Abstract
THF-diols (9,12-oxy-10,13-dihydroxyoctadecanoic and 10,13-oxy-9,12-dihydroxyoctadecanoic acids) are endocrine disrupters in rats and mitogens in breast cancer cells. Microarray analyses and real-time PCR analyses on RNA from THF-treated MCF-7 cells revealed a number of genes (Caveolin 1, Heat Shock Protein 90α and 90β, vascular endothelial growth factor, ATPase, Ca++ transporting, ubiquitous) in the Nitric Oxide Pathway (NOP) were targets for THF-diols. Chromatin immunoprecipitation studies suggest THF-diols modify of histone H4 acetylation at the Caveolin 1 promoter via an epigenetic mechanism. These findings are consistent with the well-known involvement of NOP genes in cell proliferation and sexual behavior.
Key Terms: Tetrahydrofurandiol, Nitric Oxide, Human Breast Cancer, Growth Regulation, Microarray, Reproduction, Endocrine Disruptor
Introduction
The corncob bedding commonly used to house rats and mice has an adverse effect on their reproductive functions [1, 2]. Our laboratory extracted and identified a mitogen from corncob, also detected in commercially available fresh corn and corn tortillas, which blocks adult male rat sexual behavior (including reduced mounting and intromission frequencies, increased mount, intromission, and ejaculatory latencies and a diminished ejaculatory response) [3]. The corn mitogen also reduces adult female rat reproductive behavior (lordosis), disrupts the estrous cycle and cause causes persistent metestrus [3]. While it is well known that environmentally derived phytoestrogens also adversely affect reproductive function via ER binding interactions, the corncob mitogen does not bind to the estrogen receptor, and is capable of stimulating ER positive (MCF-7) and ER negative (MDA-MB-231) breast cancer cells [3]. Similarly, the effects of the corncob mitogen on MCF-7 cells are not blocked by a pure antiestrogen (ICI-182,780) [3], suggesting its effects are independent of classical estrogen control pathways.
High Pressure Liquid Chromatography (HPLC) purification of the corncob extract showed two peaks of mitogenic activity. Peak I was identified by gas chromatography-mass spectrometry (GC-MS) as an isomeric mixture of 9,12-oxy-10,13-dihydroxyoctadecanoic acid and 10,13-oxy-9,12-dihydroxyoctadecanoic acid (tetrahydrofurandiols, THF-diols). Similar to the corncob extract, synthetic THF-diol stimulated MCF-7 human breast cancer cell proliferation in vitro [1], did not bind ER or the nuclear type II [3H]estradiol binding sites and blocked sexual behavior in male and female rats [4] at a very low dose (~0.30 mg/kg body weight/day). The second mitogenic HPLC peak of mitogenic activity was subsequently identified as 9,10-dihydroxy-12-octadecenoic acid (leukotoxin diol; LTX-diol), a well-known leukotoxin [5]. A synthetic mixture of LTX-diol, and its 12,13-dihydroxy-9-octadecenoic acid isomer (iso-leukotoxin diol; i-LTXdiol), were separated by HPLC. Both LTX-diol isomers blocked sexual behavior in female rats at a low dose (~0.80 mg/kg body weight/day). However, unlike the THF-diols, the LTX-diol isomers did not disrupt male sexual behavior in adult male rats. Recent studies show that THF-diols and LTX-diols act additively to disrupt endocrine function at concentrations (0.5 to 1.0 ppm) [6], far lower than those of classical phytoestrogens [7]. While the corncob mitogens are powerful endocrine disruptors, the mechanism through which these mitogens act has not been elucidated. Recently, our laboratory demonstrated that THF-diol stimulates phospholipase A2 (PLA2), lipoxygenases (LOX-5 and LOX-12) and cyclooxygenases (COX-1 and COX-2) in MCF-7 cells [8]. The products of these enzymes (prostaglandins, hydroxyeicosatetraenoic acids (HETE’s) and hydroxyoctadecenioc acids (HODE’s)) are well known regulators of cell growth [9-11]. The data presented in this manuscript suggests that the THF-diols may function by activating the production of nitric oxide.
Materials and Methods
MCF-7 Cell Growth Conditions
Stock cultures of MCF-7 human breast cancer cells were grown in T-150 flasks as previously described [12]. For microarray studies, the cells were seeded into T-75 flasks containing 10 mls of phenol red free DMEM media containing 5% charcoal-stripped fetal calf serum (FCS, Gibco, Carlsbad, CA, USA) and 1% penicillin-streptomycin and allowed to attach for 24 hours. After a media change, the cells grown for 24 hours in media containing 2 μl of ethanol (controls) or 8 μg/ml of THF-diol (synthesized in our lab as previously described [3]) in 2 μl of ethanol. After incubation, 5 million cells from triplicate flasks for each treatment group were harvested for RNA isolation. Viable, attached cell numbers were monitored by hemocytometer counts based on trypan blue dye exclusion [13].
