Abstract
Background
The etiology of cardiovascular disease (CVD) is multifactorial. Efforts to identify genes influencing CVD risk have met with limited success to date, likely due to the small effect sizes of common CVD risk alleles and the presence of gene by gene and gene by environment interactions.
Methods
The Heredity and Phenotype Intervention (HAPI) Heart Study was initiated in 2002 to measure the cardiovascular response to four short-term interventions affecting cardiovascular risk factors and to identify the genetic and environmental determinants of these responses. The measurements included blood pressure responses to the cold pressor stress test and to a high salt diet, triglyceride excursion in response to a high fat challenge, and response in platelet aggregation to aspirin therapy.
Results
The interventions were carried out in 868 relatively healthy Amish adults from large families. The heritabilities of selected response traits for each intervention ranged from 8–38%, suggesting that some of the variation associated with response to each intervention can be attributed to the additive effects of genes.
Conclusions
Identifying these response genes may identify new mechanisms influencing CVD and may lead to individualized preventive strategies and improved early detection of high-risk individuals.
Keywords: gene by environment interaction, heritability, cold pressor, salt sensitivity, post-prandial triglyceride, aspirin resistance
INTRODUCTION
Family studies have consistently demonstrated the importance of genetic factors in contributing to CVD susceptibility, acting independently as well as through known CVD risk factors 1–6. However, identification of the specific genes involved has proved challenging. While a number of single genes influencing CVD susceptibility have been identified to date, they collectively account for only a small proportion of the population attributable risk for CVD. The difficulties in identifying the genes influencing CVD have been ascribed to the very small individual effects of many gene variants and the presence of gene by gene and gene by environment interactions that lead to context dependent penetrance of susceptibility alleles. In recognition of the importance that gene by environment interactions may play in influencing cardiovascular health, the National Heart, Lung, and Blood Institute of the National Institutes of Health funded the PROgram for GENetic Interaction (PROGENI) network. This network includes five independent but complementary studies designed to assess the genetics of the cardiovascular response to controlled short-term perturbations that mimic long-term exposures known to affect cardiovascular health, including stress, diet, and anti-platelet and lipid lowering therapy.
The Heredity and Phenotype Intervention (HAPI) Heart Study was initiated in 2002. We chose to study the Old Order Amish as we hypothesized that genetic effects of complex phenotypes might be easier to discern in this genetically homogeneous population. Four interventions were performed including: (1) a high fat challenge, after which we measured the post-prandial excursion in triglycerides; (2) a cold pressor stress test (CPT), for which we measured changes in blood pressure and endothelial function; (3) a dietary salt intervention, involving 6-day ingestion of a high salt diet, followed by 6-day ingestion of a low salt diet, during which we measured changes in blood pressure; and (4) 14-days of low-dose aspirin therapy, during which we measured changes in platelet function. These interventions were performed in a highly controlled and consistent manner and the relevant CVD-related responses were accurately measured. We selected these interventions for study because it has been established that they induce responses in CVD risk factors, and there is large inter-individual variation in these responses (some of which are likely to be determined by genetic factors). Because a large set of baseline and response cardiovascular phenotypes were concurrently obtained, additional study goals were to determine the relationship of these response phenotypes to baseline CVD traits and to each other and to identify genes influencing individual phenotypes or sets of phenotypes.
METHODS
Study Population
Participants for the HAPI Heart Study were recruited from the Amish community of Lancaster County, PA. The Amish immigrated from central Europe (mainly Switzerland) to the United States to escape religious persecution over a 50 year period beginning in 1727 7. The earliest immigrants settled in Lancaster County, Pennsylvania, while later groups settled in the midwest. Approximately 25,000 Old Order Amish currently live in Lancaster County today 8, and virtually all can trace their ancestry back to the approximately 200 original founding individuals 9;10.
Subjects were initially identified through prior participation in one of our previous studies, word of mouth, advertisements, a community-wide mailing, and referrals from local physicians. Only subjects aged 20 years and older were eligible to participate and age-eligible family members of all participating subjects were encouraged to enroll. In the planning stages of the study, support was obtained from the Amish community by the principal investigator (Dr. Shuldiner) presenting the study objectives and procedures to community leaders and gaining their support to proceed. The study was approved by the Institutional Review Board of the University of Maryland, Baltimore and other participating institutions and was monitored by an external Data Safety and Monitoring Board. Informed consent, including permission to contact relatives, was obtained before participation.
