Abstract
Background
The mechanism of HIV-1 mother-to-child transmission (MTCT) is not well described.
Methods
Of 328 HIV-infected mother-infant pairs, we identified discordant ACE and GSTM1 alleles in 91. Maternal alleles in cord blood were quantified with real-time PCR as indicators of microtransfusions.
Results
HIV-1 infected infants had more maternal DNA in cord blood than their uninfected counterparts. Increased cord blood maternal DNA was associated with preterm delivery, low birth weight, and maternal immunosuppression.
Conclusion
Intrapartum MTCT was associated with placental microtransfusions. The associations among placental microtransfusion, in utero MTCT, immunosuppression and poor birth outcome should be further investigated.
Keywords: HIV-1 mother-to-child transmission, placental microtransfusions, Malawi
Introduction
Although HIV-1 mother-to-child transmission (MTCT) has been significantly reduced in the developed world, in 2006 an estimated 530,000 children were newly HIV-1 infected [1]. In populations where replacement feeding is unfeasible, it has been estimated that 12% of MTCT occurs in utero before 36 weeks gestation, 29% occurs between 36 weeks and delivery, 20% occurs during delivery, and the remaining 39% occurs during prolonged breastfeeding [2]. Remarkably, despite this wealth of knowledge on the timing of HIV-1 MTCT, the mechanism of HIV-1 MTCT remains unclear.
For in utero and intrapartum transmission, suggested routes of HIV-1 transmission include ascending infection, exposure to HIV-1 in the birth canal, and passage through the placental barrier. Through the measurement of placental alkaline phosphatase in umbilical cord blood, our group has previously found an association between placental microtransfusions and intrapartum MTCT [3]. In order to better understand microtransfusions, we now assess microtransfusions by quantifying the amount of maternal DNA in umbilical cord blood.
Materials and Methods
Sample collection
Participants presenting to the Antenatal Ward at Queen Elizabeth Central Hospital, in Blantyre, Malawi were enrolled into a prospective cohort study designed to assess the relationship between malaria and HIV-1 MTCT[1–3]. Women and their newborn infants received single-dose nevirapine according to the HIVNET 012 protocol [4]; no women received ongoing antiretroviral treatment. Mother-infant pairs were included in this sub-study if they delivered a live, singleton child by vaginal delivery and if matched umbilical cord and maternal peripheral blood were available. Infant HIV status was determined by real-time PCR against HIV-1 DNA as described in reference [5], according to the methods of Luo et al.[6]. Infants were considered HIV-1-infected in utero if they were HIV-1 DNA positive within 48 hrs. of birth; considered HIV-1-infected intrapartum if they were both HIV-1 DNA negative at birth and HIV-1 DNA positive at 6 weeks; and considered HIV-1 negative if they were HIV-1 DNA negative at 6 weeks (NT)[7]. This study was approved by the UNC-Chapel Hill IRB and the Malawi College of Medicine Research Ethics Committee; informed consent was obtained from all participants.
Sample preparation
Immediately after delivery the umbilical cord was clamped and cut. The umbilical cord was cleaned with saline, the umbilical vein was located, and blood was aspirated by needle and syringe. Whole blood was put into tubes containing EDTA, centrifuged in a clinical centrifuge at 1800 RPM for 5 minutes, and the plasma was removed. Whole blood pellets were stored at −80°C until processed. Genomic DNA was isolated from maternal peripheral blood and venous umbilical cord blood pellets using the Qiagen DNA Mini Kit according to the manufacturer’s instructions. Matched umbilical cord and maternal genomic DNA were PCR amplified for the polymorphic glutathione S-transferase M1 (GSTM1) and angiotensin-converting enzyme intron 16 (ACE) genes according to the methods of Lo et al. [4] using their primers, which are listed in the Supplemental Table. When the maternal sample had an ACE allele not present in the infant, this was defined as an informative ACE pair. Matched pairs uninformative for ACE were screened for a GSTM1 insertion, and samples were considered GSTM1 informative when the maternal sample was GSTM1 insertion-positive and the infant was GSTM1 insertion-null; only GSTM1-null infants whose umbilical cord blood also supported the amplification of an ACE allele were included.