RNA and Protein Preparation
Cells from EtOH controls or THF-diol-treated flasks were washed with PBS and collected with 0.25% trypsin-EDTA (4 mls). Following 5 minute incubation, the trypsin was inactivated by the addition of 10 mls of media containing 10% FCS. Approximately 5.0 × 106 cells from each flask were centrifuged (2000 rpm × 5 minutes) in RNAse/DNAse free tubes, resuspended in 1 ml of PBS and 4 mls of RNAlater (Qiagen, Valencia, CA, USA) and stored at −20oC. The frozen cells were thawed on ice, collected by centrifugation and lysed by resuspension in 0.6 mls of RTL buffer (Qiagen) containing β-mercaptoethanol. The lysed cells were homogenized by centrifugation through a Qiashredder (18,000 × g × 2 minutes). The pass through from the Qiashredder containing nucleic acids was diluted with an equal amount of 70% EtOH and loaded onto a RNeasy spin column. The column was washed with RW1 followed by RNase-free DNase digestion to remove residual DNA and further washed with RPE buffers according to the manufacturer’s instructions. Purified total RNA was eluted with 50 μl of RNAse-free water following 5 minute RT incubation. RNA integrity was verified on an Agilent 2100 Bioanalyzer in the Baylor College of Medicine Microarray Core Laboratory.
Microarray Analyses
RNA from ethanol controls or THF-diol treated MCF-7 cells was subjected to oligo(deoxythymidine)-Reverse Transcription to generate cDNA, followed by in vitro transcription and biotin-labeling of to generate cRNA (Enzo Biochem, Farmingdale, NY, USA). The fragmented, biotin-labeled cRNA was hybridized to Human Genome U133 Plus 2.0 oligonucleotide arrays (Affymetrix, Santa Clara, CA, USA) containing approximately 54,000 probe sets (38,500 genes). Each transcript is represented on the chip as 11 probe pairs and each pair contained a perfect match and mismatch. Microarrays were stained with strepavidin antibody and streptavidin-phycoerythrin in an Affymetrix Fluidics station. Arrays were scanned at 3 μM with a GeneArray scanner (Affymetrix). All experiments were performed in triplicate with independent pools of cRNA from EtOH controls and THF-diol-treated MCF-7 cells using six separate microarray chips. Following low-level quantification of the scanned data using GeneChip Operating System (GCOS, version 1.4, Affymetrix), data were further analyzed with dChip 2006 (Harvard University, Cambridge, MA, USA) to adjust the arrays to a common baseline and to estimate expression using the PM-only model [14, 15]. All 6 Genechips were normalized to the same baseline (in this case, the Genechip with the median average intensity was ethanol Chip #3) and all were modeled together. Data quality was reviewed using present call rates from GCOS (average 40.16%, range 39.0% to 41.2%), ratios of 3′ to 5′ glyceraldehydes 3-phosphate dehydrogenase probe sets from GCOS (average 1.25: range 1.14 to 1.31) and array outlier rates from dChip (average 0.21%: range 0.0% to 0.80%). Differentially expressed genes in the THF-diol treated groups relative to ethanol controls were selected using a two-sample comparison with a lower boundary 90% confidence interval of fold change greater than 1.2 and a value difference between group means of >50. The medium number of detected genes in 50 permuted samples was used to estimate the false discovery rate. Ingenuity Pathway Analysis (Version 5.1, Ingenuity Systems, Redwood City, CA, USA) identified differentially expressed gene pathways according to canonical pathway.
Real-Time Quantitative Polymerase Chain Reaction (qPCR)
Pre-validated commercially available primers (Qiagen) for the Nitric Oxide Pathway genes were used. qPCR was performed using the MyiQ SYBR Green Supermix and quantified on MyiQ Single Color Real-Time PCR Detection System using MyiQ Optical System Software, version 2.0 (Bio-Rad, Hercules, CA, USA). Validation of each primer pair was accomplished by generating standard serial dilution and melt curves on cDNA from MCF-7 cells to ensure that reaction efficiencies of 90–110% and correlation coefficients of >0.995 are obtained. Melt curves demonstrating a single product with an appropriate melting temperature confirmed that primer dimerization was not problematic. Results from qPCR on triplicate pools of RNA from EtOH controls or THF-diol-treated cells were normalized to 18S RNA and analyzed statistically by Instat (Graphpad Software, Inc, San Diego, CA) with a suitable t-test on the treatment means. Products of the optimized reactions will be analyzed by agarose gel electrophoresis to ensure that the size of the amplicon corresponds to the data provided by Qiagen for each primer pair.
SDS PAGE Analyses
Cells were grown as described above and treated with THF-diol for 2, 4, 8 or 12 days. The cells lysed with Qiagen RTL buffer and pass through Qiashredders. Proteins from the extract were collected by precipitation with 4 volumes of ice cold acetone and centrifugation. The pellets were dried under nitrogen, re-dissolved in sample extraction buffer (0.05M Tris, pH 7.4, 8M urea, 1% SDS, 0.1% β-mercaptoethanol) containing 0.02% cetyltrimethylammonium bromide (CTAB; Aldrich, Milwaukee, WI) and diluted 2:1 with sample loading buffer (0.0625M Tris, pH 6.8, 20% glycerol + bromophenol blue). The solubilized samples were centrifuged and heated for 3 min at 90oC. The cooled samples were resolved on 4–12% NuPAGE Bis–Tris gradient gels using MES running buffer (Invitrogen).
Western Blot Analyses
The electrophoresed proteins were transferred to nitrocellulose membrane (Trans- Blot Transfer Medium, Bio-Rad), washed, and blocked in Tris-buffered saline (1X TBS) with 3.0% non-fat dried milk. Rinsed membranes were incubated 1–16 hours with primary antibody (1:200 dilution). Primary antibodies (HSP90α, HSP90β, ATP2A3, CAV1, β-actin) suitable for Western blots for human nitric oxide genes were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA, USA) Following a 60 minute incubation with the appropriate species-specific horse-radish peroxidase-conjugated secondary antibody (Millipore, Billerica, MA, USA), specific antigens were detected by the Visualizer Western Blot Detection Kit (Millipore) on X-OMAT Scientific Imaging Film (Eastman Kodak Company, Rochester, New York, USA). All blots were quantified with UN-SCAN-IT Gel Software (Silk Scientific Software, Orem, UT, USA). In order for the protein bands to be normalized with β-actin, the nitrocellulose membrane was stripped with Stripping Buffer II (0.4 M glycine, 0.2% SDS, Tween-20, pH 2.2) according to the manufacturer’s instructions prior to re-probing.