Eligibility Criteria and Recruitment
Prior to enrollment, potential study participants were visited at home by a study nurse and Amish liaison team, who performed a screening examination to determine eligibility. Although most subjects qualified for all interventions, there were separate exclusion criteria for each to enable subjects to participate in only those interventions for which they qualified. A summary of the global exclusion criteria and intervention-specific exclusion criteria is provided in Table I.
Table I.
Exclusion criteria for HAPI Heart Study
Study-wide exclusion criteria:
|
Intervention-specific exclusion criteria:
|
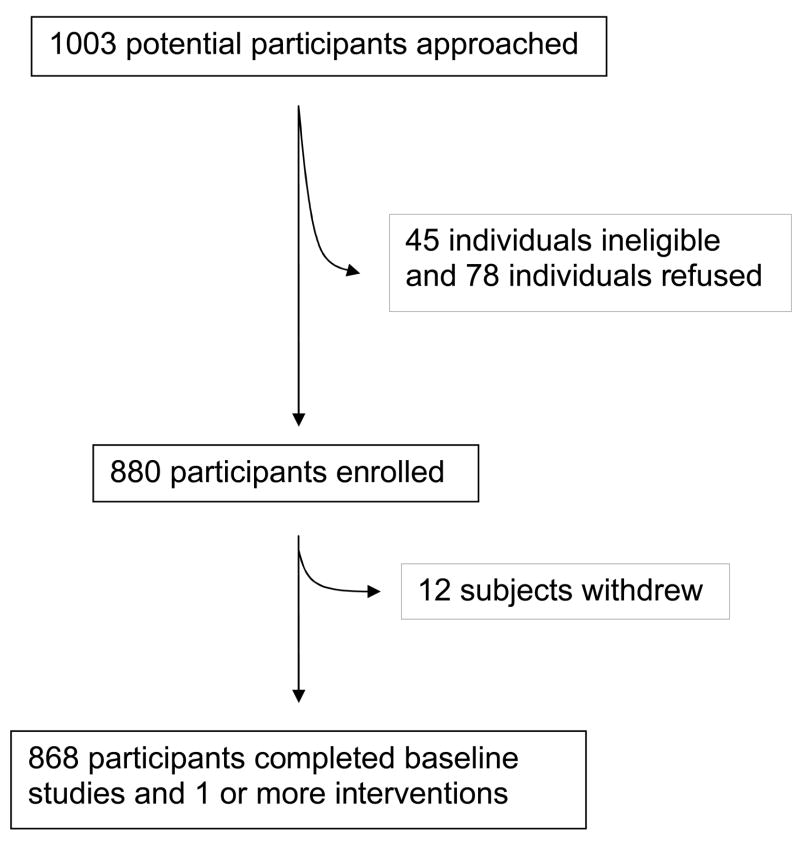
A total of 1,003 individuals were identified for recruitment and received home visits to establish eligibility. Of these, 78 (78/1003=7.8%) refused to participate and 45 (45/1003=4.5%) did not meet one or more of the global eligibility criteria. Twelve eligible and consented individuals withdrew prior to completing any of the interventions, leaving a final sample size of 868 participants (Figure I).
Figure I.
Recruitment of subjects into the HAPI Heart Study
Study Protocol and Interventions
The study protocol required a four-week period to complete all four interventions (see Figure II). After obtaining permission from their prescribing physicians, all medications, vitamins and supplements were discontinued for 7 days prior to Clinic Visit 1 and for the duration of the study. Since Amish do not drive cars, transportation to the Amish Research Clinic (Lancaster, PA) was provided. Subjects were instructed to fast for 12 hours prior to their appointment, to abstain from excessive physical activity on the morning of the visit, and to bring a first morning void urine sample. Clinic Visit 1 lasted for 8 to 10 hrs, during which time the following were performed: physical examination, urine pregnancy test, fasting blood sample for pre-aspirin analyses of platelet aggregation and inflammatory markers, blood pressure monitoring, CPT, high fat challenge, electrocardiogram, ultrasound measurement of carotid intima media thickness (IMT), measurement of pulse wave velocity (PWV) and ankle-brachial index (ABI), and echocardiogram. All blood samples were obtained through an indwelling angiocatheter.