Real-time PCR
Quantitative real-time polymerase chain reaction ([qPCR]; PE Biosystems 7300) was performed on the umbilical cord genomic DNA against the informative maternal ACE or GSTM1 allele using the primer sets and probes listed in the Supplemental Table. The thermal profile consisted of an initial incubation at 50°C for 2 minutes, then 95°C for 10 minutes, followed by 50 cycles of 95°C for 15 seconds and 56°C for 1 minute, in a reaction volume of 25μl (5 μL template DNA, 2 μM of each primer, 0.8 μM probe, and 12μl of Taqman Universal PCR Master Mix [Applied Biosystems]). All reactions were run in duplicate along with no-template control reactions. Serial dilutions of GSTM1, ACE insertion or ACE deletion containing maternal DNA demonstrated that the assay was linear over 3 logs (R2=0.99). In order to control for the total amount of DNA in the umbilical cord blood, all qPCR reactions were normalized to Rnase P concentration using the TaqMan RNase P Control Reagent Kit (Applied Biosystems).
The efficiencies of each assays was calculated using the slope of each assays’ calibration curve, according to the methods of Dorak [8]. Because the efficiencies of the target and reference genes were all within 5% of each other (data not shown), the comparative threshold (Ct) method was used to quantify the amount of maternal-specific allele in the umbilical cord DNA [8] using the following equation:
To account for any differences between the GSTM1, ACE-insertion and ACE-deletion qPCR assays, we identified 3 maternal DNA samples positive for all three target alleles. Serial dilutions of these samples were run in duplicate, against all three assays, to determine inter-assay accuracy. The GSTM1 and ACE deletion reactions gave similar dilution profiles, while the ACE insertion profile consistently resulted in lower Ct. values than the other two assays. This discrepancy was corrected by multiplying all ACE-insertion values by 0.895, which normalized the three informative-gene assays (Supplemental Figure).
Samples were excluded from data analysis if there was no detectable amplification in both the informative gene and the RNase P assay (n=4). Samples with detectable amplification in the RNase P assay, but lacking detectable amplification in the informative gene assay (n=42), were rerun using 4-times more genomic DNA. The limit of detection for the informative gene assays was estimated to be Ct.=40; samples below this limit were all assigned ct. values of 40 (n=42).
Data Analysis
Owing to likely differences in the mechanism of transmission, in utero and intrapartum transmission were analyzed as independent outcomes. DNA admixture data are presented as a fractional concentration, defined as the ratio of the normalized target gene (ACE or GSTM1) to RnaseP, as described above. The relationship between DNA admixture and dichotomous variables was tested with the Wilcoxon rank-sum test. Binary coding for the following variables was created by dichotomization at the median: duration of labor (10 hrs.), placental delivery time (5 min.), high HIV-1 RNA concentration (>4.2 log10 copies/ml, n=56), and low placental weight (<540g). Women were tested for syphilis using the Rapid Plasma Reagin test ([RPR]), and all RPR reactive sera were tested with the Treponema pallidum Hemagglutination Assay (TPHA). Women with a reactive RPR followed by a reactive TPHA were considered syphilis seroreactive. Placental malaria parasitemia and chorioaminionitis were determined via placental histology, as described in Mwapasa et al. [9]; only 47 of the 91 participants were examined for chorioamnionitis. All other clinical characteristics were determined by the study nurse at the time of delivery. Associations with p<0.2 are noted. All analyses were run on Stata version 10.0 (Stata Corp., College Station, TX).
