure 3. We detected 741 protein spots in gels of total protein prepared from the HBx3′-30-transfected Huh7 cells. Of these, 147 were found to have significantly different values in the HBx3′-30-transfected Huh7 cells compared with the cells transfected with full-length HBx. We were able to detect 549 protein spots in gels of total protein prepared from the HBx3′-40-transfected Huh7 cells. Of these, 135 were found to have significant differences in their expression compared with the cells transfected with full-length HBx. Of the differentially expressed proteins, most are involved in pathways associated with metabolism, transcriptional regulation, cell junctions and protein degradation. Selected differentially expressed proteins are listed in supplementary material 4 (available at Carcinogenesis Online). Differentially expressed proteins in hepatoma cells expressing HBx3′-30 or HBx3′-40 determined by mass chromatographic analysis are shown in supplementary material 4 (available at Carcinogenesis Online).
Fig. 3.
Two-dimensional gel electrophoresis of protein expression in Huh7 cells transfected with the HBx3′-30 or HBx3′-40 mutant, or with full-length HBx. The red arrow indicates overexpressed protein, and green arrow indicates decreased protein expression.
In addition, the differentially expressed genes and proteins were analyzed in HBx mutant-transfected Huh7 cells and compared with cells transfected with full-length HBx. The results were in agreement with the cDNA microarray and 2D gel data (data not shown). However, the expression of a few genes, such as spectrin and ubiquitin-activating enzyme E1, demonstrated differences in results at the mRNA and protein levels in the HBx mutant-transfected Huh7 cells.
Wnt-5a expression in HBx mutant-transfected Huh7 cells and in HCC tissues
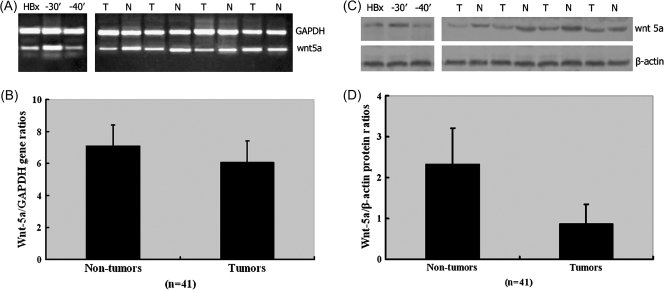
To validate our microarray results and to further demonstrate the difference in the expression pattern for Wnt-5a in cells with mutant and full-length HBx, we carried out semiquantitative RT–PCR, western blotting and immunohistochemistry analyses with HCC tissues and Huh7 cells. The Wnt-5a mRNA and protein expression levels in Huh7 cells transfected with the HBx3′-30 or HBx3′-40 mutant and in human clinical HCC and corresponding non-cancerous liver tissue samples are shown in Figure 4. The levels of these transcripts in Huh7 cells transfected with the HBx3′-30 or HBx3′-40 mutant, and the cancer tissues and their corresponding non-tumorous liver tissues by RT–PCR are shown in Figure 4A. The relative expression levels of Wnt-5a to GAPDH genes in HBx3′-30 and full-length HBx-transfected Huh7 cells by using real-time RT–PCR were 25.8 ± 0.47 and 3.15 ± 0.59, respectively. There are statistically significant differences between of them (P < 0.01). In contrast, the expression was downregulated in HBx3′-40 when compared with full-length HBx-transfected Huh7 cells (0.45 ± 0.52 versus 3.15 ± 0.59, P < 0.05).
Fig. 4.