Figure II.

Schema of HAPI Heart Study intervention schedules
The CPT was conducted by having the subject immerse his/her right hand and wrist in ice water for a period of 2 ½ minutes. Prior to immersion, blood pressure measurements were taken every 5 minutes for at least 20 minutes. Nine additional blood pressure measurements were taken during and after the cold pressor stimulus at minutes 1, 2, 3, 4, 5, 7.5, 10, 15, and 20 following immersion. A brachial artery reactivity test (BART) was performed during the course of the cold pressor stimulus to measure changes in brachial artery diameter during and after the CPT stimulus.
The high fat challenge was administered one hour after completion of the CPT. Prior to the beginning of the fat challenge, blood was drawn to measure fasting lipids and brachial artery flow-mediated vasodilation (FMD) was measured to assess fasting endothelial function. The high fat challenge, prepared in the form of a whipping cream milk shake, was standardized to consist of 782 calories per m2 of body surface with 77.6% of calories from fat, 19.2% from carbohydrate, and 3.1% from protein. Following ingestion, blood was drawn at 1, 2, 3, 4, and 6 hours to assess the triglyceride excursion. FMDs were also performed at 2, 4, and 6 hrs following the fat challenge to assess post-prandial effects on endothelial function. The subject rested and remained fasting during the 6 hours post-fat challenge.
The aspirin intervention was begun the day after Clinic Visit 1; for the next 14 consecutive days the subject took 81 mg aspirin every day. One to three days prior to the second clinic visit a nurse and liaison performed a home visit to ensure compliance. On the 14th day the subject took his/her aspirin shortly before arriving at the Amish Research Clinic for Clinic Visit 2. Fasting blood was drawn to measure post-aspirin platelet aggregation. The subject also collected and brought his/her first morning urine sample for measurement of 11-dehydrothromboxane B2 (TXB2). To assess compliance, a pill count was performed and each subject kept a diary. A subject was permitted to miss up to four aspirin doses over the two-week period and still be included in the final analysis, provided that he/she took the aspirin for at least three consecutive days prior to Clinic Visit 2. The aspirin intervention could be extended for up to three days (17 days total) to meet this criterion.
The Monday following Clinic Visit 1 subjects began six days of a high salt diet, followed by a 6–14 day wash-out period, and then six days of a low salt diet. The six day course for the high/low salt interventions was chosen so that the special diet would not interfere with their Sunday church meal. All meals were culturally appropriate and prepared by an Amish caterer and provided to the subjects by home delivery. The high and salt diets contained 280 meq and 40 meq sodium/day, respectively. Both diets contained 140 meq potassium/day). The diets provided approximately 2,900 kilocalories/day with 55% from carbohydrates, 15% from protein and 30 % from fats. On the sixth day of each diet, the subject wore an ambulatory blood pressure monitor to assess his/her blood pressure during a 24-hour period. A nurse and Amish liaison visited the subject on the third and fifth days of each diet to ensure that she/he was not adversely affected by the diets and to check her/his food diary for compliance. In addition, the subject was instructed to collect the first morning urine sample on days 4, 5, and 6 for measurement of sodium and creatinine as an independent assessment of compliance to the salt diets. The 3-day means of the urine sodium/creatinine ratios were 162.1 ± 60.8 meq/mg (range: 20.4 – 444.1) and 33.5 ± 18.6 meq/mg (range: 6.9 – 164.9) for the high and low salt diets, respectively, indicating a >4-fold decrease in urinary sodium excretion from high- to low-salt diets. During the high and low salt interventions, subjects also recorded into their diaries the times they went to bed and woke up in order to define active and inactive periods. Direct measurements of physical activity were obtained during this period by having subjects wear accelerometers (Actical, Mini Mitter/Respironics, Bend, OR) for 6 consecutive days.
DNA Collection Protocol and Genotyping
Genomic DNA was isolated from whole blood for genetic analyses on all participants. The quality of DNA was assessed by single SNP TaqMan genotyping (ABI) as well as by agarose gel electrophoresis as part of the protocol for whole genome genotyping with Affymetrix 500K SNP chips.