Chromatin Immunoprecipitation Assay
Following treatment of the MCF-7 cells as described above, the monolayers were fixed with formaldehyde to crosslink protein to DNA. The chromatin was then digested (17.5′) with Enzymatic Shearing Cocktail (ChIP-IT Kit, Active Motif) to generate DNA fragments averaging 200 bp in length. Aliquots of the digested chromatin were pre-cleared with Protein A-Sepharose and incubated with (+Ab) or without (−Ab) anti-acetylated (Lys 5, 8, 12, 16) histone H4 antibody. Following incubation with Protein A-Sepharose beads and cross-link reversal, the immunoprecipitated DNA was de-proteinized and analyzed by PCR. Primers for the promoter region of CAV1 (−216 to +18 bp) were as follows: forward primer: 5′-GAACCTTGGGGATGTGCC-3′ and reversed primer: 5′-GCCTACCTCCGAGTCTACG-3′ [16]. PCR products from the Input DNA and DEPC water were additional controls
Results
Microarray Hybridization Analysis of THF-diol Effects on Gene Expression in MCF-7 Breast Cancer Cells
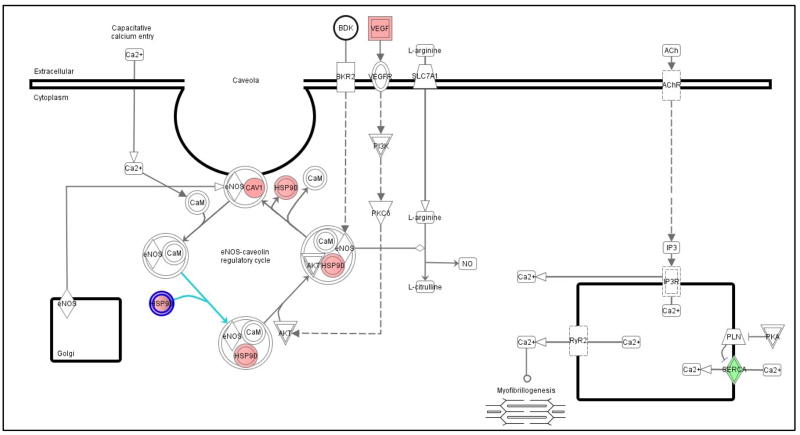
Genes regulated by THF-diol treatment were identified by comparing the mRNA expression pattern of MCF-7 human breast cancer cells treated with EtOH or THF-diol at a 8 μg/ml, a level previously shown to stimulate MCF-7 cells [1]. Microarray analysis utilizing the HG_U133 Plus 2.0 chips was performed as described in Materials and Methods. Of the 38,500 genes represented on the array, 39–41.2% of the genes were present in the MCF-7 cells. Of the present genes, 604 genes were expressed differentially between EtOH controls and THF-diol treated cells (Table I). Further analysis of the differentially expressed genes with Ingenuity Pathway Analysis software identified 7 regulatory pathways significantly (p value <0.05) modulated by THF-diol treatment (Table II). Due to the well known effects of nitric oxide on cellular growth and reproduction [17–19], the nitric oxide pathway was chosen for further examination. Five genes in the Nitric Oxide Pathway (Fig 1, Table III) were up-regulated (pink) or down-regulated (green) in MCF-7 cells following THF-diol treatment. Of these genes, only ATPase, Ca++ transporting, ubiquitous (ATP2A3) was down-regulated. Several other key genes in the pathway including caveolin 1 (CAV1), heat shock 90kDa protein 1, alpha (HSP90α), heat shock 90kDa protein 1, beta (HSP90β) and vascular endothelial growth factor (VEGF) were up-regulated by THF-diol treatment relative to the ethanol controls.
Table I.
Number of significantly increased or decreased MCF-7 cell genes 24 hours following treatment.
Percentage out of total on array (54,000 probe sets)
Table II.
MCF-7 pathways significantly affected by THF-diol treatment
| Pathway | Changed Genes | Genes in Pathway | Ratio | P Value |
|---|---|---|---|---|
| Hypoxia Signaling | 7 | 44 | 0.16 | 3.33E-04 |
| Ephrin Receptor Signaling | 9 | 113 | 0.08 | 7.57E-03 |
| Nitric Oxide Signaling | 5 | 41 | 0.12 | 7.98E-03 |
| PI3K/AKT Signaling | 7 | 89 | 0.08 | 1.92E-02 |
| Protein Ubiquination | 11 | 181 | 0.06 | 2.28E-02 |
| PDGF Signaling | 5 | 54 | 0.09 | 2.46E-02 |
| Propanoate Metabolism | 5 | 58 | 0.09 | 3.23E-02 |
Figure 1. Ingenuity Diagram of Nitric Oxide Pathway Genes Significantly Increased or Decreased by THF-diol Treatment.
D-Chip output was analyzed by Ingenuity to generate the diagram. The results represent mean fold-change in gene expression in response to THF-diol treatment relative to ethanol control. Significantly (p<0.05) up-regulated genes are in pink and down-regulated genes are in green.
Table III.
