Abstract
The annexin A2 (A2) heterotetramer, consisting of two copies of A2 and two copies of S100A10/p11, promotes fibrinolytic activity on the surface of vascular endothelial cells by assembling plasminogen and tissue plasminogen activator (tPA) and accelerating the generation of plasmin. In humans, overexpression of A2 by acute promyelocytic leukemia cells is associated with excessive fibrinolysis and hemorrhage, whereas anti-A2 autoantibodies appear to accentuate the risk of thrombosis in patients with anti-phospholipid syndrome. Complete deficiency of A2 in mice leads to a lack of tPA cofactor activity, accumulation of intravascular fibrin, and failure to clear arterial thrombi. Within the endothelial cell, p11 is required for Src kinase-mediated tyrosine phosphorylation of A2, which signals translocation of both proteins to the cell surface. Here we show that p11 is expressed at very low levels in the absence of A2 both in vitro and in vivo. We demonstrate further that unpartnered p11 becomes polyubiquitinated and degraded via a proteasome-dependent mechanism. A2 stabilizes intracellular p11 through direct binding, thus masking an autonomous p11 polyubiquitination signal that triggers proteasomal degradation. This interaction requires both the p11-binding N-terminal domain of A2 and the C-terminal domain of p11. This mechanism prevents accumulation of free p11 in the endothelial cell and suggests that regulation of tPA-dependent cell surface fibrinolytic activity is precisely tuned to the intracellular level of p11.
Vascular endothelial cells modulate blood fluidity through the elaboration of secreted products and through expression of membrane-associated thromboresistance molecules (1). Fluid phase products include endothelin, nitric oxide, tissue plasminogen activator, and prostacylin, whereas cell surface regulators include tissue factor pathway inhibitor, heparan sulfate-bound antithrombin III, and the thrombomodulin-endothelial cell protein C receptor complex (2). In addition, endothelial cells assemble fibrinolytic proteins through expression of cell surface receptors such as the urokinase receptor and annexin A2. The latter systems activate plasmin and limit fibrin accumulation in response to vascular injury (3).
Annexin A2 (A2)2 is one of a dozen annexin family members expressed in humans (4, 5). The annexins are calcium-regulated, membrane-binding proteins that possess a C-terminal phospholipid-binding core domain as well as an N-terminal ligand-interacting domain. Although cytoplasmic A2 is mainly monomeric, membrane-associated A2 is part of a heterotetrameric complex in which the N termini of two copies of A2 bind to dual copies of the S100 family protein, S100A10 (p11) (6). Complex formation with p11 increases the affinity of A2 for calcium and phospholipid, thereby directing it to membrane surfaces (7). In addition, the A2 heterotetramer specifically binds tissue plasminogen activator as well as its substrate, plasminogen, and strongly accelerates the catalytic efficiency of plasmin generation in vitro (8, 9).
Several lines of evidence indicate that the A2 complex regulates fibrin homeostasis in vivo, both under basal conditions and in response to vascular injury. First, A2–/– microvascular endothelial cells were found to lack tPA cofactor activity, and the fibrin content of highly perfused A2–/– tissues was ∼2-fold greater than that observed in wild type controls (10). Second, arterial injury in A2–/– mice was followed by a 2-fold increase in the degree of thrombotic vascular occlusion with an equivalent reduction in blood flow recovery. Third, injury-induced carotid artery thrombosis in rats was averted by pretreatment with recombinant A2 (11). Fourth, cerebral infarct size was reduced and cerebral blood flow increased upon infusion of recombinant A2 in a rat model of embolic stroke (12). Fifth, A2 was strikingly overexpressed in blast cells and associated with hyperfibrinolysis and hemorrhage in patients with acute promyelocytic leukemia (13). Sixth, autoantibodies directed against A2 were highly prevalent and highly associated with clinical thrombosis in patients with the anti-phospholipid syndrome (14). These findings suggest that the A2-p11 complex is a significant regulator of in vivo fibrin balance.
Protein p11 (also known as S100A10) belongs to the S100 family of proteins, which consists of some 19 separate gene products ranging in molecular mass from 9 to 14 kDa (15, 16). Members are characterized by two EF-hand type calcium-binding motifs, each represented by a helix-loop-helix structure, defined by a calcium-binding loop flanked by two α-helices. At least three different S100 proteins (S100A11, S100A10, and S100A6) bind annexins (annexins 1, 2, and 11, respectively) through amphipathic helices located in the annexin N-terminal interaction domains (4). The crystal structure of p11 in complex with the N-terminal 13 amino acids of A2 supports this model and suggests that the basic unit of p11 structure is a noncovalently linked homodimer, each component of which binds the A2 tail peptide to form a heterotetramer (17). Upon binding to p11, the A2 tail peptide assumes an amphipathic helix that introduces key hydrophobic residues (Val3, Ile6, and Leu10) within a hydrophobic cleft formed by loop 2 and helix IV of one p11 monomer, and helix I of the other (17, 18). Within the C-terminal helix IV region of p11 (85YFVVHM90), hydrophobic residues, such as Tyr85 and Phe86, contribute additional contact points for binding to A2 (19).
In canonical S100 proteins, calcium binding induces a conformational change that places helix III in a more perpendicular orientation relative to helix IV, forming a cleft that can more readily accept target proteins (15). However, p11 is an exception to the calcium activation rule, as it permanently assumes a “calcium-on” state, due to replacement of the monodentate Asp56 with Cys61 and the bidentate Glu65 with Ser70 (20, 21). Pairing of intracellular p11 and A2 thus occurs constitutively and consequently is regulated by the concentration of each binding partner, rather than by calcium flux. Among the few agents known to selectively increase p11 without altering cellular A2 are interferon-γ (22) and thrombin (23).
p11 appears to function in the trafficking and membrane anchorage of several membrane proteins (24). For example, the tetrodotoxin-resistant sodium channel (Nav 1.8) (25, 26), the acid-sensitive potassium channel (TASK-1) (27, 28), the transient receptor potential channels (TRPV5 and TRPV6) (29), the acid-sensing ion channel (ASIC1a) (30), and the plasma membrane-resident serotonin receptor (5-HT1B) (31) are all integral membrane proteins that appear to require p11 binding for their cell surface presentation and activity. Similarly, we have shown that expression of the annexin A2-p11 complex on the surface of endothelial cells is a dynamic process that is stimulated by cellular injury such as that seen in heat shock. In addition to Src-mediated tyrosine 23 phosphorylation of A2, translocation of A2 to the endothelial cell surface requires expression of p11, because silencing of p11 expression specifically prevents the appearance of new A2 on the cell surface (32).