Description of Genetic Analysis Approaches
The primary phenotypes collected in this study are listed in Table II. The primary outcomes are the responses to the different interventions. These can be parameterized in a number of ways. For example, biologically relevant parameters that can be derived from the CPT include maximal blood pressure change from baseline, total blood pressure excursion [incremental area under the curve (iAUC)], or divided into reactivity (iAUC during the first 2 minutes) and recovery (iAUC from 3–5 minutes) 11. More detailed descriptions of response phenotypes will be the subject of future reports. Three types of genetic analyses will be carried out to detect genetic effects of response phenotypes in the HAPI Heart Study: heritability analysis to quantify the contribution of unmeasured additive genetic effects11, linkage analysis to detect genetic effects attributable to specific measured loci 12;13, and association analyses to detect measured genotype-phenotype associations. Power to detect these types of genetic effects in the HAPI Heart Study sample was estimated by simulating genotypes for founding members of the pedigrees, dropping alleles down through the pedigrees according to Mendel’s laws, and assigning phenotypic values for the 868 examined individuals under various genetic models 12;13.
Table II.
Phenotypic measurements in the HAPI Heart Study
| Category | Measurements |
|---|---|
| Demographics | Age, sex, marital status, parity, occupation |
| Medical history | Hypertension, diabetes, cancer, myocardial infarction, stroke, heart or carotid surgery, medication use |
| Lifestyle | Smoking, physical activity |
| Anthropometry and resting BP and HR | Height, weight, waist circumference, blood pressure (BP) and heart rate (HR) (three measures) |
| Baseline blood analyses | Complete blood count (CBC), fasting serum glucose, insulin, adiponectin, AST, ALT, TSH, BUN, creatinine, lipid profile and subfractions, lipid particle size, measures of inflammation |
| Baseline urine analyses | Sodium, creatinine |
| Other | Carotid intima media thickness (IMT), pulse wave velocity (PWV), ankle-brachial index (ABI), echocardiogram |
| Intervention-specific: | |
| High fat challenge | Lipids: fasting blood sample (just prior to the test) and at 1, 2, 3, 4, and 6 hours after the test |
| Brachial artery flow-mediated vasodilation (FMD): fasting and at 2, 4, and 6 hours after the test | |
| Cold pressor stress test | BP: stable baseline and during and after the test at 1, 2, 3, 4, 5, 7.5, 10, 15, and 20 minutes |
| HR: along with BP measurements | |
| Brachial artery reactivity (BART): before and during the test and after withdrawal from ice water | |
| High/low salt diets | Ambulatory BP and HR monitoring: on the 6th day of each diet for 24 hours |
| Urine sodium and creatinine: first morning void collected on days 4, 5, and 6 of each diet | |
| Aspirin | Platelet aggregation and platelet function assays: whole blood samples before and after 14 days of aspirin therapy using collagen, ADP and arachidonic acid as agonists |
| Urinary 11-dehydrothromboxane B2 (TXB2): first morning void before and after 14 days of aspirin therapy | |
RESULTS
Composition and Family Structures of the HAPI Heart Study
A total of 868 subjects were enrolled into the HAPI Heart Study between May 2003 and August 2006. Participants included members of 184 sibships, ranging in size from 2 – 10, plus an additional 344 individuals with no siblings in the study. A total of 425 of these subjects could be combined into a single pedigree by adding in the parents and grandparents of all examined individuals. Relationship types among the examined individuals included 633 sib pairs, 327 parent-offspring pairs, 443 avuncular (aunt/uncle-niece/nephew) pairs, and 191 first cousin pairs.
Clinical Characteristics of HAPI Heart Study Participants
Clinical characteristics of the 868 HAPI Heart Study participants are described in Table III. The mean age (± SD) of study subjects was 43.7 (± 14.0) yrs with a range of 20 – 80 yrs. Mean body mass was 25.6 kg/m2 and 27.8 kg/m2 in men and women, respectively. HDL cholesterol levels were relatively high and triglyceride levels low in this physically active Amish population.
Table III.