Nitric Oxide Pathway genes regulated by THF-diol
| Affy ID | Gene Title | Gene Symbol | Entrez Gene | Fold Change | Adjusted P Value |
|---|---|---|---|---|---|
| 207522_s_at | ATPase, Ca++ transporting, ubiquitous | ATP2A3 | 489 | −1.26 | 0.02849 |
| 203065_s_at | caveolin 1, caveolae protein, 22kDa | CAV1 | 857 | 1.45 | 0.001962 |
| 211968_s_at | heat shock 90kDa protein 1, alpha | HSP90α | 3320 | 1.20 | 0.00191 |
| 1557910_at | heat shock 90kDa protein 1, beta | HSP90β | 3326 | 1.45 | 0.039156 |
| 210512_s_at | vascular endothelial growth factor | VEGF | 7422 | 1.40 | 0.028475 |
Evaluation of THF-diol Effects on Nitric Oxide Genes in MCF-7 Cells by qPCR and Western Blot
To further confirm the microarray data (Fig 1) presented above, MCF-7 cells were treated with THF-diol or EtOH (controls) for 24 hours as described for the microarray studies. qPCR experiments on cDNA made from the total RNA from these cells were performed. Studies of the nitric oxide genes (Fig 2) show that THF-diol treatment inhibited ATP2A3 gene expression, as indicated by the microarray analysis (Table III) and stimulated the expression of CAV1, HSP90α, HSP90β and VEGF. In order to further confirm the affects of THF-diol gene expression in MCF-7 cells at the protein level, a series of western blots was performed. Protein extracted from treated cells was subjected to Western Blot analysis as described in Materials and Methods. VEGF is secreted and therefore not examined in this assay. The immunoblot bands were scanned with UN-SCAN-IT gel to determine the individual intensity of each band. HSP90α and HSP90β protein expression was increased by 4 days while CAV1 was increased by 8 days. In contrast, ATP2A3 protein expression was decreasing by 8 days. The qPCR (Fig 2) and Western Blot (Fig 3) data clearly demonstrate that THF-diol has a significant effect on genes in the nitric oxide pathway.
Figure 2. QPCR Analysis of Alteration of Nitric Oxide Gene Expression by THF-diol.
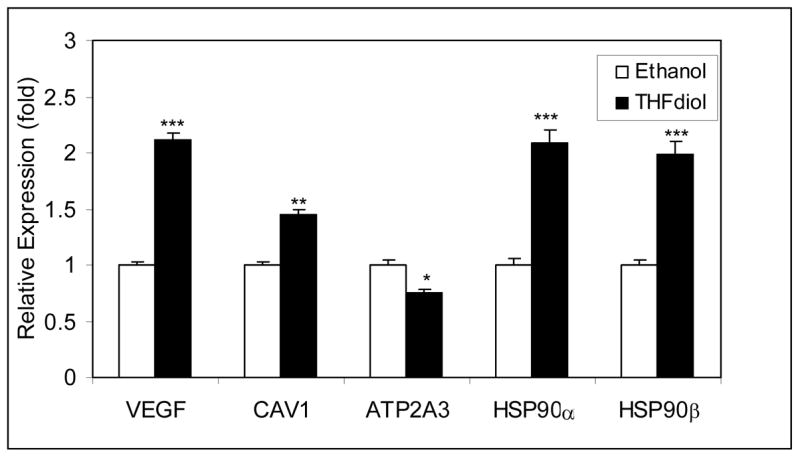
MCF-7 cells were treated for 24 Hours with 2 μl EtOH (controls) or THF-diol in 2 μl EtOH (final concentration of 8 μg/ml). RNA was extracted and analyzed by qPCR as described in Methods. The results represent the mean SEM for three independent RNA sets normalized to 18S RNA. Significantly different from control; *P<0.05, **p<0.01, ***p.0.001
Figure 3. Western Blot Analyses of Nitric Oxide Protein Expression Altered by THF-diol.
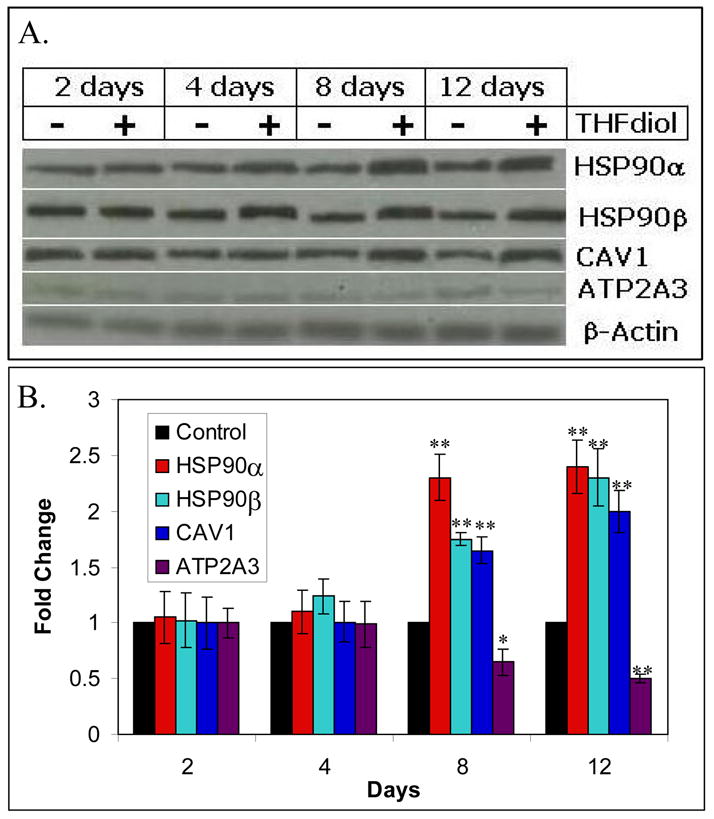
Panel A. Protein extracted from MCF-7 cells treated for 2, 4, 8 or 12 days with THF-diol were subjected to western blot analysis with EtOH-treated cells as a control. Panel B. Bands were analyzed by UNSCAN-IT gel to determine band intensity, normalized to β-actin and graphed according to the change in fold expression relative to the EtOH controls. Significantly different from EtOH control; *P<0.05, **p<0.001.