Wnt-5a mRNA and protein expression in Huh7 cells transfected with the HBx3′-30 or HBx3′-40 mutant, and in human clinical HCC and corresponding non-cancerous liver tissue samples. (A) RT–PCR. Wnt-5a mRNA was upregulated in HBx3′-30-transfected Huh7 cells but downregulated in HBx3′-40-transfected Huh7 cells, compared with cells expressing full-length HBx. Wnt-5a mRNA expression varies in human HCCs when compared with their corresponding non-cancerous liver tissues. (B) Real-time RT–PCR. No statistically significant difference in Wnt-5a mRNA expression was found between HCC and the corresponding non-cancerous liver tissues. (C) Western blotting. The expression of Wnt-5a protein was upregulated in HBx3′-30-transfected Huh7 cells, but was not significantly different in HBx3′-40-transfected Huh7 cells, compared with cells transfected with full-length HBx. (D) The protein was present at significantly lower levels in the tumor tissues than in the corresponding non-cancerous liver tissue (P < 0.01); N, non-tumor; T, HCC; Bars, mean; columns, standard deviation.
Among the 41 HCC tissue samples, Wnt-5a mRNA was overexpressed in 16 samples (39%) and showed decreased expression in 11 samples (26.8%) compared with their corresponding non-cancerous liver tissues. The relative expression levels of Wnt-5a to GAPDH gene in HCC and the corresponding non-cancerous liver tissues are shown in Figure 4B. The relative expression levels of Wnt-5a to GAPDH gene in HCC compared with the corresponding non-cancerous liver tissues by using real-time RT–PCR were 7.13 ± 1.28 and 6.08 ± 1.33, respectively (P = 0.608). The relative expression levels of Wnt-5a to GAPDH gene in moderately differentiated and poorly differentiated cells were 7.74 ± 1.56 and 6.69 ± 1.47, respectively (P = 0.066). However, there was no significant difference between Wnt-5a expression and other clinicopathological findings such as age, serum alpha-fetoprotein concentration, cirrhosis, tumor capsule and intrahepatic metastasis.
The expression pattern of the Wnt-5a gene was further investigated at the protein level, as measured semiquantitatively by immunoblotting. As shown in Figure 4C and D The expression of the Wnt-5a protein was upregulated in HBx3′-30-transfected Huh7 cells compared with the cells transfected with full-length HBx (4.23 ± 0.77 versus 0.35 ± 0.21, P < 0.01). In contrast, the expression was not significantly different in HBx3′-40-transfected Huh7 cells compared with the cells expressing full-length HBx (0.30 ± 0.25 versus 0.35 ± 0.21, P > 0.01). Wnt-5a protein was generally present in significantly lower levels in the tumor tissues than in the corresponding non-cancerous liver tissues. The relative expression levels of Wnt-5a to β-actin in HCC compared with the corresponding non-cancerous liver tissues by using western blot were 2.34 ± 0.87 and 0.87 ± 0.48, respectively (P < 0.01).
To determine whether there is a positive or negative correlation between the mRNA and protein expression levels, Spearman’s bivariate correlation analysis was performed; however, no statistical correlation was found between the mRNA and protein expression levels in HCC and corresponding non-cancerous liver tissues.
Strongly positive Wnt-5a immunoreactivity was observed in 29 of 41 (70.7%) non-cancerous liver tissues. In hepatocytes, the Wnt-5a protein was strongly and uniformly stained in both the cytoplasm and nucleus; all of the surrounding connective tissues, including blood vessels and bile ducts, were negative. The HCC cells were stained in various patterns, being categorized as diffusely to focally positive in 12 patients (29.3%) and negative in 29 patients (70.7%) (supplementary material 5 is available at Carcinogenesis Online).
The correlation between HBx mutations and Wnt-5a expression in HCC tissues
To demonstrate the status of HBx mutations and their correlation with Wnt-5a expression in HCC tissues, HBx-Alu PCR and sequencing were performed for each sample. The results showed that deletions in HBx gene were significantly higher in HCC than that of non-cancerous liver tissues (33/41 versus 10/41, P < 0.001). Among them, the deletion of HBx C-terminal was detected in 28 HCC tissues. Spearman’s chi-square analysis results showed that there was a negative correlation between the expression of Wnt-5a immunoreactivity and the HBx COOH mutations in HCC tissues (r = −0.368, P = 0.018). However, no obviously correlation between the upexpression of Wnt-5a mRNA and the HBx COOH mutations in HCC tissues was found (Table II).