Previously, Puisieux and Ozturk (33) reported that transfection of HepG2 cells with an A2 expression construct leads to new expression of A2, as well as up-regulation of p11 through a post-translational mechanism. In this study, we report that in the absence of A2, endothelial cell p11 is rapidly polyubiquitinated and degraded via a proteasome-dependent pathway both in vitro and in vivo. We have identified an autonomous degradation determinant, or “degron,” on p11 that is specifically masked upon association of p11 with A2. We have mapped its location to the C-terminal extension of p11. In the absence of sufficient A2, we suggest that polyubiquitin-mediated degradation of p11 prevents accumulation of free p11, thus maintaining a precise compartmentalization of A2 monomer and tetramer. Because p11 is required for translocation of A2 to the cell surface (32), our data suggest that A2-mediated endothelial cell fibrinolysis is a dynamic function, subject to regulation by ambient p11 levels.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—N-Carbobenzoxyl-l-leucinyl-l-norleucinal (MG132), lactacystin, leupeptin, trans-epoxysuccinyl-l-leucylamido-4-guanidino butane, N-α-p-tosyl-l-lysine chloromethyl ketone were obtained from Calbiochem-Novabiochem. Bortezomib was from Millennium. Other chemicals were purchased from Sigma. The following antibodies were purchased from the indicated vendors: anti-actin, anti-GAPDH, anti-Myc 9E10 (Santa Cruz Biotechnology); anti-β-tubulin (Sigma); anti-A2 and anti-p11 (BD Transduction Laboratories); anti-hemagglutinin (HA) (Roche Applied Science or Zymed Laboratories Inc.); anti-FLAG M2 (Sigma); anti-GFP (Zymed Laboratories Inc. or Clontech). Polyclonal antibodies against a peptide sequence of mouse p11 were produced and affinity-purified at Covance.
Mice—A2–/– were generated as described previously (10). All animal experiments were according to IACUC guidelines.
Plasmid Constructs—The human p11 pET23 expression vector was a generous gift from Professor Volker Gerke (University of Meunster, Germany). C-terminal truncations and N-terminal FLAG-tagged (DYKDDDDK) versions of the p11 cDNA were constructed in the mammalian expression vector pcDNA3.1 (Invitrogen). Full-length or truncated cDNA fragments encoding human A2 were generated by standard PCR methods and subcloned into the pcDNA3.1(+)MycHis vector (Invitrogen). HA-ubiquitin wild type and K48R and K29R mutant plasmids were provided by Dr. Ze'ev Ronai (Mt. Sinai School of Medicine) (34). The fusion plasmid GFP-p11(86–97) was constructed by appending p11 C terminus sequence at amino acids 86–97 to the C-terminal end of GFP (Clontech).
Endothelial Cells and Cell Lines—A2+/+ and A2–/– mouse cardiac microvascular endothelial cells (CMEC) were obtained as described previously (10). First passage subconfluent CMEC were immortalized via sequential transduction with highly efficient DNA flap lentiviral vectors encoding human telomerase (100 ng p24 units/ml) or SV40 large T antigen (20 ng p24 units/ml) in EGM-2 medium (35, 36). Individually transduced cells were isolated by limiting dilution, and clones were selected on the basis of cobblestone morphology, contact inhibition at confluence, and uptake of acetylated low density lipoprotein. Cells transduced in this manner, as well as their conditioned media, were shown to be free of detectable retroviral particles, using a sensitive enzyme assay for p24 in the first and second passages. Human umbilical vein endothelial cells were harvested and propagated as described (8).
Transfections—HEK 293 cells were transiently transfected using DNA-calcium phosphate precipitation and 10–20 μg of plasmid (37). Mouse A2–/– CMEC were electroporated with 20–30 μg of plasmid using a Gene Pulser (Bio-Rad) at 250 V and 975 microfarads.
Flow Cytometry and Fluorescence-activated Cell Sorting—Mouse A2–/– CMEC, cotransfected with A2 and GFP, were subjected to flow cytometry and cell sorting using a FACStar Plus or FACSVantage cell sorter (BD Biosciences) equipped with an INNOVA 70-4 argon laser tuned to 488 nm. Nonviable cells were excluded by gating on forward and side scatter. GFP signals were detected with a 530/30-nm bandpass filter. Cells were sorted according to GFP positivity with >90% efficiency at 2500–3000 events/s using forward scatter as a triggering signal. Data acquisition and analysis (≥10,000 events per analysis) were performed with CellQuest software.
Immunoblotting—Proteins resolved by SDS-PAGE were transferred to nitrocellulose membranes, blocked (5% nonfat milk in 50 mm Tris-HCl, pH 7.5, 280 mm NaCl, 0.02% Tween 20), incubated with primary antibodies (1 h), exposed to peroxidase-conjugated goat anti-mouse or anti-rabbit IgG (Amersham Biosciences), and visualized by enhanced chemiluminescence (ECL kit, PerkinElmer Life Sciences).
Reverse Transcription-PCR—Total RNA was extracted from A2+/+ and A2–/– mouse tissues using TRIzol reagent (Invitrogen). Total RNA (1 μg) was reverse-transcribed using the one-step reverse transcription-PCR kit (Qiagen) for 30 cycles according to the manufacturer's protocol, using mouse p11, annexin A2, and glyceraldehyde-3-phosphate dehydrogenase gene-specific primers. The products were analyzed by 2% agarose gel electrophoresis.
Coimmunoprecipitation—Cells were washed with cold phosphate-buffered saline and lysed (15 min, 4 °C) in 0.5 ml of lysis buffer (50 mm Tris-HCl, pH 7.5, 280 mm NaCl, 0.1% Nonidet P-40, 0.5 mm EDTA, 10 μg/ml aprotinin and leupeptin, 1 mm phenylmethylsulfonyl fluoride). Lysates were clarified (4 °C, 15 min, 18,000 × g), and aliquots (500 μg) incubated with 5 μg of anti-HA or anti-FLAG IgG (18 h, 4 °C) with gentle rocking. Immune complexes were collected with 20 μl of protein G-Sepharose, washed three times with lysis buffer, denatured with Laemmli buffer, and immunoblotted.
A2 Silencing—siRNA directed at A2, as well as control oligonucleotides (100 nm, Qiagen, or targeted to 146–165 of A2, from Dharmacon), were transiently transfected according to the manufacturer's instructions into HUVECs propagated in 6-well plates using Oligofectamine (Invitrogen). At 72 h after transfection, cells were harvested and tested for p11 or A2 expression.