THF-diol Modification of Histone H4 Acetylation at the CAV1 Promoter
Following the discovery of NO regulation by THF-diol, we developed methodology for analyzing the acetylation state of histone H4 associated with the CAV1 promoter. MCF-7 cells treated with EtOH (control) or THF-diol as described above were harvested and chromatin prepared as described in Methods. The enzymatically digested chromatin consisting of 200 base pair fragments (Fig 4A) was immunoprecipitated with anti-acetyl histone H4 antibodies and analyzed by PCR with primers against the CAV1 promoter (Fig 4B). THF-diol treatment caused a significant increase in the intensity of the PCR signal. Quantitation of the signal with Un-Scan-It gel and normalization of the signal to the Input DNA revealed a 30% increase in signal of THF-diol treated cells compared to control (Fig 4C). These findings suggest that THF-diol stimulation of CAV1 gene expression occurred as a result from an alteration in the binding of histone acetyltransferases and/or histone deacetylases.
Figure 4. ChIP Assay for Caveolin-1 Promoter.
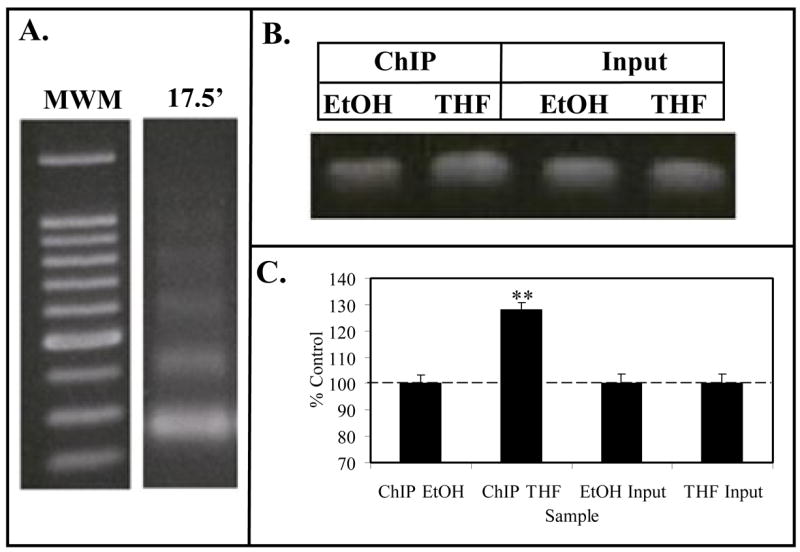
Panel A. Formaldehyde fixed MCF-7 cell chromatin was digested (17.5′) with Enzymatic Shearing Cocktail (Active Motif). Following cross-link reversal and proteinase K treatment, sheared DNA was electrophoresed on 1% agarose and stained with ethidium bromide. The gel contains a 100 bp molecular weight ladder (MWM), and DNA that yielded 100–200 bp fragments suitable for ChIP assays. Panel B (ChIP Assay). Following the immunoprecipitation chromatin from EtOH (controls) or THF-diol treated MCF-7 cells, the DNA was subjected to PCR analysis (36 cycles) with primers for the CAV1 promoter. PCR products from the Input DNA (1:10 dilution) are shown as additional controls. Results from 3 PCR reactions (as in panel B) for EtOH controls or THF-diol treated cells were quantified with UN-SCAN-IT (Silk Scientific Software), normalized to Input DNA and analyzed with Instat (panel C).
Discussion
Previous studies in our laboratory identified THF-diols [1] and LTX-diols [5] as endocrine disruptors from corncob animal bedding with mitogenic activity in breast and prostate cancer cells. LTX-diols and THF-diols block sexual behavior and cyclicity in adult female rats [1, 5]. Although the THF-diols also block male sexual behavior, LTX-diols apparently lack this biological activity [4]. While the exact mechanism of action of these compounds remains unknown, the regulation of genes in the nitric oxide pathway by THF-diols described by the present studies is consistent with the well-known effects of these compounds on reproductive function [3, 4, 6] and their mitogenic activities in tumor cells [1, 3, 8].
