Abstract
The SNF1/AMP-activated protein kinase (AMPK) family is required for adaptation to metabolic stress and energy homeostasis. The γ subunit of AMPK binds AMP and ATP, and mutations that affect binding cause human disease. We have here addressed the role of the Snf4 (γ) subunit in regulating SNF1 protein kinase in response to glucose availability in Saccharomyces cerevisiae. Previous studies of mutant cells lacking Snf4 suggested that Snf4 counteracts autoinhibition by the C-terminal sequence of the Snf1 catalytic subunit but is dispensable for glucose regulation, and AMP does not activate SNF1 in vitro. We first introduced substitutions at sites that, in AMPK, contribute to nucleotide binding and regulation. Mutations at several sites relieved glucose inhibition of SNF1, as judged by catalytic activity, phosphorylation of the activation-loop Thr-210, and growth assays, although analogs of the severe human mutations R531G/Q had little effect. We further showed that alterations of Snf4 residues that interact with the glycogen-binding domain (GBD) of the β subunit strongly relieved glucose inhibition. Finally, substitutions in the GBD of the Gal83 β subunit that are predicted to disrupt interactions with Snf4 and also complete deletion of the GBD similarly relieved glucose inhibition of SNF1. Analysis of mutant cells lacking glycogen synthase showed that regulation of SNF1 is normal in the absence of glycogen. These findings reveal novel roles for Snf4 and the GBD in regulation of SNF1.
The SNF1/AMP-activated protein kinase (AMPK)3 family plays a central role in responses to metabolic stress and regulation of energy homeostasis in eukaryotes (1, 2). In mammals, AMPK regulates many metabolic processes, notably glucose and lipid metabolism, and controls transcription and protein synthesis to maintain energy balance. AMPK is activated by hormones and by stresses that deplete cellular ATP, thereby elevating the AMP:ATP ratio, and in humans, it is a target of drugs used in treatment of type 2 diabetes. AMPK is a heterotrimer comprising the catalytic α subunit and two regulatory subunits, β and γ, which exist as different isoforms (α1, α2, β1, β2, γ1, γ2, and γ3). The kinase is activated by phosphorylation on Thr-172 in the activation loop of the α subunit by upstream kinases, including LKB1 (3–6), Ca2+/calmodulin-dependent protein kinase kinase (7–10), and possibly TAK1 (11). AMP activates AMPK allosterically, inhibits dephosphorylation, and may enhance phosphorylation, whereas ATP is inhibitory (4, 12–16).
The effects of nucleotides are mediated by the γ subunit, which comprises four cystathionine β-synthase (CBS) repeats (17) (Fig. 1). CBS repeats occur in a variety of proteins as tandem pairs, termed Bateman domains (18), and bind adenosine derivatives (19). Biochemical studies suggested that each γ subunit can bind two AMP or ATP molecules (19). Crystallographic analysis of a partial heterotrimer of rat AMPK, comprising the C-terminal domains of α1 and β2 and the complete γ1 subunit, revealed three nucleotide-binding sites, one of which contained tightly bound, nonexchangeable AMP (20). The first identification of a γ subunit mutation was the substitution R200Q in γ3 of pig, which increases glycogen levels in skeletal muscle (21). In humans, mutations in the γ2 subunit cause cardiac hypertrophy, glycogen accumulation, and ventricular pre-excitation associated with Wolff-Parkinson-White syndrome (19, 22–26). Such mutations decrease the affinity of the γ subunit for AMP and ATP, reduce allosteric AMP activation, and despite some controversy, appear to increase the basal phosphorylation and activity of AMPK in cells expressing LKB1 (18, 19, 26–28). Many of the altered residues are located close to nucleotide-binding sites in the mammalian enzyme structure (20).
FIGURE 1.
Sequence alignment of CBS motifs of Snf4 and human AMPK γ1, γ2, and γ3 subunits. Equivalent amino acid residues in the four CBS motifs are aligned vertically. Identical residues within each CBS motif are shaded black. Residues altered by mutation are indicated, with those in the γ subunits labeled as such, and marked (arrows). The residues are indicated by single-letter code.