RESULTS
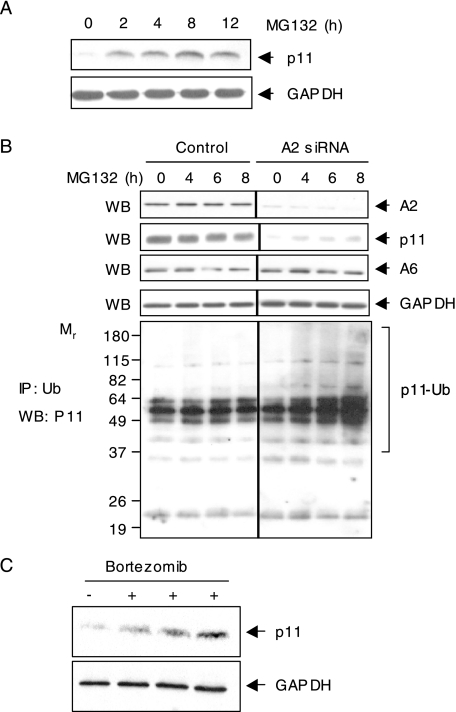
A2–/– Tissues Express Low Levels of p11 Protein—Tissues from A2–/– and A2+/+ mice were analyzed by RT-PCR for steady state p11 mRNA levels (Fig. 1A). p11 mRNA was detected uniformly in tissues from mice of both genotypes, whereas A2 mRNA was only detected in A2+/+ tissues and in variable quantities. On the other hand, p11 protein was readily observed by immunoblot analysis in A2+/+ tissues but not observed in extracts of lung, heart, spleen, kidney, and liver from A2–/– mice (Fig. 1B). To determine whether p11 protein expression was linked to that of A2, we cotransfected A2-myc and GFP expression plasmids into A2–/– CMEC, which express low levels of p11 (Fig. 1C). In CMEC selected for GFP positivity by fluorescence-activated cell sorting, reconstitution of A2 expression was associated with a significant increase in p11 by immunoblot analysis. This result suggested a role for A2 in stabilizing p11 protein expression.
FIGURE 1.
A2–/– tissues express low levels of p11 protein. A, total RNA (1 μg) isolated from lung, heart, liver, kidney, and spleen tissue from A2+/+ and A2–/– mice was subjected to reverse transcription-PCR analysis using gene-specific mouse A2, p11, or GAPDH primers. B, protein extracts (40 μg) from lung, heart, liver, kidney, and spleen from A2+/+ and A2–/– mice were resolved on 15% SDS-polyacrylamide gels and immunoblotted with IgG directed against A2, p11, or GAPDH. C, A2-myc- and GFP-encoding expression plasmids were cotransfected into mouse A2–/– CMEC by electroporation. GFP-positive cells were selected by fluorescence-activated cell sorter, and lysates were immunoblotted for A2, p11, and β-tubulin (β-tub).
We next addressed whether endogenous endothelial cell p11 is degraded via a proteasomal mechanism. Treatment of A2–/– CMEC with the proteasomal inhibitor, MG132, resulted in increased expression of p11 within 2 h. Expression peaked at 8–12 h, at which point it showed a 4–6-fold increase relative to levels of GAPDH, which remained unchanged (Fig. 2A). Because polyubiquitination typically predetermines proteins for degradation in the proteasome, we next employed A2 siRNA to deplete A2 in nonimmortalized, low passage human umbilical vein endothelial cells (HUVEC), and then further treated the cells with the proteasomal inhibitor MG132 to enable detection of polyubiquitinated species (Fig. 2B). By immunoblot analysis, depletion of A2 reduced intracellular levels of both A2 and p11, whereas levels of A6 and GAPDH were not appreciably altered. In cells treated with an irrelevant siRNA, both A2 and p11 were easily detected. Immunoblot analysis of proteins precipitated with anti-ubiquitin revealed the accumulation of high molecular weight species with increasing time of proteasomal inhibition. This effect was not observed in cells in which A2 was not depleted. Similarly injection of bortezomib, a proteasomal protease inhibitor, into A2–/– mice (Fig. 2C), significantly rescued p11 protein expression in the lung, without interfering with GAPDH expression. Taken together, these data establish that p11 is polyubiquitinated in low passage human cells when A2 is depleted or absent and that p11 can be rescued by inhibition of proteasomal protease activity both in vitro and in vivo.
FIGURE 2.
Proteasomal degradation of p11 in the absence of A2. A, A2–/– CMECs were treated with or without MG132 (1 μm) for the indicated time periods. Cell lysates were immunoblotted (WB) for p11 and GAPDH. B, HUVECs were transiently transfected with control or A2-directed siRNA oligonucleotides. After 72 h, cells were treated with or without MG-132 (10 μm) for the indicated time periods, and whole cell lysates were either immunoblotted with anti-A2, anti-p11, anti-A6, or GAPDH IgG (upper panels) or immunoprecipitated (IP) with anti-ubiquitin (Ub) IgG (lower panel) and blotted with anti-p11 IgG. C, A2–/– mice were sacrificed 6 h following tail vein injection of either saline (–) or bortezomib (+, 1 mg/kg). Lung tissue was harvested, and p11 expression analyzed by immunoblot. GAPDH served as a loading control.
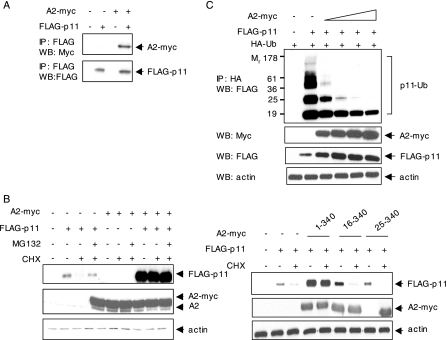
p11 Is Polyubiquitinated in an A2-sensitive Manner—We next developed a transient transfection system to study the fate of p11 in the absence of A2. We transfected HEK 293 cells, which express very low levels of p11 and A2, with FLAG-p11 (Fig. 3A). After 48 h, protein synthesis was arrested with cycloheximide, and p11 expression was examined by immunoblot at 2–8 h. In the absence of exogenous A2, expression of p11 decreased with a t½ of ∼5.6 h. In the presence of the proteasome inhibitor MG132, however, p11 was stabilized. Treatment with lactacystin, another proteasome inhibitor, also stabilized p11, whereas inhibitors of lysosomal enzymes and calpains failed to do so (supplemental Fig. 1).