Nitric oxide (NO) is synthesized from L-arginine by nitric oxide synthase (NOS), an enzyme present in three isoforms. Neuronal NOS (NOS-1 or nNOS) and endothelial NOS (NOS-3 or eNOS, studied in this manuscript) require calcium/calmodulin for activation to produce a continuous basal release of NO [20, 21]. The third isoform is a calcium-independent, inducible form (iNOS or NOS-2), which is expressed in response to pro-inflammatory cytokines [22]. Apart from calcium, several other factors can regulate the activity of the NOS isoforms. eNOS possesses site for myristylation, which links it to the cell membrane [23]. In addition, eNOS activity is dependent on substrate concentration as well as the presence of co-factors NADPH, flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD) and (6R)-5, 6, 7, 8-tetrahydro-L-biopterin (THB) [24]. Phosphorylation at Serine 1177 in near the carboxy-terminal is required for eNOS activation by calcium influx. Phosphorylation of Serine 633 in the flavin mononucleotide (FMN) increases eNOS activity and maintenance of NO synthesis [25]. Several proteins are involved in the regulation of eNOS. HSP90 facilitates the binding of calmodulin to eNOS and increases eNOS activity at low and high calcium levels [26]. VEGF signaling leads to the activation of AKT, which binds to eNOS in a complex with HSP90 and calmodulin. AKT phosphorylation of eNOS is required for the NO synthesis at low calcium levels [27]. An increase in CAV1 expression leads to activation of eNOS [28]. In addition, the suppression of ATP2A3 in breast cancer cells leads to a release of calcium, which stimulates eNOS [29]. Thus, the regulation of these genes by THF-diol suggests a possible role for THF-diol in the control of the synthesis of nitric oxide.
The identification of genes specifically regulated by THF-diol provides targets that will aid in the elucidation of the underlying mechanism of action of these endocrine disrupter/mitogens. As seen in Figure 4, treatment of MCF-7 cells with THF-diol directly affects the acetylation state of lysines 5, 8, 12 and 16 of histone H4 associated with the CAV1 promoter. Acetylation of these four lysines is typically seen in active euchromatin while histones in heterochromatin are hypoacetylated [30]. In particular, active transcription coincides with acetylation of lysines 8 and 16, while histone disposition is typically associated with acetylation of lysines 5 and 12 [31]. Therefore, it is not surprising that THF-diol up-regulation of CAV1 correlates with elevated acetylation histones at the promoter. Utilization of ChIP assays and the knowledge of what promoters are affected will allow us to identify other post-translational nucleosome modifications associated with THF-diol. Studies on the mechanism of action of THF-diol and related metabolites may lead to a new understanding of the effects of these compounds on both cancer cell growth and proliferation and reproductive function.
NO plays an important role in a variety of physiological systems and pathways, such asvasodilation in the vasculature, neurotransmission in neurons and as an antimicrobial agent from macrophages [32]. However, it has a profound role in cancer. NOS activity has been detected in a variety of tumor cells and is associated with tumor grade, proliferation rate and expression of important signaling components in cancer development and progression. High levels of NOS expression are cytostatic for tumor cells while a low level promotes tumor growth [17]. In the murine mammary tumor model, NO promoted tumor growth and metastasis by enhancing invasive, angiogenesis and migratory capacities in tumor cells [18]. NO-induced p53 mutations can result in inactivation of p53 and loss of regulatory control. In addition, NO has a demonstrated role in cancer cell growth and progression as inhibition of NOS leads to a reduction of tumor cell growth [33]. The role of NO in tumor cell growth may explain the mitogenic action of the THF-diols in human breast cancer and prostate cancer cells.
In addition to NO’s role in cancer, this molecule is also involved in reproductive function [19]. All three isoforms of NOS are present in the rat cervix, while the uterus expresses iNOS and eNOS [34]. During labor, nNOS and iNOS expression increases in the cervix and iNOS decreases in the uterus, while eNOS levels remain constant. NOS inhibition by NG-nitro-L-arginine methyl ester demonstrates a prolonged duration of delivery and decreased cervical extensibility. NO plays an important role during fertilization in the oviduct. iNOS and eNOS expression regulates the transport of gametes and embryos in addition to providing a specific microenvironment that facilitates the fertilization process [35]. In the ovary, increases in NO correlate with increases in estrogen, regulates follicular development [36] and ovulation [37]. In the male rabbits, NO induces penile erection [38]. NOS activity localized to the rat penile neurons innervate the corpora cavernosa and eNOS present in the endothelium of penile vasculature act as a physiological mediator of erectile function [39]. The activity of eNOS in the testis regulates blood flow, cell permeability and contractile function of myofibroblasts, leading to steroid synthesis and transport [40]. NO also plays a role in sperm mobility with low levels (physiological) enhancing mobility and high levels (pathological) inhibiting mobility [41]. In addition to the above effects, NO regulates the release of gonadotropin hormone releasing hormone (GnRH) from the hypothalamus by stimulating phospholipase A2 and prostaglandin E2 [42]. GnRH then stimulates the production of sex steroids in the gonads leading to spermatogenesis, ovulation, mating behavior, lordosis and penile erection. THF-diol likely acts on many or all of these pathways through its regulation of NO. A recent study by our laboratory is consistent with these findings and has shown that THF-diols modulate the expression of genes controlling phospholipase A2, lipoxygenase and cyclooxygenase in breast cancer cells [8]. Thus, the endocrine disruptive and mitogenic properties of THF-diols appear closely coupled to nitric oxide controlled pathways regulating both cellular proliferation and sexual behavior.