Clinical characteristics of HAPI Heart Study participants (n = 868), according to sex†
| Characteristic | Men
(n = 460) |
Women
(n = 408) |
|---|---|---|
| Age (yrs) | 42.2 ± 0.6 | 45.4 ± 0.7*** |
| BMI (kg/m2) | 25.6 ± 0.1 | 27.8 ± 0.3*** |
| TCHOL (mg/dl) | 202.5 ± 2.1 | 215.7 ± 2.5** |
| HDL (mg/dl) | 52.6 ± 0.6 | 59.5 ± 0.8*** |
| TG (mg/dl) | 63.9 ± 1.7 | 73.8 ± 2.3*** |
| SBP (mmHg) | 121.5 ± 0.6 | 121.4 ± 0.8 |
| DBP (mmHg) | 77.6 ± 0.4 | 75.8 ± 0.4*** |
| Diabetes (%) | 0.9 | 1.0 |
| Current smokers (%)§ | 20.0 | 0.0*** |
| % on lipid-lowering meds‡ | 1.1 | 1.0 |
| % on anti-hypertensive meds‡ | 0.2 | 0.3 |
values represent means ± SE (or percentiles)
p < 0.001;
p < 0.01, men vs. women, adjusted for age
medication use assessed at the time of recruitment (prior to participants being asked to discontinue their medication use)
includes use of cigarettes, cigars and pipes
Of the 868 subjects enrolled in the study, 466 completed all 4 interventions, 796 completed at least 3 interventions, 857 completed at least 2 interventions, and 867 completed at least 1 intervention. The salt intervention could not be offered throughout the entire study period, including during the wedding season each year (Nov-Dec); it was carried out in only 473 subjects. Table IV provides mean (± standard deviations) for several of the primary response traits and their heritabilities. The traits shown include the TG excursions following the high fat challenge, expressed as iAUC, the systolic and diastolic blood pressure responses to the CPT, computed as iAUC from 0 to 2 min during the CPT, the systolic and diastolic blood pressure responses to the high salt diet during the active (awake) period, computed as the differences in the means between the high and low salt diets, and the percent change in platelet aggregation to a 1 mcg/ml collagen agonist in response to 14 days of aspirin therapy. As expected, there is great inter-individual variation in responses to the same standardized interventions. Significant differences in mean response between men and women were observed for the triglyceride response to the high fat challenge (p < 0.001); there were no significant sex differences in the other response phenotypes.
Table IV.
Mean (± SE) values and heritabilities of selected primary response phenotypes†
| Phenotype | Men
(n = 460) |
Women
(n = 408) |
Heritability ± SE |
|---|---|---|---|
| High Fat Challenge | |||
| Triglyceride iAUC (6 hrs) | 552.0 ± 15.9 | 369.7 ± 19.3*** | 0.38 ± 0.08*** |
| Cold pressor stress test reactivity | |||
| SBP iAUC (2 min) | 27.3 ± 0.8 | 29.3 ± 1.2 | 0.16 ± 0.07** |
| DBP iAUC (2 min) | 19.9 ± 0.6 | 15.5 ± 0.9 | 0.24 ± 0.08*** |
| Salt Sensitivity (active period) | |||
| SBP | 0.5 ± 6.0 | 0.1 ± 0.5 | 0.08 ± 0.11 |
| DBP | −1.9 ± 0.3 | −1.4 ± 0.4 | 0.19 ± 0.12* |
| Aspirin | |||
| Collagen (1 ug/ml) | 6.8 ± 0.2 | 6.5 ± 0.3 | 0.19 ± 0.08** |
iAUC = incremental area under the curve
Sex differences adjusted for age; all heritability analyses adjusted for age, sex, and BMI (cold pressor analyses additionally adjusted for baseline heart rate)
p<0.05;
p<0.01;
p<0.001
The heritabilities of all response traits, after adjustment for age, sex, and BMI, differed significantly from zero, with the exception of the SBP response to the high salt diet during the active period. The heritabilities for the remaining traits ranged from 0.16 for the reactivity of SBP to the CPT (p = 0.007) to 0.38 for the triglyceride response to the high fat challenge (p < 0.001). More detailed heritability analyses will be the subject of future studies.