FIGURE 3.
p11 is polyubiquitinated in the absence of A2. A, HEK 293 cells were transiently transfected with FLAG-p11 for 48 h. The cells were pretreated with cycloheximide (CHX) (10 μg/ml, 30 min) and then incubated with or without MG132 (10 μm) as indicated. Lysates were immunoblotted for FLAG-p11, A2, and GAPDH. B, HEK 293 cells were cotransfected with plasmids encoding FLAG-p11 and HA-ubiquitin (HA-Ub). Cell lysates (500 μg) were either immunoprecipitated (IP) with anti-HA and immunoblotted (WB) with anti-FLAG IgG (left panel) or immunoprecipitated with anti-FLAG and blotted with anti-HA IgG (right panel). C, dominant negative ubiquitin mutants, K48R and K29R, or empty control vectors were cotransfected with FLAG-p11, and cell lysates were subjected to immunoblotting with anti-p11 and anti-actin IgG after 36 h. D, FLAG-p11 was transfected with either HA-wt- or HA-K48R-ubiquitin into HEK 293 cells. Cell lysates were coimmunoprecipitated with anti-FLAG IgG and either probed with anti-HA antibody (top panel) or immunoblotted with anti-p11 (lower panel).
To determine whether ubiquitin marks unpartnered p11 for proteasomal degradation, we cotransfected HEK 293 cells with plasmids encoding FLAG-p11 and HA-ubiquitin (Fig. 3B). After 48 h, cell lysates were immunoprecipitated with anti-HA IgG and immunoblotted with anti-FLAG IgG. Ubiquitinated p11 conjugates were readily detected in a pattern typical for polyubiquitinated proteins, even in the absence of proteasome inhibitors, indicating the presence of high levels of p11-polyubiquitin conjugates (Fig. 3B, left panel). Similarly, immunoprecipitation of the same lysates with anti-FLAG IgG and immunoblotting with anti-HA IgG also revealed the association of ubiquitin with p11 (Fig. 3B, right panel). These data indicate that exogenously expressed FLAG-p11 is subject to ubiquitination in HEK 293 cells.
Although lysine 48 of ubiquitin is the primary site for polyubiquitination, ubiquitin polymers can also be linked to target proteins through lysine 29 (38, 39). K48R and K29R ubiquitin mutants act as dominant negative inhibitors of polyubiquitination by inducing premature termination of ubiquitin chains and accumulation of incompletely ubiquitinated species that are not properly targeted for degradation by the proteasome (40). To determine whether polyubiquitination is required for the degradation of p11, we cotransfected HEK 293 cells with plasmids encoding FLAG-p11, as well as K48R and K29R ubiquitin mutants. Expression of either mutant resulted in accumulation of FLAG-p11, suggesting that disruption of the polyubiquitin chain elongation stabilizes p11 (Fig. 3C). In addition, polyubiquitination of p11 was diminished upon cotransfection of FLAG-p11 with K48R (Fig. 3D). We conclude that polyubiquitination of p11 is necessary for its degradation in transfected HEK 293 cells.
The N-terminal Domain of A2 Is Required for Rescue of p11—A2 spontaneously forms a heterotetramer with p11 (41). Coimmunoprecipitation experiments confirmed the association of p11 with A2 in our cotransfection system, as immune complexes precipitated with anti-FLAG IgG were also specifically recognized by anti-Myc IgG (Fig. 4A). To determine whether association with A2 blocks p11 degradation, HEK 293 cells were transiently transfected with FLAG-p11 with or without A2-myc, pretreated with cycloheximide, and then treated with MG132 (Fig. 4B). By immunoblot analysis, FLAG-p11 was stabilized by either treatment with MG132 or cotransfection with A2-myc.
FIGURE 4.
The N-terminal tail domain of A2 protects p11 from ubiquitin-dependent proteolysis. A, HEK 293 cells were cotransfected with A2-myc and either FLAG-p11 or empty vector. Immune complexes were precipitated with anti-FLAG IgG and immunoblotted anti-Myc IgG (upper panel). The same blot was stripped and reprobed with anti-FLAG IgG (lower panel). B, HEK 293 cells were cotransfected with plasmids encoding FLAG-p11 and C-terminal Myc-tagged A2. After 48 h, cells were pretreated with cycloheximide (CHX) (10μg/ml, 30 min) and then incubated with or without MG132 (10 μm, 8 h). Lysates were immunoblotted with anti-FLAG, anti-A2, and anti-actin. C, HEK 293 cells were cotransfected with HA-ubiquitin, plus A2-myc and FLAG-p11 at molar ratios of 1:1, 2:1, 3:1, and 4:1. Lysates were either immunoprecipitated (IP) with anti-HA IgG and blotted with anti-FLAG IgG (upper panel) or immunoblotted (WB) with anti-Myc, anti-FLAG, and anti-actin IgG (lower panels). D, HEK 293 cells were transfected for 36 h with Myc-tagged full-length or N-terminal truncation mutants of A2, as well as FLAG-p11. Cells were treated with cycloheximide (10 μg/ml, 30 min) in the presence or absence of MG132 (10 μm, 8 h), and lysates were immunoblotted with anti-FLAG, anti-Myc, and anti-actin IgG.
To confirm the regulatory role of A2 in p11 degradation, we transfected an HA-ubiquitin construct, together with FLAG-p11 and increasing molar ratios of A2-myc (Fig. 4C). Ubiquitin-conjugated FLAG-p11 species were assessed by immunoprecipitation with anti-HA IgG and immunoblotting with anti-FLAG IgG. Although mono- and polyubiquitinated p11 species were detected in cells transfected with p11 alone, cotransfection with A2 efficiently blocked p11 ubiquitination in a dose-related manner. Interestingly, the most highly ubiquitinated species disappeared first. These data strongly imply that A2 blocks polyubiquitin-dependent proteasomal degradation of p11. Furthermore, the regulation of p11 degradation by A2 was selective; in parallel experiments, we observed no difference in TNF-induced IκBα phosphorylation and degradation in A2–/– versus A2+/+ CMEC, indicating that A2 has no effect on ubiquitination of IκBα (not shown).
The first 14 amino acids of A2 interact with p11 to form the functional heterotetramer (42, 43). To test whether protection of p11 is controlled by the N-terminal tail domain of A2, two Myc-tagged N-terminal truncation mutants of A2 (residues 16–340 and 25–340, respectively) were expressed in HEK 293 cells at levels similar to the full-length protein. After 48 h, the cells were treated with cycloheximide (8 h), and cell lysates were immunoblotted with anti-FLAG, anti-Myc, and anti-actin IgG. Expression of p11 was preserved in the presence full-length A2, whereas neither N-terminal deletion mutant protected p11 (Fig. 4D). Further coimmunoprecipitation experiments on HEK 293 cells triply transfected with A2-myc, FLAG-p11, and HA-ubiquitin showed that the two truncated A2 mutants failed to interact with p11 and allowed it to be highly ubiquitinated (supplemental Fig. 2). Collectively, these data reveal that residues within the first 15 p11-binding amino acids of A2 are required to prevent p11 ubiquitination.