Acknowledgments
This research is supported by grants from the National Institute for Environmental Health Sciences (ES-009964), the Office of Research on Women’s Health and National Cancer Institute (CA 35480).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Markaverich BM, Alejandro MA, Markaverich D, Zitzow L, Casajuna N, Camarao N, Hill J, Bhirdo K, Faith R, Turk J, Crowley JR. Identification of an endocrine disrupting agent from corn with mitogenic activity. Biochem Biophys Res Commun. 2002;291(3):692–700. doi: 10.1006/bbrc.2002.6499. [DOI] [PubMed] [Google Scholar]
- 2.Port CD, Kaltenbach JP. The effect of corncob bedding on reproductivity and leucine incorporation in mice. Lab Anim Care. 1969;19(1):46–9. [PubMed] [Google Scholar]
- 3.Markaverich B, Mani S, Alejandro MA, Mitchell A, Markaverich D, Brown T, Velez-Trippe C, Murchison C, O’Malley B, Faith R. A novel endocrine-disrupting agent in corn with mitogenic activity in human breast and prostatic cancer cells. Environ Health Perspect. 2002;110(2):169–77. doi: 10.1289/ehp.02110169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mani SK, Reyna AM, Alejandro MA, Crowley J, Markaverich BM. Disruption of male sexual behavior in rats by tetrahydrofurandiols (THF-diols) Steroids. 2005;70(11):750–4. doi: 10.1016/j.steroids.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Markaverich BM, Crowley JR, Alejandro MA, Shoulars K, Casajuna N, Mani S, Reyna A, Sharp J. Leukotoxin diols from ground corncob bedding disrupt estrous cyclicity in rats and stimulate MCF-7 breast cancer cell proliferation. Environ Health Perspect. 2005;113(12):1698–704. doi: 10.1289/ehp.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markaverich BM, Alejandro M, Thompson T, Mani S, Reyna A, Portillo W, Sharp J, Turk J, Crowley JR. Tetrahydrofurandiols (THF-diols), leukotoxindiols (LTX-diols), and endocrine disruption in rats. Environ Health Perspect. 2007;115(5):702–8. doi: 10.1289/ehp.9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markaverich BM, Webb B, Densmore CL, Gregory RR. Effects of coumestrol on estrogen receptor function and uterine growth in ovariectomized rats. Environ Health Perspect. 1995;103(6):574–81. doi: 10.1289/ehp.95103574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markaverich BM, Crowley J, Rodriquez M, Shoulars K, Thompson T. Tetrahydrofurandiol stimulation of phospholipase A2, lipoxygenase, and cyclooxygenase gene expression and MCF-7 human breast cancer cell proliferation. Environ Health Perspect. 2007;115(12):1727–31. doi: 10.1289/ehp.10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nolan RD, Danilowicz RM, Eling TE. Role of arachidonic acid metabolism in the mitogenic response of BALB/c 3T3 fibroblasts to epidermal growth factor. Mol Pharmacol. 1988;33(6):650–6. [PubMed] [Google Scholar]
- 10.Glasgow WC, Eling TE. Structure-activity relationship for potentiation of EGF-dependent mitogenesis by oxygenated metabolites of linoleic acid. Arch Biochem Biophys. 1994;311(2):286–92. doi: 10.1006/abbi.1994.1239. [DOI] [PubMed] [Google Scholar]
- 11.Glasgow WC, Eling TE. Epidermal growth factor stimulates linoleic acid metabolism in BALB/c 3T3 fibroblasts. Mol Pharmacol. 1990;38(4):503–10. [PubMed] [Google Scholar]
- 12.Markaverich BM, Alejandro MA. Type II [3H]estradiol binding site antagonists: inhibition of normal and malignant prostate cell growth and proliferation. Int J Oncol. 1998;12(5):1127–35. doi: 10.3892/ijo.12.5.1127. [DOI] [PubMed] [Google Scholar]
- 13.Markaverich BM, Shoulars K, Alejandro MA. Nuclear type II [3H]estradiol binding site ligands: inhibition of ER-positive and ER-negative cell proliferation and c-Myc and cyclin D1 gene expression. Steroids. 2006;71(10):865–74. doi: 10.1016/j.steroids.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2(8):RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98(1):31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tirado OM, Mateo-Lozano S, Villar J, Dettin LE, Llort A, Gallego S, Ban J, Kovar H, Notario V. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Res. 2006;66(20):9937–47. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 17.Xu W, Liu LZ, Loizidou M, Ahmed M, Charles IG. The role of nitric oxide in cancer. Cell Res. 2002;12(5–6):311–20. doi: 10.1038/sj.cr.7290133. [DOI] [PubMed] [Google Scholar]
- 18.Jadeski LC, Chakraborty C, Lala PK. Role of nitric oxide in tumour progression with special reference to a murine breast cancer model. Can J Physiol Pharmacol. 2002;80(2):125–35. doi: 10.1139/y02-007. [DOI] [PubMed] [Google Scholar]
- 19.Rosselli M. Nitric oxide and reproduction. Mol Hum Reprod. 1997;3(8):639–41. doi: 10.1093/molehr/3.8.639. [DOI] [PubMed] [Google Scholar]
- 20.Griffith OW, Stuehr DJ. Nitric oxide synthases: properties and catalytic mechanism. Annu Rev Physiol. 1995;57:707–36. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 21.Dixit VD, Parvizi N. Nitric oxide and the control of reproduction. Anim Reprod Sci. 2001;65(1–2):1–16. doi: 10.1016/s0378-4320(00)00224-4. [DOI] [PubMed] [Google Scholar]