Power to Detect Genetic Effects
Power calculations indicated that this sample provides 69% power to detect a heritability of 10% and 89% power to detect a heritability of 15%, at α = 0.01. For linkage, our simulations indicated that we would have 57% power to detect a LOD score of 3.0 and 85% power to detect a LOD score of 2.0 for a quantitative trait locus (QTL) accounting for 20% of the total trait variation. Power to detect associations in this sample is excellent, with our simulations indicating 80% power to detect a QTL explaining 1–2% of phenotypic variation at α = 0.01, 2–3% of phenotypic variation at α = 0.0001, and 4–5% of phenotypic variation at α = 0.000001.
DISCUSSION
The HAPI Heart Study was designed to facilitate the mapping of genes that influence response to four different environmental perturbations relevant to CVD. Our design is an efficient way of studying gene by environment interactions since the between person variation across different environments is eliminated. Indeed, the heritability analyses indicate that for nearly all traits, there is statistical evidence that some of this variation can be attributed to the additive effects of genes. Furthermore, since the four interventions were performed in the same individuals, we may discern genes that have pleiotropic effects, e.g., blood pressure response to CPT and blood pressure response to a high salt diet.
The Amish in Lancaster County are predominantly Old Order Amish, the most conservative of the Amish sects in terms of adherence to strict social norms and resistance to using modern technologies. All Amish are rural-living and most earn their living by farming. Marriages between Amish and those of non-Amish descent are rare. Because of their unique ancestral background, there is a high degree of consanguinity. First cousin marriages are uncommon, but on average, any two Amish individuals are more closely related than second cousins, but less related than first cousins once removed, who share 1/64 and 1/32 of their genes in common, respectively.
This Amish population has several unique characteristics that make it particularly well suited for genetic studies. First, the population is a closed founder population that is genetically homogeneous. There are extensive genealogical records spanning 12–14 generations that allow researchers to trace all living Amish to their founding ancestors in the mid-1700s with minimal pedigree errors and/or non-paternity 9;14. Second, Amish families are characterized by very large sibships averaging 7–9 siblings each who are geographically localized. This, combined with extraordinary cooperation from the Amish community, greatly facilitates high rates of participation, remarkable adherence to complicated intervention protocols, and reliable follow-up. Third, the homogeneity of socioeconomic status and lifestyle (e.g., minimal smoking and alcohol consumption, abundant and consistent physical activity, limited medication use, and similar dietary routines) reduces sources of non-genetic variation. Despite the unique ancestral background of the Amish, it seems reasonable to expect that most common disease-predisposing alleles found in other European Caucasian populations will also be found in the Amish. Indeed, classical population genetic theory and our15 and others’ empirical data show that populations with a modest number of founders like the Amish have allele frequencies (on average) that are similar to those in the ancestral population, particularly for common alleles (e.g., minor allele frequencies > 5%).
The family-based design of the HAPI Heart Study provides additional opportunities for analyses that are not possible in conventional population-based studies. For example, heritability and linkage analysis contrast phenotypic similarities between related and unrelated individuals. Family-based tests of association that test for evidence of association in the presence of linkage, such as the TDT or FBAT, are also possible, although the added value of such tests is debatable in genetically homogenous isolate populations with minimal population stratification, such as the Amish. Lastly, the unique family structure of the HAPI Heart Study cohort facilitates investigations of genetic imprinting, maternal effects, and other non-classical genetic phenomenon that might play a role in complex diseases.
Conventional genetic association tests can also be carried out in families, with the caveat that the nonindependence of family members must be taken into account since failure to do so will inflate the variance of the genotypic effect size 13. Analytic approaches for doing this include accounting for family structure as a random effect in a mixed effect model or through generalized estimating equations (GEE) modeling or by specifying explicitly the covariance structure as a random effect using variance component modeling. Because of the unique ancestral background of this population with its relatively small number of founders, an additional analytic approach potentially useful in the Amish is to map recessively acting genes with rare alleles through homozygosity by descent mapping 16.
In conclusion, through its use of standardized interventions relevant to CVD, the HAPI Heart Study employs a powerful approach for assessing how genes modify the response to particular controlled environments. Identifying these response genes may identify new mechanisms influencing the regulation of cardiovascular health, leading eventually to improved and individualized preventive strategies and treatments and to improved early identification of high-risk individuals.