Structural analysis of AMPK from the fission yeast Schizosaccharomyces pombe suggested that the ability of the γ subunit to bind nucleotide is broadly conserved (29, 30). The crystal structure of a partial heterotrimeric AMPK, lacking the catalytic domain, defined a binding site for AMP and ATP in the C-terminal Bateman domain of the γ subunit (29). This site corresponds to the site in the mammalian enzyme that contains nonexchangeable AMP and is not involved in AMP/ATP sensing (20). Additional studies of the S. pombe partial heterotrimer revealed a binding site for ADP in the N-terminal Bateman domain (30). However, S. pombe AMPK has not been well characterized genetically or biochemically, and it is not known whether it is regulated by nucleotides.
In the budding yeast Saccharomyces cerevisiae, SNF1 protein kinase is the ortholog of AMPK (for review, see Ref. 31). SNF1 is required for the adaptation of cells to metabolic stress, notably carbon stress (32), but also starvation for other nutrients (33–35) and various environmental stresses (36–40). Carbon metabolism is particularly important for yeast, and SNF1 is required for the adaptation of yeast cells to glucose limitation and for growth on carbon sources that are less preferred than glucose; the initial snf1 mutation was named for the sucrose-nonfermenting phenotype (41). The kinase is required to activate transcription of genes involved in the metabolism of sucrose and other carbon sources (42, 43); it is not required for growth on glucose, which inhibits its activity. In addition to regulating transcription, SNF1 controls the activity of metabolic enzymes involved in fatty acid metabolism and carbohydrate storage (44–46).
Like other members of the AMPK family, SNF1 protein kinase is heterotrimeric, comprising the Snf1 catalytic (α) subunit, one of three β subunit isoforms (Gal83, Sip1, or Sip2), and the Snf4 (γ) subunit. Snf1 is phosphorylated on Thr-210 in the activation loop by the upstream kinases Sak1, Tos3, and Elm1 (3, 47, 48) and is dephosphorylated by Reg1-Glc7 protein phosphatase 1 (49, 50). SNF1 is activated by glucose limitation (46, 50–52), but the regulatory signal(s) is not known. Acute glucose limitation depletes ATP and elevates the AMP:ATP ratio (51), but AMP does not allosterically activate SNF1 in vitro (45, 46, 51). The possibility remains that AMP or ATP controls SNF1 activity in vivo.
The β subunits control subcellular localization of SNF1 (53) and interactions with substrates (54). Gal83 is the major isoform during growth on glucose (53) and contributes the most to SNF1 activity in response to acute glucose limitation (55). Gal83 and Sip2 contain a conserved glycogen-binding domain (GBD) and bind glycogen in vitro (56).
The Snf4 subunit, like mammalian γ subunits, consists of four CBS repeats (Fig. 1). Snf4 interacts directly with Snf1 and the β subunits (52, 57, 58) and is important for SNF1 function in vivo and in vitro; very little catalytic activity is detected in snf4 mutant cells, which exhibit a phenotype similar to that of snf1 cells (46, 59, 60). In contrast, Snf4 is not required for function of the truncated Snf1 catalytic domain (residues 1–309) (61) or of Snf1 lacking residues 381–488 (62), and Snf4 interacts directly with the C terminus of Snf1 (52, 58), suggesting that Snf4 counteracts autoinhibition by a C-terminal autoinhibitory sequence. However, several lines of evidence indicated that Snf4 is dispensable for glucose regulation. Various Snf4-independent mutant Snf1 proteins conferred glucose-regulated gene expression in the absence of Snf4 function (52, 61, 62), and the phosphorylation of Thr-210 on full-length Snf1 remained glucose-regulated in snf4Δ mutants (50, 63). These data indicate that at least one glucose regulatory mechanism operates independent of Snf4. The question remains: does Snf4 have any direct role in mediating regulatory signals?