RESULTS
Matched maternal and umbilical cord genomic DNA was available from 328 of the 565 HIV-1 eligible mother-infant pairs. Of the 328 samples available, 91 (28%) were ACE or GSTM1 informative. The percentage of informative pairs is consistent with the findings of Lo et al., who detected 32% informative pairs in their population of 156 matched pairs[10]. If we dichotomize the fractional concentration of maternal DNA at 10−2, where there is a natural inflection of the data, 39 (43%) of the infants had an appreciable amount of maternal DNA in their umbilical cord blood; this is similar to the 40% reported by Lo et al. [10].
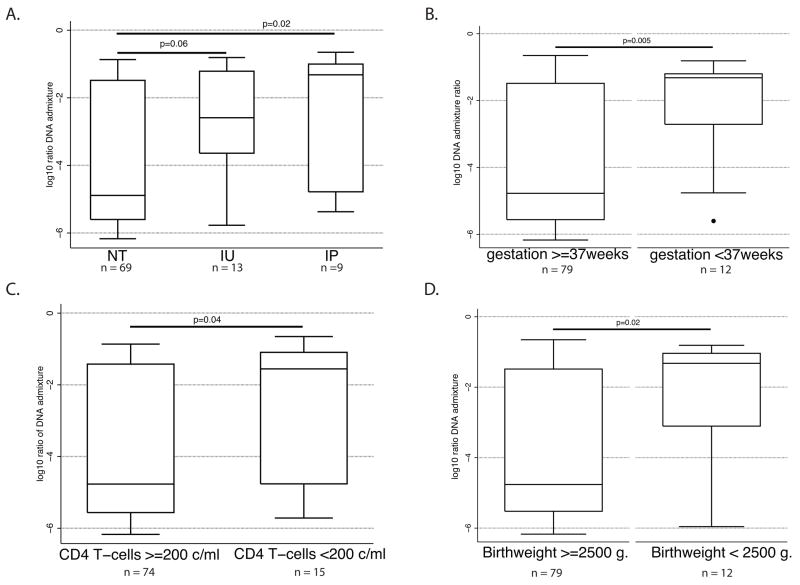
Among the 91 informative mother-infant pairs, there were 69 exposed, but uninfected infants, 13 in utero-infected infants, and 9 intrapartum-infected infants. The median log10 fractional concentrations of maternal DNA in umbilical cord blood were −4.9, −2.6, and −1.3, for uninfected, in utero and intrapartum infants, respectively (Figure 1A). As expected, DNA admixture was significantly higher in the umbilical cord blood of intrapartum-infected infants than of uninfected infants (p=0.02). The difference between in utero and unifected infants was not as robust (p=0.06).
Figure 1.
Box and whisker plots of factors associated with log10 transformed maternal-fetal DNA admixture: (A) HIV-1 vertical transmission status, (B) preterm delivery (<37 weeks gestation), (C) low CD4 T-cell counts (<200 cells/ml), (D) low birth weight (<2500 grams). NT=uninfected infant; IU= in utero-infected infant; IP=intrapartum infected infant. Line represents median value, box edges represent 25th and 75th percentile, whisker edges represent upper adjacent values, and dots represent outside values. P-values were determined by the Wilcoxon rank-sum test.
DNA admixture was also associated with poor birth outcomes. Among all 91 umbilical cord DNA isolates, increased amounts of maternal-fetal DNA admixture were associated with low birth weight (median log10 fractional concentration during low birth weight = −1.3 versus −4.8, p=0.02) and preterm delivery (median log10 fractional concentration during preterm delivery= −1.3 versus −4.8, p=0.005).
Immunological status also appeared to influence microtransfusions. A higher fractional DNA concentration was found in umbilical cord blood of infants from mothers with low CD4 T-cell count (<200 c/ml; p=0.04) than those with high CD4 counts. However, no difference in the fractional concentration of maternal DNA was found between women with high versus low HIV-1 concentration (data not shown).