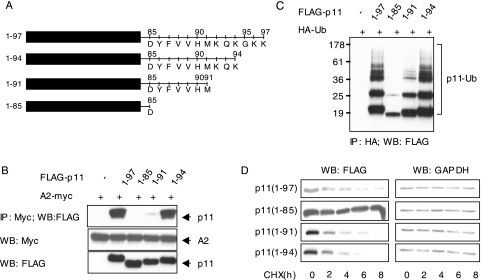
The C Terminus of p11 Contains a Polyubiquitination and Degradation Determinant—The A2 heterotetramer forms as the N-terminal α-helix of one p11 and the C-terminal helix and C-terminal extension (KQKGKK) of an adjacent p11 bind to the tail domain of A2 (19). We therefore expressed three p11 truncation mutants (residues 1–85, 1–91, and 1–94) in HEK 293 cells (Fig. 5A), and we tested their ability to interact with intact A2. Both full-length (1–97) and 1–94 deletion mutants bound to A2, as judged by immunoprecipitation with anti-Myc IgG and immunoblot with anti-FLAG IgG (Fig. 5B). In contrast, mutants lacking the C-terminal 6 (1–91) or 12 (1–85) residues of p11 bound to A2 to a lesser extent (1–91) or not at all (1–85). These data indicate that a C-terminal mutant of p11 (1–85) that lacks the final 12 amino acids failed to associate with p11.
FIGURE 5.
The C-terminal region of p11 contains a polyubiquitination and degradation determinant. A, schematic representation of FLAG-tagged p11 C-terminal truncation plasmids. B, FLAG-tagged C-terminal truncation mutants of p11 were cotransfected with A2-myc into HEK 293 cells. Whole cell extracts were either immunoprecipitated (IP) with anti-Myc and blotted with anti-FLAG IgG (upper panel) or immunoblotted (WB) with anti-Myc or anti-FLAG IgG (lower panels). C, lysates from cells cotransfected with FLAG-tagged p11 truncation mutants and HA-ubiquitin (HA-Ub) were immunoprecipitated with anti-HA and blotted with anti-FLAG IgG. D, HEK 293 cells, transfected with p11 truncation mutants, were treated with cycloheximide at 48 h after transfection (CHX, 10 μg/ml) for the indicated time periods. Cell lysates were immunoblotted with anti-FLAG (left panels) or anti-GAPDH (right panels).
To test whether amino acids 86–97 of p11 contain a polyubiquitination determinant, we cotransfected HEK 293 cells with each of the three mutants and HA-ubiquitin. Although deletion of lysines 96 and 97 did not perturb p11 ubiquitination at all, the polyubiquitinated p11 complex was markedly diminished upon deletion of residues 86–97 and partially reduced upon deletion of residues 92–97 (Fig. 5C). We also examined the stability of these truncated p11 mutants by treating transfected HEK 293 cells with cycloheximide for 0, 2, 4, 6, and 8 h. In agreement with their polyubiquitination pattern, the deletion of C-terminal 12 residues (1–85) protected p11, whereas deletion of the last 3 and 6 amino acids (1–94 and 1–91 mutants, respectively) had no effect on its disappearance (Fig. 5D). Decay curves depicting the ratio of each mutant to the GAPDH level confirmed that deletion of the last 12 residues (1–85) increased the half-life of p11 by ∼7-fold compared with the full-length protein (t½ = 44.3 versus 6.4, 4.2, and 3.9 h for 1–85, 1–97, 1–91, and 1–94 mutants, respectively) (supplemental Fig. 3). These data suggest that residues 86–95 (YFVVHMKQKG) of p11 harbor a degradation determinant, or degron, that specifies p11 polyubiquitination and proteasomal degradation.
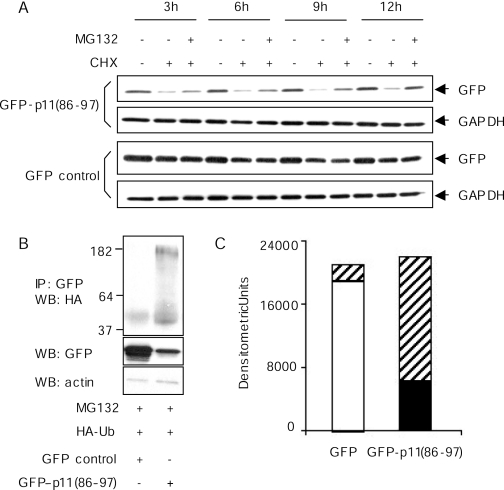
The p11 Degron Functions Autonomously—To determine whether the C terminus of p11 directs protein degradation independently of protein context, we fused residues 86–97 of p11 to the C terminus of GFP, creating a GFP-p11 fusion protein. In HEK 293 cells treated with cycloheximide, the expression level of this chimeric protein decreased by ∼80% over 12 h, whereas unmodified GFP remained stable (Fig. 6A). In the presence of the proteasome inhibitor, MG132, steady state levels of GFP-p11 were stabilized, suggesting that the p11 C-terminal sequence targeted GFP for proteasomal degradation and thus functioned as an autonomous degron.
FIGURE 6.
The p11 degron functions autonomously. A, 20 h after transfection with GFP control or GFP-p11(86–97) plasmids, HEK 293 cells were pre-treated with cycloheximide (CHX, 10 μg/ml, 30 min) and then incubated with or without MG132 (10 μm) for the indicated time periods. Lysates were immunoblotted for GFP and GAPDH. B, HEK 293 cells were cotransfected with an HA-ubiquitin-encoding plasmid and either a GFP- or GFP-p11(86–97)-encoding plasmid. Forty eight hours later, the cells were treated with 10 μm MG132 for 8 h. Whole cell extracts (500 μg) were then immunoprecipitated (IP) with anti-GFP and immunoblotted with anti-HA IgGs (upper panel). The same samples were immunoblotted with anti-GFP (middle panel) and anti-actin IgG (lower panel). C, bands in the upper two panels shown in B were scanned. Densitometric units for nonubiquitinated (middle panel) and ubiquitinated (upper panel) GFP and GFP-p11(86–97) were calculated using ScionImage software. The open bar represents nonubiquitinated GFP; the closed bar represents nonubiquitinated GFP-p11; and the cross-hatched bars represent the respective ubiquitinated species.