Table II.
The correlation between the Wnt-5a expression and the HBx COOH mutations in HCC tissues
| HBx COOH mutation | P value | ||
| Wnt-5A | + | − | |
| Immunoreactivity | |||
| + | 5 | 7 | 0.018 |
| − | 23 | 6 | |
| mRNA upexpression | |||
| + | 9 | 7 | 0.194 |
| − | 19 | 6 | |
Discussion
Although epidemiological and experimental data support a link between HBV infection and HCC, the underlying mechanisms are not fully understood. Several efforts have been devoted to exploring the role of the HBx protein in liver carcinogenesis (3). We have previously analyzed the structure of the HBx in malignant and normal non-cancerous liver tissues obtained from HCC patients who were serum HBsAg positive, demonstrating a high rate of HBx mutation in both tumor and non-tumor tissues (22). The deletion of amino acids at the C-terminal end is a major feature identified in tumor tissues by sequence analysis (5,21,32). Tu et al. (32) reported that the C-terminal region of HBx is crucial to its transcriptional function and that, in tumor tissues, most HBx natural mutants have lost their capacity for controlling cell proliferation, viability and transformation. Our results further confirmed that a variety of mechanisms are involved in the functions of the HBx C-terminus and that mutations within this region may directly affect cell proliferation and invasion by regulating cell cycle and apoptosis. These results suggest that mutations of HBx exist in the clonally expanding hepatocytes, supporting the hypothesis that HBx C-terminal deletions have a critical role in liver carcinogenesis.
In the present study, the hypothesis that different deletions at the C-terminal end, such as HBx3′-30 and HBx3′-40, may have different biological consequences in regulating cell proliferation and invasion was investigated by genomic and proteomic expression profiling using cDNA microarray, two-dimensional electrophoresis and mass spectrometry fingerprinting assays. The results demonstrate that the majority of the differentially expressed genes and proteins are mainly involved in key cellular functions, such as transcriptional regulation, oncogenesis, cell junction formation, signal transduction, metabolism and the immune response.
Unexpectedly, we found that the expression of Wnt-5a differed significantly and in opposite directions after cells were transfected with two different HBx mutants. Wnt-5a was upregulated by the HBx3′-30 mutant, but downregulated by the HBx3′-40 mutant. These changes, to the best of our knowledge, have not been described previously and may explain the different patterns of Wnt-5a expression observed in human HCC tissues. The importance of Wnt-signaling pathway in liver diseases has been indicated by several reports (17,33–35). β-catenin is central in the Wnt/β-catenin-signaling pathway and can be targeted for proteasome-mediated degradation (36). The ultimate result of Wnt/β-catenin pathway activation is the formation of a free, signaling pool of β-catenin in the cell that then enters the nucleus and forms a complex with members of the Lymphoid enhancer factor/T cell factor proteins family of transcription factors to regulate the expression of target genes (36–38). In addition, dysfunction of the Wnt-signaling pathway can cause developmental defects and cancer (39,40). Wnt-5a, a member of the Wnt family, has been shown to be upregulated in many human cancers (41–44). Wnt-5a antagonizes the canonical Wnt pathway by promoting the degradation of β-catenin, possibly suppressing tumor formation. This pathway is seven in-absentia homolog 2 and anaphase-promoting complex dependent, but glycogen synthase kinase-3 and β-TrCP independent (41). Cha et al. (45) found that HBx is involved in activating Wnt/β-catenin signaling in hepatoma cells. Ectopic expression of HBx along with Wnt-1 activates the Wnt/β-catenin signaling in Huh7 cells by stabilizing cytoplasmic β-catenin, which is achieved by suppressing glycogen synthase kinase 3 activity via activation of Src kinase. Our results in this study demonstrate that there is abnormal expression of Wnt-5a mRNA in ∼65% of HCC tissues. There was a tendency toward an inverse relationship between Wnt-5a expression and the tumor histological grades. Low HBx protein expression and a high rate of HBx mutation, particularly deletions of amino acids at the C-terminal end, were found in the tumor tissues. However, no statistically significant difference was found between HCC and the corresponding non-cancerous liver tissues, and no statistical correlation was found between the mRNA and protein expression levels (46). Thus, the biological consequence and the mechanisms with respect to the role Wnt-5a in HBV-associated HCC need further investigation.