Obstetric factors such as delayed placental delivery, visible placental tears, sex of the newborn, vaginal/vulval tears, episiotomy, rupture of membranes >4 hrs., labor >10 hrs., primigravidity, or low placental weight were not associated with DNA admixture (p>0.2). In addition, other infections such as chorioamnionitis (16 cases), syphilis (1 case) and placental malaria (5 cases) were also not associated with DNA admixture, although the sample sizes were small. The relationship between these infections and HIV-1 MTCT in the parent cohort is described elsewhere[5].
Discussion
In both breastfeeding and non-breastfeeding populations, a majority of HIV-1 MTCT occurs at or around the time of delivery through unknown mechanisms [2]. The two most likely routes of intrapartum HIV-1 infection are birth canal exposure or transplacental passage, and in this study, we observed that maternal-fetal DNA admixture was associated with intrapartum MTCT; this observation is consistent with our previously reported findings linking elevated umbilical cord placental alkaline phosphatase with intrapartum MTCT [3]. Like umbilical cord placental alkaline phosphatase activity, we interpret maternal-fetal DNA admixture as a measurement of placental microtransfusions, and therefore we conclude that microtransfusions increase the risk of intrapartum HIV-1 MTCT.
In a previous study by Lo et al., separation of the buffy coat from the plasma allowed the investigators to determine the amount of both cell-associated and cell-free maternal DNA in the umbilical cord. Because we extracted genomic DNA from pelleted whole blood, we were unable to determine whether the maternal DNA was cell-derived, which would have provided an important clue to the nature of the microtransfusions (i.e. large enough to allow maternal cells to pass through). A second limitation of these data is that the associations detected between MTCT and DNA admixture are derived from transmission groups with small sample sizes, and therefore they should be interpreted with caution. While this presents difficulty in the interpretation of the non-significant association between DNA admixture and in utero MTCT, the significant association with intrapartum MTCT is consistent with our previously published findings [3]. Finally, also as a result of the small sample size, we were unable to do multivariate modeling, which precluded adjustment of the data for covariates known to be associated with HIV-1 MTCT.
In conclusion, when combined with our previous microtransfusion findings [3], this study supports placental microtransfusions as a mode of intrapartum HIV-1 MTCT. In an attempt to synthesize these data with the existing literature, we propose the following model of MTCT: low CD4 T-cell counts, through an unknown mechanism, compromise the maternal-fetal placenta barrier, which, in turn, leads to both poor birth outcomes and placental microtransfusions; the resulting placental microtransfusions allow passage of the HIV-1 directly into the infant bloodstream, leading to increased rates of HIV-1 MTCT. The nature of the placental-insult remains ill-defined, but it could be either through the physical stress of labor and delivery, or via specific infections such as malaria, chorioamnionitis, or syphilis; alternatively, the nature of the insult could be less important than the inflammation itself, which may provoke a common inflammatory response, such as tumor necrosis factor alpha production [11,12]. The novel association between placental microtransfusions, in utero MTCT, low CD4 count and poor birth outcome is intriguing and it, along with our model, should be evaluated in a larger study.
Supplementary Material
Acknowledgments
We thank the Malawian women and their infants for their consent and participation; the study nurses and laboratory technicians for their excellent support; and the UNC Center for AIDS Research for their financial assistance.
Funding. This research was supported by NIH awards to JJK (K99-HD056586; R00-HD056586), VM (R01-TW007305), SRM (R01-AI49084; R21-AI065369), and by the UNC Center For AIDS Research (P30-AI50410); SJR is supported by a Senior Fellowship from The Wellcome Trust. The funding agencies had no role in study design, data collection, data analysis, decision to publish, or preparation of the manuscript, and the content does not necessarily represent the official view of the National Institute of Child Health and Human Development, the National Institute of Allergy and Infectious Diseases, or the National Institutes of Health. This work has been submitted to the 15th Conference on Retroviruses and Opportunistic infections for discussion (Boston, MA, February 2008).