In a separate experiment, HEK 293 cells were cotransfected with GFP-p11 and HA-ubiquitin. In lysates of these cells, anti-GFP precipitated an array of high molecular mass species that also reacted with anti-HA (Fig. 6B). These bands were not observed in cells transfected with HA-ubiquitin and unmodified GFP. Although steady state quantities of GFP were severalfold greater than those of GFP-p11 in the transfected cells, total anti-GFP reactive protein (nonubiquitinated plus ubiquitinated) was equivalent, indicating complete inhibition of degradation by MG132 (Fig. 6C). Together, these data show that the C-terminal region of p11 acts autonomously and confers the capacity for polyubiquitination and degradation to an otherwise stable protein.
DISCUSSION
S100 family members have been postulated to regulate a variety of cellular processes, including cytoskeletal assembly, channel dynamics, and enzymatic activities (15). Of the 13 S100 gene products identified in humans, 3 (S100A11, S100A10/p11, and S100A6) have been found to bind annexins (annexins 1, 2, and 11, respectively); in the case of the A22p112 heterotetramer, this association confers increased phospholipid-binding affinity, redistributing A2 away from the cytoplasmic pool and toward membrane surfaces (4). Although p11 has been reported to chaperone a number of other proteins to the plasma membrane (25, 27, 29, 31, 44), A2 is, to date, its only identified endothelial cell binding partner.
Here we examined the fate of p11 in the absence of A2. Although p11 mRNA levels were indistinguishable among A2–/– and A2+/+ tissues, we found dramatically reduced levels of p11 protein in A2–/– tissues compared with wild type tissues (Fig. 1). Moreover, in cultured A2–/– endothelial cells, either reconstitution of A2 expression by transient transfection (Fig. 1) or treatment with the proteasomal protease inhibitor, MG132 (Fig. 2), replenished p11 protein levels. In addition, “knockdown” of A2 in low passage human endothelial cells led to depletion of unmodified p11 and accumulation of the polyubiquitinated species (Fig. 2). Treatment of A2–/– mice with bortezomib, an inhibitor of proteasomal chymotrypsin-like activity, however, rapidly restored p11 expression in the lung (Fig. 2). Although others have reported an increase in p11 expression in cells of a cultured human bronchial epithelial cell line treated with the proteasome inhibitor lactacystin (45), to our knowledge our report is the first to demonstrate proteasomal regulation of p11 in vivo.
In investigating the mechanism by which A2 controls cellular p11 levels, we developed a model transfection system in which we observed that unpartnered p11 was rapidly ubiquitinated and destroyed in an A2-sensitive manner (Fig. 3). Rescue of p11 by A2 required the presence of the p11-binding, N-terminal region of A2 (Fig. 4), whereas ubiquitination of p11 required residues 86–95 (YFVVHMKQKG), its A2-binding C-terminal motif (Fig. 5). The C-terminal degron of p11 finally promoted the polyubiquitination of unrelated protein, GFP, and attenuated its half-life (Fig. 6). These data reveal that A2 prevents ubiquitin-dependent proteasomal degradation of p11 by masking an autonomous polyubiquitination determinant.
The polyubiquitin-proteasome system is a primary regulator of protein turnover (38, 46). Targeting of cellular proteins for proteolysis by the proteasome is a complex, tightly regulated process involving a cascade of enzyme activation reactions culminating in the covalent attachment of ubiquitin, an evolutionarily conserved 76-amino acid protein. Protein ubiquitination occurs as the result of a multistep, ATP-dependent pathway, in which ubiquitin first forms a thioester linkage with the ubiquitin-activating enzyme. Ubiquitin is next transferred to a ubiquitin-conjugating enzyme and then further transferred to the ε-amino group of a lysine residue within the target protein, usually through the action of an ubiquitin-protein ligase. Repeat cycles add additional ubiquitin moieties, typically to Lys48 of the preceding ubiquitin, thereby generating the polyubiquitin chain. In most instances, a tetra-ubiquitin chain is a minimal requirement for proteasomal degradation. However, because some nonubiquitinated proteins are degraded by the proteasome, and because ubiquitination may serve functions other than degradation, ubiquitination alone does not predict proteasomal degradation (47, 48). In this study, however, we show that the dominant negative ubiquitin mutants K48R and K29R significantly increase the stability of p11, confirming a link between ubiquitination and proteolysis of p11.
Most ubiquitin-proteasome systems require the presence of both a recognition motif for ubiquitin ligase, and one or more nearby lysine residues for ubiquitin conjugation (49–53). Ligase recognition sites can consist of sequences containing phosphorylatable serine residues, such as DS*GLDS* (* = phosphate), found in the NFκB inhibitor IκB, specific N-end rule amino acids, or particular C-terminal domains such as the yeast RNA polymerase II sequence SPTSPSY (54). In addition, pre-folded surface structures may provide conformation-dependent ligase docking sites, as has been observed for MATα2 (49). Because the region in p11 that is essential for polyubiquitination, 86YFVVHMKQK94, overlaps significantly with residues critical for A2 binding, we hypothesize that A2 binding to p11 masks residues required for the interaction between p11 and a ubiquitin ligase. Because p11 can form a stable complex with A2, even when amino acids 92–97 of p11 are absent (17), we postulate that A2 blocks a ubiquitin ligase interaction site, which our data would place within residues 86–91, rather than the actual site of ubiquitin ligation. Our data further predict that Lys92 or Lys94 may be sites for p11-ubiquitin conjugation.
Within cells, effector proteins may associate with binding partners that regulate their subcellular localization and activity. Stimulus-induced dissociation of the complex often leads to relocation of the effector and rapid proteasomal destruction of the binding partner. For example, caveolin-2, which normally associates with caveolin-1 in the plasma membrane, is retained in a Golgi compartment in caveolin-1-deficient cells and subsequently undergoes proteasomal degradation (55). Likewise, the inhibitor of nuclear factor-κB(IκB) maintains nuclear factor-κB (NF-κB) in a cytoplasmic location until phosphorylation of IκB by IκB kinase results in complex dissociation, translocation of NF-κB to the nucleus, and IκB ubiquitination and degradation (56). Similarly, oxygen prevents translocation of hypoxia-inducible factor-α (HIF-1α) to the nucleus by stimulating its dissociation from p300/CBP, its subsequent polyubiquitination by the von Hippel-Lindau ubiquitin ligase complex, and its proteasomal destruction (57).