Our results also indicate that there is a correlation between HBx gene expression and cell proliferation. In HBx3′-30-transfected Huh7 cells, the expression of genes related to cell growth-suppressive capacity, such as DHRS4L2, MTA2 and SERPINB5, was upregulated, and myc, a key regulator of cell proliferation, cell growth, differentiation and apoptosis, was downregulated. The upregulated expression of oncogene Ets (47,48) (4.376849), LOC221091 (10.87552), MMP10 (49) and MMP13 (50) was observed in the HBx3′-40-transfected Huh7 cells. These genes have been linked to cell growth, angiogenesis, cell junction disruption and extracellular matrix degradation. These results suggest that the 40 aa deletion at the C-terminal end would promote cell growth and invasion. These changes have not yet been found in human HCC samples.
In conclusion, differential expression of genes and proteins has been observed following transfection of Huh7 cells with HBx C-terminal deletion mutants. Most of the differentially expression genes and proteins are involved in transcriptional regulation, oncogenesis, cell junction maintenance, signal transduction, metabolism and the immune response. Changes in Wnt-5a expression were found in some of the analyzed HCC tissues. These results support the hypothesis that mutant forms of HBx gene are involved in liver carcinogenesis, possibly through Wnt-5a-signaling pathways, providing a better understanding of the function of HBx in hepatocarcinogenesis.
Supplementary material
Supplementary materials can be found at http://carcin.oxfordjournals.org/
Funding
National Nature Science Foundation of China (30070344 and 30070839) to M.Z.; National Institutes of Health (CA104025 and CA111427) to M.A.F.
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- cDNA
complementary DNA
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HBV
hepatitis B virus
- HBx
hepatitis B virus x gene
- HCC
hepatocellular carcinoma
- mRNA
messenger RNA
- MMTV
murine-mammary-tumour virus
- PCR
polymerase chain reaction
- RT–PCR
reverse transcription–polymerase chain reaction
- Wnt-5a
wingless-type MMTV integration site family, member 5A
References
- 1.Farazi PA, et al. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 2.Thorgeirsson SS, et al. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 3.Feitelson MA, et al. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Feitelson MA, et al. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am. J. Pathol. 1997;150:1141–1157. [PMC free article] [PubMed] [Google Scholar]
- 5.Sirma H, et al. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18:4848–4859. doi: 10.1038/sj.onc.1202867. [DOI] [PubMed] [Google Scholar]
- 6.Feitelson MA, et al. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene. 1993;8:1109–1117. [PubMed] [Google Scholar]
- 7.Terradillos O, et al. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene. 1998;17:2115–2123. doi: 10.1038/sj.onc.1202432. [DOI] [PubMed] [Google Scholar]
- 8.Koike K, et al. Compensatory apoptosis in preneoplastic liver of a transgenic mouse model for viral hepatocarcinogenesis. Cancer Lett. 1998;134:181–186. doi: 10.1016/s0304-3835(98)00252-3. [DOI] [PubMed] [Google Scholar]
- 9.Kim CM, et al. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 10.Yu DY, et al. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J. Hepatol. 1999;31:123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- 11.Schaefer S, et al. Properties of tumour suppressor p53 in murine hepatocyte lines transformed by hepatitis B virus X protein. J. Gen. Virol. 1998 doi: 10.1099/0022-1317-79-4-767. 79 (Pt 4), 767–777. [DOI] [PubMed] [Google Scholar]