Structural analysis has not yet resolved the issue of whether Snf4 binds nucleotide. The crystal structure of the C-terminal Bateman (Bateman2) domain revealed a dimer with a pocket at its center, and residues corresponding to those altered by disease-causing mutations were located near this pocket (64). The structure of a partial heterotrimer of SNF1, containing C-terminal fragments of Snf1 and Sip2 and full-length Snf4, was reported (58). This structure lacked bound nucleotide, although AMP was included in the crystallization solution, and comparison with the S. pombe structure revealed side chain differences in Snf4 that would interfere with AMP binding; however, another crystal appeared to have AMP bound to Snf4 at a similar position as in the S. pombe subunit (58).
In this study, we used genetic analysis to address the regulatory role of Snf4. We first introduced substitutions at sites that, in mammalian γ subunits, contribute to nucleotide binding, including alterations analogous to those causing human disease, and assayed the effects of each mutant Snf4 protein on regulation of Thr-210 phosphorylation and activity of SNF1 in yeast cells. Some of these mutations relieved glucose inhibition of phosphorylation and activity, providing evidence that Snf4 participates in a glucose regulatory mechanism, which may involve ligand binding. Additional substitutions that lack mammalian counterparts and alter residues that are distant from putative nucleotide-binding sites also relieved glucose inhibition of SNF1. Inspection of the crystal structure suggested that interactions of Snf4 with the GBD of the β subunit were perturbed. We further showed that substitutions in the GBD and deletion of the GBD similarly relieve glucose inhibition of SNF1. Finally, we showed that glucose inhibition of SNF1 is normal in cells lacking glycogen. These findings provide evidence for novel roles of Snf4 and the GBD in glucose regulation of SNF1.
EXPERIMENTAL PROCEDURES
Expression of Mutant Snf4 Proteins—Centromeric plasmid pOV75 expresses Snf4 from the native promoter; this plasmid contains a NotI site at the C terminus of the open reading frame and differs from pOV76 (53) in lacking green fluorescent protein sequence. Mutations were introduced into pOV75 by using the QuikChange II XL site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. Wild-type and mutant Snf4 proteins were expressed from pOV75 and its mutant derivatives in snf4Δ mutant cells of S. cerevisiae strain MCY2634 (MATa snf4Δ2 his3 leu2 ura3), which has the S288C genetic background. The elc1Δ::KanMX4 mutant strain was a derivative of BY4741 (MATa his3 leu2 met15 ura3; Research Genetics).
Expression of Mutant Gal83 Proteins—Centromeric plasmid pRT12 expresses Gal83 fused to green fluorescent protein from the native promoter (53). pOV81 and pHW39 (56) are derivatives of pRT12 that express Gal83G235R and Gal83W184A,R214Q, respectively. The GBD (codons 153–243) was deleted from pRT12 by site-directed mutagenesis, yielding pMM66. Wild-type and mutant Gal83 proteins were expressed from these plasmids in S. cerevisiae strain MCY4099 (MATα gal83Δ::TRP1 sip1Δ::kanMX6 sip2Δ::kanMX4 ade2 can1 his3 leu2 trp1 ura3) (56), which has the W303 genetic background.
Assay of SNF1 Catalytic Activity by Phosphorylation of SAMS Peptide—Cultures were grown to exponential phase (A600 of 0.7–1.0) in selective synthetic complete (SC) medium containing 2% glucose. Cells (100 ml) were collected by rapid filtration and were either scraped from the filter and frozen in liquid nitrogen or resuspended in 0.05% glucose for 10 min, collected, and frozen. For some experiments, after incubation in 0.05% glucose, an aliquot of cells was incubated in 2% glucose for 10 min, collected, and frozen. Cell extracts were prepared from two to three independent cultures as described (55). SNF1 was partially purified by chromatography on DEAE-Sepharose (Amersham Biosciences) and assayed for phosphorylation of a synthetic peptide (HMRSAMSGLHLVKRR, called the SAMS peptide) (65) as described (46, 55), using different protein concentrations to confirm linearity. Protein concentrations were determined by Bio-Rad assay. Kinase activity is expressed as nanomoles of phosphate incorporated into the peptide/min/mg of protein (65).