Our data suggest a similar mechanism for A2 and p11. Mutational analyses have revealed that the binding region of A2 for p11 is limited to its first 14 amino acids (42), and that N-acetylation of the N-terminal serine residue is also required (18). In an A2 tail 2p112 crystal structure analyzed at 2.4 Å resolution, the N-terminal peptide of A2 formed an amphipathic helix and occupied a groove formed by the second interhelical loop and helix IV of one p11 and helix I of the second p11 (17). The hydrophobic face of the A2 tail helix interacted with hydrophobic residues of the twin p11 molecules, namely Phe13 of helix I, Phe38 and Phe41 of helix II, and Leu78, Tyr85, and Met90 of helix IV. Our findings, which indicate that the first 14 p11-binding amino acids of A2, as well as 10 of the final 12 A2-binding amino acids of p11, are required for stabilization of p11, support the hypothesis that a direct physical association between A2 and p11 is necessary for stabilization of p11.
The finding of a p11-specific degron, which can be masked by A2, suggests that the subcellular distribution of A2 is regulated by intracellular levels of p11. In the endothelial cell, p11 expression levels may be selectively increased by agents associated with vascular injury, such as interferon-γ and thrombin (22, 23). Because p11 exists in a perpetually “on” state with respect to intracellular calcium, elevated p11 results in increased A2 heterotetramer, which would then be directed to the plasma membrane and poised for tyrosine phosphorylation and translocation (32). Excess p11, on the other hand, would be polyubiquitinated by virtue of its exposed degron, directed to the proteasome, and eliminated. Within such a system, the cytoplasmic level of p11 would be the primary determinant of cell surface A2, allowing p11 to establish the “set point” for A2-mediated cell surface fibrinolytic potential.
Supplementary Material
Acknowledgments
We thank Amy Lam for critical reading of the manuscript and Emil Lev for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants HL42493, HL46403, and HL67839. This work was also supported by March of Dimes Grant 6-FY05-94. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
Footnotes
The abbreviations used are: A2, annexin A2; siRNA, short interfering RNA; HUVEC, human umbilical vein endothelial cell; CMEC, cardiac microvascular endothelial cell; GFP, green fluorescent protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HA, hemagglutinin.
References
- 1.Pober, J. S., and Sessa, W. C. (2007) Nat. Rev. Immunol. 7 803–815 [DOI] [PubMed] [Google Scholar]
- 2.Hajjar, K. A., Esmon, N. L., Marcus, A. J., and Muller, W. A. (2006) in Williams Hematology (Lichtman, M. A., Beutler, E., Kipps, T. J., Seligsohn, U., Kaushansky, K., and Prchal, J. T., eds) pp. 1715–1739, McGraw-Hill, New York
- 3.Hajjar, K. A., and Francis, C. W. (2006) in Williams Hematology (Lichtman, M. A., Beutler, E., Kipps, T. J., Seligsohn, U., Kaushansky, K., and Prchal, J. T., eds) pp. 2089–2115, McGraw-Hill, New York
- 4.Gerke, V., Creutz, C. E., and Moss, S. E. (2005) Nat. Rev. Mol. Cell Biol. 6 449–461 [DOI] [PubMed] [Google Scholar]
- 5.Moss, S. E., and Morgan, R. O. (2004) Genome Biol. 5 219.1–219.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman-Maus, G., and Hajjar, K. A. (2005) Br. J. Haematol. 129 307–321 [DOI] [PubMed] [Google Scholar]
- 7.Powell, M. A., and Glenney, J. R. (1987) Biochem. J. 247 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajjar, K. A., Jacovina, A. T., and Chacko, J. (1994) J. Biol. Chem. 269 21191–21197 [PubMed] [Google Scholar]
- 9.Kassam, G., Choi, K. S., Ghuman, J., Kang, H. M., Fitzpatrick, S. L., Zackson, T., Zackson, S., Toba, M., Shinomiya, A., and Waisman, D. M. (1998) J. Biol. Chem. 273 4790–4799 [DOI] [PubMed] [Google Scholar]
- 10.Ling, Q., Jacovina, A. T., Deora, A. B., Febbraio, M., Simantov, R., Silverstein, R. L., Hempstead, B. L., Mark, W., and Hajjar, K. A. (2004) J. Clin. Invest. 113 38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii, H., Yoshida, M., Hiraoka, M., Hajjar, K. A., Tanaka, A., Yasukochi, Y., and Numano, F. (2001) Circ. Res. 89 1240–1245 [DOI] [PubMed] [Google Scholar]
- 12.Tanaka, Y., Ishii, H., Hiraoka, M., Miyasaka, N., Kuroiwa, T., Hajjar, K. A., Nagaoka, T., Duong, T. Q., Ohno, K., and Yoshida, M. (2007) Brain Res. 1165 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menell, J. S., Cesarman, G. M., Jacovina, A. T., McLaughlin, M. A., Lev, E. A., and Hajjar, K. A. (1999) N. Engl. J. Med. 340 994–1004 [DOI] [PubMed] [Google Scholar]
- 14.Cesarman-Maus, G., Rios-Luna, N. P., Deora, A. B., Huang, B., Villa, R., Cravioto, M. C., Alarcon-Segovia, D., Sanchez-Guerrero, J., and Hajjar, K. A. (2006) Blood 107 4375–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donato, R. (2001) Int. J. Biochem. Cell Biol. 33 637–668 [DOI] [PubMed] [Google Scholar]
- 16.Schafer, B. W., and Heizmann, C. W. (1996) Trends Biochem. Sci. 21 134–140 [DOI] [PubMed] [Google Scholar]
- 17.Rety, S., Sopkova, J., Renouard, M., Osterloh, D., Gerke, V., Tabaries, S., Russo-Marie, F., and Lewit-Bentley, A. (1999) Nature Struct. Biol. 6 85–89 [DOI] [PubMed] [Google Scholar]
- 18.Becker, T., Weber, K., and Johnsson, N. (1990) EMBO J. 9 4204–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kube, E., Becker, T., Weber, K., and Gerke, V. (1992) J. Biol. Chem. 267 14175–14182 [PubMed] [Google Scholar]
- 20.Glenney, J. R. (1985) FEBS Lett. 192 79–82 [DOI] [PubMed] [Google Scholar]