- 12.Tarn C, et al. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J. Biol. Chem. 1999;274:2327–2336. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- 13.Gottlob K, et al. Hepatitis B virus X protein transcription activation domains are neither required nor sufficient for cell transformation. Cancer Res. 1998;58:3566–3570. [PubMed] [Google Scholar]
- 14.Birrer RB, et al. Hepatocellular carcinoma and hepatitis virus. Ann. Clin. Lab. Sci. 2003;33:39–54. [PubMed] [Google Scholar]
- 15.Groisman IJ, et al. Downregulation of DNA excision repair by the hepatitis B virus-x protein occurs in p53-proficient and p53-deficient cells. Carcinogenesis. 1999;20:479–483. doi: 10.1093/carcin/20.3.479. [DOI] [PubMed] [Google Scholar]
- 16.Qiao L, et al. Hepatitis B virus X protein increases expression of p21(Cip-1/WAF1/MDA6) and p27(Kip-1) in primary mouse hepatocytes, leading to reduced cell cycle progression. Hepatology. 2001;34:906–917. doi: 10.1053/jhep.2001.28886. [DOI] [PubMed] [Google Scholar]
- 17.Miyazawa J, et al. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004;64:2024–2029. doi: 10.1158/0008-5472.can-03-1855. [DOI] [PubMed] [Google Scholar]
- 18.Ng RK, et al. cDNA microarray analysis of early gene expression profiles associated with hepatitis B virus X protein-mediated hepatocarcinogenesis. Biochem. Biophys. Res. Commun. 2004;322:827–835. doi: 10.1016/j.bbrc.2004.07.188. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, et al. The transactivation and p53-interacting functions of hepatitis B virus X protein are mutually interfering but distinct. Cancer Res. 1997;57:5137–5142. [PubMed] [Google Scholar]
- 20.Wang Y, et al. Characterization of HBV integrants in 14 hepatocellular carcinomas: association of truncated X gene and hepatocellular carcinogenesis. Oncogene. 2004;23:142–148. doi: 10.1038/sj.onc.1206889. [DOI] [PubMed] [Google Scholar]
- 21.Iavarone M, et al. Characterisation of hepatitis B virus X protein mutants in tumour and non-tumour liver cells using laser capture microdissection. J. Hepatol. 2003;39:253–261. doi: 10.1016/s0168-8278(03)00217-4. [DOI] [PubMed] [Google Scholar]
- 22.Liu XH, et al. COOH-terminal deletion of HBx gene is a frequent event in HBV-associated hepatocellular carcinoma. World J. Gastroenterol. 2008;14:1346–1352. doi: 10.3748/wjg.14.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lian Z, et al. The translation initiation factor, hu-Sui1 may be a target of hepatitis B X antigen in hepatocarcinogenesis. Oncogene. 1999;18:1677–1687. doi: 10.1038/sj.onc.1202470. [DOI] [PubMed] [Google Scholar]
- 24.Feitelson MA, et al. X antigen polypeptides in the sera of hepatitis B virus-infected patients. Virology. 1990;177:367–371. doi: 10.1016/0042-6822(90)90492-a. [DOI] [PubMed] [Google Scholar]
- 25.DeRisi J, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 26.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, et al. Gene expression patterns in human liver cancers. Mol. Biol. Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeindl-Eberhart E, et al. Detection and identification of tumor-associated protein variants in human hepatocellular carcinomas. Hepatology. 2004;39:540–549. doi: 10.1002/hep.20060. [DOI] [PubMed] [Google Scholar]
- 29.Zeindl-Eberhart E, et al. Proteome analysis of rat hepatomas: carcinogen-dependent tumor-associated protein variants. Electrophoresis. 2001;22:3009–3018. doi: 10.1002/1522-2683(200108)22:14<3009::AID-ELPS3009>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 30.Hamilton S, et al. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Digestive System. Lyon: IARC Press; 2000. pp. 167–169. [Google Scholar]