Acknowledgments
This work supported by the NIH Institutional Training Grant in Cardiac and Vascular Cell Biology (T32HL072751), and by grants U01 HL72515, the University of Maryland General Clinical Research Center (GCRC)(M01 RR 16500), the Johns Hopkins University GCRC (M01 RR 000052), National Center for Research Resources, the Clinical Nutrition Research Unit of Maryland (P30 DK072488), and the Paul Beeson Physician Faculty Scholars in Aging Program of the American Federation of Aging Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, de Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247–54. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 2.Wienke A, Herskind AM, Christensen K, Skytthe A, Yashin AI. The heritability of CHD mortality in danish twins after controlling for smoking and BMI. Twin Res Hum Genet. 2005;8:53–59. doi: 10.1375/1832427053435328. [DOI] [PubMed] [Google Scholar]
- 3.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–46. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 4.Hawe E, Talmud PJ, Miller GJ, Humphries SE. Family history is a coronary heart disease risk factor in the Second Northwick Park Heart Study. Ann Hum Genet. 2003;67:97–106. doi: 10.1046/j.1469-1809.2003.00017.x. [DOI] [PubMed] [Google Scholar]
- 5.Greenland P, Knoll MD, Stamler J, Neaton JD, Dyer AR, Garside DB, et al. Major risk factors as antecedents of fatal and nonfatal coronary heart disease events. JAMA. 2003;290:891–97. doi: 10.1001/jama.290.7.891. [DOI] [PubMed] [Google Scholar]
- 6.Khot UN, Khot MB, Bajzer CT, Sapp SK, Ohman EM, Brener SJ, et al. Prevalence of conventional risk factors in patients with coronary heart disease. JAMA. 2003;290:898–904. doi: 10.1001/jama.290.7.898. [DOI] [PubMed] [Google Scholar]
- 7.McKusick VA. Medical Genetic Studies of the Amish. Baltimore, MD: Johns Hopkins University Press; 1978. [Google Scholar]
- 8.Church Directory of the Lancaster County Amish. Gordonville, PA: Pequea Publishers; 2002. [Google Scholar]
- 9.Agarwala R, Biesecker LG, Hopkins KA, Francomano CA, Schäffer AA. Software for constructing and verifying pedigrees within large genealogies and an application to the Old Order Amish of Lancaster County. Genome Res. 1998;8:211–21. doi: 10.1101/gr.8.3.211. [DOI] [PubMed] [Google Scholar]
- 10.Cross HE. Population studies and the Old Order Amish. Nature. 1976;262:17–20. doi: 10.1038/262017a0. [DOI] [PubMed] [Google Scholar]
- 11.Roy-Gagnon MH, Weir MR, Sorkin JD, Ryan KA, Sack PA, Hines S, et al. Genetic influences on the blood pressure response to stress: Results from the HAPI Heart Study. J Hypertens. 2007 doi: 10.1097/HJH.0b013e3282f524b4. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsueh W-C, Mitchell BD, Schneider JL, Wagner MJ, Bell CJ, Nanthakumar E, et al. QTL influencing blood pressure maps to the region of PPH1 on chromosome 2q31–34 in Old Order Amish. Circulation. 2000;101:2810–16. doi: 10.1161/01.cir.101.24.2810. [DOI] [PubMed] [Google Scholar]
- 13.McArdle PF, O’Connell JR, Pollin TI, Baumgarten M, Shuldiner AR, Peyser PA, et al. Accounting for relatedness in family based genetic association studies. Hum Hered. 2007;64:234–42. doi: 10.1159/000103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwala R, Schäffer AA, Tomlin JF. Towards a complete North American Anabaptist Genealogy II: Analysis of inbreeding. Hum Biol. 2001;73:533–45. doi: 10.1353/hub.2001.0045. [DOI] [PubMed] [Google Scholar]
- 15.Newman DL, Hoffjan S, Bourgain C, Abney M, Nicolae RI, Profits ET, et al. Are common disease susceptibility alleles the same in outbred and founder populations? Eur J Hum Genet. 2004;12:584–90. doi: 10.1038/sj.ejhg.5201191. [DOI] [PubMed] [Google Scholar]
- 16.Abney M, Ober C, McPeek MS. Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum- insulin level in the Hutterites. Am J Hum Genet. 2002;70:920–34. doi: 10.1086/339705. [DOI] [PMC free article] [PubMed] [Google Scholar]