Footnotes
Conflict of Interest. None reported.
References
- 1.UNAIDS/WHO. 2006 Report on the Global AIDS Epidemic:May 2006. 2006 Available: www.unaids.org via the Internet.
- 2.Kourtis AP, Lee FK, Abrams EJ, Jamieson DJ, Bulterys M. Mother-to-child transmission of HIV-1: timing and implications for prevention. Lancet Infect Dis S. 2006;6:726–732. doi: 10.1016/S1473-3099(06)70629-6. [DOI] [PubMed] [Google Scholar]
- 3.Kwiek JJ, Mwapasa V, Milner DAJ, Alker AP, Miller WC, Tadesse E, Molyneux ME, Rogerson SJ, Meshnick SR. Maternal-fetal microtransfusions and HIV-1 mother-to-child transmission in Malawi. PLoS Med. 2006;3:e10. doi: 10.1371/journal.pmed.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guay LA, Musoke P, Fleming T, Bagenda D, Allen M, Nakabiito C, Sherman J, Bakaki P, Ducar C, Deseyve M, Emel L, Mirochnick M, Fowler MG, Mofenson L, Miotti P, Dransfield K, Bray D, Mmiro F, Jackson JB. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 5.Mwapasa V, Rogerson SJ, Kwiek JJ, Wilson PE, Milner D, Molyneux ME, Kamwendo DD, Tadesse E, Chaluluka E, Meshnick SR. Maternal syphilis infection is associated with increased risk of mother-to-child transmission of HIV in Malawi. AIDS. 2006;20:1869–1877. doi: 10.1097/01.aids.0000244206.41500.27. [DOI] [PubMed] [Google Scholar]
- 6.Luo W, Yang H, Rathbun K, Pau C-P, Ou C-Y. Detection of HIV-1 DNA in Dried Blood Spots Using a Duplex Real-Time PCR Assay. J Clin Microbiol. 2005;43:1851–1857. doi: 10.1128/JCM.43.4.1851-1857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryson YJ, Luzuriaga K, Sullivan JL, Wara DW. Proposed definitions for in utero versus intrapartum transmission of HIV-1. N Engl J Med. 1992;327:1246–1247. doi: 10.1056/NEJM199210223271718. [DOI] [PubMed] [Google Scholar]
- 8.Dorak MT. Real-time PCR. Oxford: Taylor & Francis; 2006. [Google Scholar]
- 9.Mwapasa V, Rogerson SJ, Molyneux ME, Abrams ET, Kamwendo DD, Lema VM, Tadesse E, Chaluluka E, Wilson PE, Meshnick SR. The effect of Plasmodium falciparum malaria on peripheral and placental HIV-1 RNA concentrations in pregnant Malawian women. AIDS. 2004;18:1051–1059. doi: 10.1097/00002030-200404300-00014. [DOI] [PubMed] [Google Scholar]
- 10.Lo YM, Lau TK, Chan LY, Leung TN, Chang AM. Quantitative analysis of the bidirectional fetomaternal transfer of nucleated cells and plasma DNA. Clin Chem. 2000;46:1301–1309. [PubMed] [Google Scholar]
- 11.Kfutwah AK, Mary JY, Nicola MA, Blaise-Boisseau S, Barre-Sinoussi F, Ayouba A, Menu E. Tumour necrosis factor-alpha stimulates HIV-1 replication in single-cycle infection of human term placental villi fragments in a time, viral dose and envelope dependent manner. Retrovirology. 2006;3:36. doi: 10.1186/1742-4690-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogerson SJ, Brown HC, Pollina E, Abrams ET, Tadesse E, Lema VM, Molyneux ME. Placental tumor necrosis factor alpha but not gamma interferon is associated with placental malaria and low birth weight in Malawian women. Infect Immun. 2003;71:267–270. doi: 10.1128/IAI.71.1.267-270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.