- 31.Xian ZH, et al. Expression of aspartyl beta-hydroxylase and its clinicopathological significance in hepatocellular carcinoma. Mod. Pathol. 2006;19:280–286. doi: 10.1038/modpathol.3800530. [DOI] [PubMed] [Google Scholar]
- 32.Tu H, et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803–7810. [PubMed] [Google Scholar]
- 33.Thompson MD, et al. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 34.Shackel N. Zebrafish and the understanding of liver development: the emerging role of the Wnt pathway in liver biology. Hepatology. 2007;45:540–541. doi: 10.1002/hep.21543. [DOI] [PubMed] [Google Scholar]
- 35.Zeng G, et al. Wnt'er in liver: expression of Wnt and frizzled genes in mouse. Hepatology. 2007;45:195–204. doi: 10.1002/hep.21473. [DOI] [PubMed] [Google Scholar]
- 36.Werth M, et al. Hepatic expression of glutamine synthetase in rats is controlled by STAT5 and TCF transcription factors. Hepatology. 2006;44:967–975. doi: 10.1002/hep.21322. [DOI] [PubMed] [Google Scholar]
- 37.Lee TK, et al. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition. Cancer Res. 2006;66:9948–9956. doi: 10.1158/0008-5472.CAN-06-1092. [DOI] [PubMed] [Google Scholar]
- 38.Oyama T, et al. Further upregulation of beta-catenin/Tcf transcription is involved in the development of macroscopic tumors in the colon of ApcMin/+ mice. Carcinogenesis. 2008;29:666–672. doi: 10.1093/carcin/bgn001. [DOI] [PubMed] [Google Scholar]
- 39.Cha I, et al. Angiomyolipoma of the liver in fine-needle aspiration biopsies: its distinction from hepatocellular carcinoma. Cancer. 1999;87:25–30. [PubMed] [Google Scholar]
- 40.Kim I, et al. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kremenevskaja N, et al. Wnt-5a has tumor suppressor activity in thyroid carcinoma. Oncogene. 2005;24:2144–2154. doi: 10.1038/sj.onc.1208370. [DOI] [PubMed] [Google Scholar]
- 42.Kurayoshi M, et al. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 2006;66:10439–10448. doi: 10.1158/0008-5472.CAN-06-2359. [DOI] [PubMed] [Google Scholar]
- 43.Dejmek J, et al. Wnt-5a protein expression in primary dukes B colon cancers identifies a subgroup of patients with good prognosis. Cancer Res. 2005;65:9142–9146. doi: 10.1158/0008-5472.CAN-05-1710. [DOI] [PubMed] [Google Scholar]
- 44.Pukrop T, et al. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc. Natl Acad. Sci. USA. 2006;103:5454–5459. doi: 10.1073/pnas.0509703103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha MY, et al. Hepatitis B virus X protein is essential for the activation of Wnt/beta-catenin signaling in hepatoma cells. Hepatology. 2004;39:1683–1693. doi: 10.1002/hep.20245. [DOI] [PubMed] [Google Scholar]
- 46.Liu XH, et al. Expression of Wnt-5a and its clinicopathological significance in hepatocellular carcinoma. Dig. Liver Dis. 2008 doi: 10.1016/j.dld.2007.12.011. doi:10.1016/j.dld.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 47.Sementchenko VI, et al. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 48.Mavrothalassitis G, et al. Proteins of the Ets family with transcriptional repressor activity. Oncogene. 2000;19:6524–6532. doi: 10.1038/sj.onc.1204045. [DOI] [PubMed] [Google Scholar]
- 49.Schnabl B, et al. Zinc finger protein 267 is up-regulated during the activation process of human hepatic stellate cells and functions as a negative transcriptional regulator of MMP-10. Biochem. Biophys. Res. Commun. 2005;335:87–96. doi: 10.1016/j.bbrc.2005.07.043. [DOI] [PubMed] [Google Scholar]
- 50.Chakraborti S, et al. Regulation of matrix metalloproteinases: an overview. Mol. Cell. Biochem. 2003;253:269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.