- 21.Johnsson, N., and Weber, K. (1990) J. Biol. Chem. 265 14464–15568 [PubMed] [Google Scholar]
- 22.Huang, X. L., Pawliczak, R., Yao, X. L., Cowan, M. J., Gladwin, M. T., Walter, M. J., Holtzman, M. J., Madara, P., Logun, C., and Shelhamer, J. H. (2003) J. Biol. Chem. 278 9298–9308 [DOI] [PubMed] [Google Scholar]
- 23.Petersen, E. A., Sutherland, M. R., Nesheim, M. E., and Pryzdial, E. L. (2003) J. Cell Sci. 116 2399–2408 [DOI] [PubMed] [Google Scholar]
- 24.Rescher, U., and Gerke, V. (2007) Pfluegers Arch. Eur. J. Physiol. 455 575–582 [DOI] [PubMed] [Google Scholar]
- 25.Okuse, K., Malik-Hall, M., Baker, M. D., Poon, W. Y. L., Kong, H., Chao, M. V., and Wood, J. N. (2002) Nature 417 653–656 [DOI] [PubMed] [Google Scholar]
- 26.Poon, W. Y., Malik-Hall M., Wood, J. N., and Okuse, K. (2004) FEBS Lett. 558 114–118 [DOI] [PubMed] [Google Scholar]
- 27.Girard, C., Tinel, N., Terrenoire, C., Romey, G., Lazdunski, M., and Borsotto, M. (2002) EMBO J. 21 4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renigunta, V., Yuan, H., Zuzarte, M., Rinne, S., Koch, A., Wischmeyer, E., Schlichthorl, G., Gao, Y., Karschin A., Jacob, R., Schwappach, B., Daut, J., and Preisig-Muller, R. (2006) Traffic 7 168–181 [DOI] [PubMed] [Google Scholar]
- 29.Van de Graaf, S. F. J., Hoenderop, J. G. J., Gkika, D., Lamers, D., Prenen, J., Rescher, U., Gerke, V., Stub, O., Nilius, B., and Bindels, R. J. M. (2003) EMBO J. 22 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donier, E., Rugiero, F., Okuse K., and Wood, J. N. (2005) J. Biol. Chem. 280 38666–38672 [DOI] [PubMed] [Google Scholar]
- 31.Svenningsson, P., Chergui, K., Rachleff, I., Flajolet, M., Zhang, X., Yacoubi, M. E., Vaugeois, J. M., Nomikos, G. G., and Greengard, P. (2006) Science 311 77–80 [DOI] [PubMed] [Google Scholar]
- 32.Deora, A. B., Kreitzer, G., Jacovina, A. T., and Hajjar, K. A. (2004) J. Biol. Chem. 279 43411–43418 [DOI] [PubMed] [Google Scholar]
- 33.Puisieux, A., Ji, J., and Ozturk, M. (1996) Biochem. J. 313 51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong, H., Li, H., Kong, H. J., Chen, Y., Zhao, J., Xiong, S., Huang, B., Gu, H., Mayer, L., Ozato, K., and Unkeless, J. C. (2005) J. Biol. Chem. 280 23531–23539 [DOI] [PubMed] [Google Scholar]
- 35.Weksler, B. B., Subileau, E. A., Perriere, N., Charneau, P., Holloway, K., Leveque, M., Tricoire-Leugnel, H., Nicotra, A., Bourdoulous, S., Turowski, P., Male, D. K., Roux, F., Greenwood, J., Romero, I. A., and Couraud, P. O. (2005) FASEB J. 19 1872–1874 [DOI] [PubMed] [Google Scholar]
- 36.Zennou, V., Serguera, C., Sarkis, C., Colin, P., Perret, E., Mallet, J., and Charneau, P. (2001) Nat. Biotechnol. 19 446–450 [DOI] [PubMed] [Google Scholar]
- 37.Wigler, M., Pellicer, A., Silverstein, S., and Axel, R. (1978) Cell 14 725–731 [DOI] [PubMed] [Google Scholar]
- 38.Ciechanover, A., and Schwartz, A. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2727–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U., and Jentsch, S. (1999) Cell 96 635–644 [DOI] [PubMed] [Google Scholar]
- 40.Ward, C. L., Omura, S., and Kopito, R. R. (1995) Cell 14 725–731 [DOI] [PubMed] [Google Scholar]
- 41.Thiel, C., Osborn, M., and Gerke, V. (1992) J. Cell Sci. 103 733–742 [DOI] [PubMed] [Google Scholar]
- 42.Johnsson, N., Marriott, G., and Weber, K. (1988) EMBO J. 7 2435–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jost, M., Zeusschner, D., Seemann, J., Weber, K., and Gerke, V. (1997) J. Cell Sci. 110 221–228 [DOI] [PubMed] [Google Scholar]
- 44.Beaton, A. R., Rodriguez, J., Reddy, Y. K., and Roy, P. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13154–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gladwin, M. T., Yao, X. L., Cowan, M., Huang, X. L., Schneider, R., Grant, L. R., Logun, C., and Shelhamer, J. H. (2000) Am. J. Physiol. 279 L1103–L1109 [DOI] [PubMed] [Google Scholar]
- 46.Pickart, C. M., and Eddins, M. J. (2004) Biochim. Biophys. Acta 1695 55–72 [DOI] [PubMed] [Google Scholar]
- 47.Pickart, C. M. (2001) Mol. Cell 8 499–504 [DOI] [PubMed] [Google Scholar]
- 48.Verma, R., and Deshaies, R. J. (2000) Cell 101 341–344 [DOI] [PubMed] [Google Scholar]
- 49.Johnson, P. B., Swanson, R., Rakhilina, L., and Hochstrasser, M. (1998) Cell 94 217–227 [DOI] [PubMed] [Google Scholar]
- 50.Laney, J. D., and Hochstrasser, M. (1999) Cell 97 427–430 [DOI] [PubMed] [Google Scholar]
- 51.Lawson, T. G., Gronros, D. L., Evans, P. E., Bastien, M. C., Michalewich, K. M., Clark, J. K., Edmonds, J. H., Graber, K. H., Werner, J. A., Lurvey, B. A., and Cate, J. M. (1999) J. Biol. Chem. 274 9904–9908 [PubMed] [Google Scholar]
- 52.Mitsui, A., and Sharp, P. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 6054–6059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogers, S., Wells, R., and Rechsteiner, M. (1986) Science 234 364–368 [DOI] [PubMed] [Google Scholar]
- 54.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 427. [DOI] [PubMed] [Google Scholar]
- 55.Razani, B., Engelman, J. A., Wang, X. B., Schubert, W., Zhang, X. L., Marks, C. B., Macaluso, K., Russell, R. G., Li, M., Pestell, R. G., Di Vizio, D., Hou, H., Kneitz, B., Lagaud, G., Christ, G. J., Edelmann, W., and Lisanti, M. P. (2001) J. Biol. Chem. 276 38121–38138 [DOI] [PubMed] [Google Scholar]
- 56.Karin, M., and Ben-Nariah, Y. (2000) Annu. Rev. Immunol. 18 621–663 [DOI] [PubMed] [Google Scholar]
- 57.Safran, M., and Kaelin, W. G. (2003) J. Clin. Invest. 111 779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.