- 22.Karpuzoglu E, Ahmed SA. Estrogen regulation of nitric oxide and inducible nitric oxide synthase (iNOS) in immune cells: implications for immunity, autoimmune diseases, and apoptosis. Nitric Oxide. 2006;15(3):177–86. doi: 10.1016/j.niox.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Busconi L, Michel T. Endothelial nitric oxide synthase. N-terminal myristoylation determines subcellular localization. J Biol Chem. 1993;268(12):8410–3. [PubMed] [Google Scholar]
- 24.Tsumori M, Murakami Y, Koshimura K, Kato Y. Endogenous nitric oxide inhibits growth hormone secretion through cyclic guanosine monophosphate-dependent mechanisms in GH3 cells. Endocr J. 1999;46(6):779–85. doi: 10.1507/endocrj.46.779. [DOI] [PubMed] [Google Scholar]
- 25.Mount PF, Kemp BE, Power DA. Regulation of endothelial and myocardial NO synthesis by multi-site eNOS phosphorylation. J Mol Cell Cardiol. 2007;42(2):271–9. doi: 10.1016/j.yjmcc.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi S, Mendelsohn ME. Calmodulin-dependent and -independent activation of endothelial nitric-oxide synthase by heat shock protein 90. J Biol Chem. 2003;278(11):9339–44. doi: 10.1074/jbc.M212651200. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi S, Mendelsohn ME. Synergistic activation of endothelial nitric-oxide synthase (eNOS) by HSP90 and Akt: calcium-independent eNOS activation involves formation of an HSP90-Akt-CaM-bound eNOS complex. J Biol Chem. 2003;278(33):30821–7. doi: 10.1074/jbc.M304471200. [DOI] [PubMed] [Google Scholar]
- 28.Pan YM, Yao YZ, Zhu ZH, Sun XT, Qiu YD, Ding YT. Caveolin-1 is important for nitric oxide-mediated angiogenesis in fibrin gels with human umbilical vein endothelial cells. Acta Pharmacol Sin. 2006;27(12):1567–74. doi: 10.1111/j.1745-7254.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 29.Baldi C, Vazquez G, Boland R. Capacitative calcium influx in human epithelial breast cancer and non-tumorigenic cells occurs through Ca2+ entry pathways with different permeabilities to divalent cations. J Cell Biochem. 2003;88(6):1265–72. doi: 10.1002/jcb.10471. [DOI] [PubMed] [Google Scholar]
- 30.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 31.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 32.Rosselli M, Keller PJ, Dubey RK. Role of nitric oxide in the biology, physiology and pathophysiology of reproduction. Hum Reprod Update. 1998;4(1):3–24. doi: 10.1093/humupd/4.1.3. [DOI] [PubMed] [Google Scholar]
- 33.Orucevic A, Lala PK. Effects of N(g)-methyl-L-arginine, an inhibitor of nitric oxide synthesis, on interleukin-2-induced capillary leakage and antitumor responses in healthy and tumor-bearing mice. Cancer Immunol Immunother. 1996;42(1):38–46. doi: 10.1007/s002620050249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buhimschi I, Ali M, Jain V, Chwalisz K, Garfield RE. Differential regulation of nitric oxide in the rat uterus and cervix during pregnancy and labour. Hum Reprod. 1996;11(8):1755–66. doi: 10.1093/oxfordjournals.humrep.a019481. [DOI] [PubMed] [Google Scholar]
- 35.Rosselli M, Dubey RK, Rosselli MA, Macas E, Fink D, Lauper U, Keller PJ, Imthurn B. Identification of nitric oxide synthase in human and bovine oviduct. Mol Hum Reprod. 1996;2(8):607–12. doi: 10.1093/molehr/2.8.607. [DOI] [PubMed] [Google Scholar]
- 36.Bonello N, McKie K, Jasper M, Andrew L, Ross N, Braybon E, Brannstrom M, Norman RJ. Inhibition of nitric oxide: effects on interleukin-1 beta-enhanced ovulation rate, steroid hormones, and ovarian leukocyte distribution at ovulation in the rat. Biol Reprod. 1996;54(2):436–45. doi: 10.1095/biolreprod54.2.436. [DOI] [PubMed] [Google Scholar]
- 37.Van Voorhis BJ, Moore K, Strijbos PJ, Nelson S, Baylis SA, Grzybicki D, Weiner CP. Expression and localization of inducible and endothelial nitric oxide synthase in the rat ovary. Effects of gonadotropin stimulation in vivo. J Clin Invest. 1995;96(6):2719–26. doi: 10.1172/JCI118339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmquist F, Stief CG, Jonas U, Andersson KE. Effects of the nitric oxide synthase inhibitor NG-nitro-L-arginine on the erectile response to cavernous nerve stimulation in the rabbit. Acta Physiol Scand. 1991;143(3):299–304. doi: 10.1111/j.1748-1716.1991.tb09236.x. [DOI] [PubMed] [Google Scholar]
- 39.Burnett AL, Nelson RJ, Calvin DC, Liu JX, Demas GE, Klein SL, Kriegsfeld LJ, Dawson VL, Dawson TM, Snyder SH. Nitric oxide-dependent penile erection in mice lacking neuronal nitric oxide synthase. Mol Med. 1996;2(3):288–96. [PMC free article] [PubMed] [Google Scholar]
- 40.Davidoff MS, Middendorff R, Mayer B, Holstein AF. Nitric oxide synthase (NOS-I) in Leydig cells of the human testis. Arch Histol Cytol. 1995;58(1):17–30. doi: 10.1679/aohc.58.17. [DOI] [PubMed] [Google Scholar]
- 41.Hellstrom WJ, Bell M, Wang R, Sikka SC. Effect of sodium nitroprusside on sperm motility, viability, and lipid peroxidation. Fertil Steril. 1994;61(6):1117–22. doi: 10.1016/s0015-0282(16)56766-1. [DOI] [PubMed] [Google Scholar]
- 42.McCann SM, Rettori V. The role of nitric oxide in reproduction. Proc Soc Exp Biol Med. 1996;211(1):7–15. doi: 10.3181/00379727-211-43950b. [DOI] [PubMed] [Google Scholar]