Immunoblot Analysis—Proteins (2–4 μg) from assayed fractions were separated on 10% SDS-PAGE, except where noted otherwise, and analyzed by immunoblotting using anti-Thr(P)-172-AMPK (Cell Signaling Technologies), anti-polyhistidine (Sigma), anti-Snf1 (32), and anti-Snf4 (66) antibodies. Before the membrane was reprobed, it was incubated in 0.2 m glycine, pH 2, for 10 min. ECL Plus or ECL Advance (Amersham Biosciences) was used for visualization.
Growth Assay for 2-Deoxyglucose Resistance—Fresh overnight cultures were grown in selective SC plus 2% glucose and diluted to A600 of 2 in selective SC. The cells were spotted with 2-fold dilutions on selective SC medium containing 2% sucrose, 2-deoxy-d-glucose (200 μg/ml), and antimycin A (1 μg/ml). The media contained the respiratory inhibitor antimycin A to increase the stringency of the assay. The control plates lacked 2-deoxyglucose. The plates were incubated at 30 °C and photographed.
RESULTS
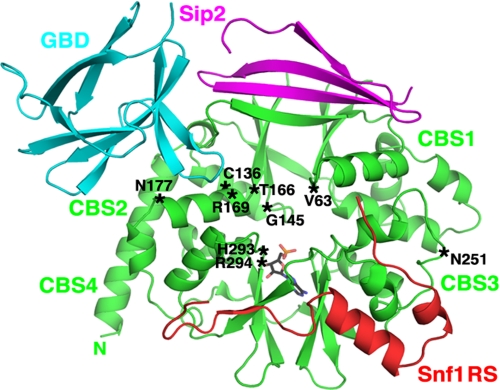
Strategy to Assess Function of SNF1 Protein Kinase Containing a Mutant Snf4 Subunit—To determine whether alteration of Snf4 at sites that cause disease in humans would affect the regulation of SNF1, we introduced mutations into the SNF4 gene, under the control of its own promoter on a centromeric plasmid, and expressed the mutant proteins in snf4Δ yeast cells. The positions of the altered residues are shown in the Snf4 structure from the partial SNF1 heterotrimer, with AMP positioned as it is in the S. pombe γ subunit (58) (Fig. 2). We used three assays to analyze the function of the mutant SNF1. First, we assessed the regulation of kinase activity in response to glucose availability. The cells were grown to mid-log phase in 2% glucose, and an aliquot was subjected to abrupt glucose depletion. The cells were collected by rapid filtration to preserve the phosphorylation state of the kinase, and SNF1 activity was assayed by phosphorylation of the SAMS peptide. In cells expressing wild-type Snf4, the kinase was inhibited by glucose and activated by glucose depletion (Fig. 3A). Second, we examined the phosphorylation of Thr-210 in the activation loop of Snf1 by immunoblot analysis of the same protein samples. Antibody against Thr(P)-172 of AMPK specifically recognizes Thr(P)-210 of Snf1 (11, 35, 47). For wild-type SNF1, phosphorylation of Thr-210 increased substantially in response to glucose limitation, whereas Snf1 and Snf4 protein levels remained constant (Fig. 3B). Finally, we used a growth assay to assess the mutant phenotype. The glucose analog 2-deoxyglucose, which is phosphorylated but not metabolized, prevents growth of wild-type cells on sucrose, and mutations that relieve glucose inhibition of the SNF1 pathway, such as those affecting Reg1-Glc7 protein phosphatase 1, confer 2-deoxyglucose resistance (67). The addition of 2-deoxyglucose to glucose-limited cells inhibits SNF1 (68). The cells expressing wild-type protein kinase did not grow, whereas cells expressing various mutant forms grew to differing extents (Fig. 3C).
FIGURE 2.
Locations of altered residues relative to the structure of S. cerevisiae SNF1. A schematic representation of the partial heterotrimer of SNF1 is shown, adapted from that of Amodeo et al. (58), including Snf4 (green), the GBD (residues 161–247) of Sip2 (cyan), residues 347–412 of Sip2 (magenta), and residues 460–498 of the Snf1 regulatory sequence (RS; red); the position of AMP (stick model) corresponds to that observed in the S. pombe structure (29). The positions corresponding to Snf4 residues that were altered in this study, indicated in single-letter code, are marked (asterisks).
FIGURE 3.
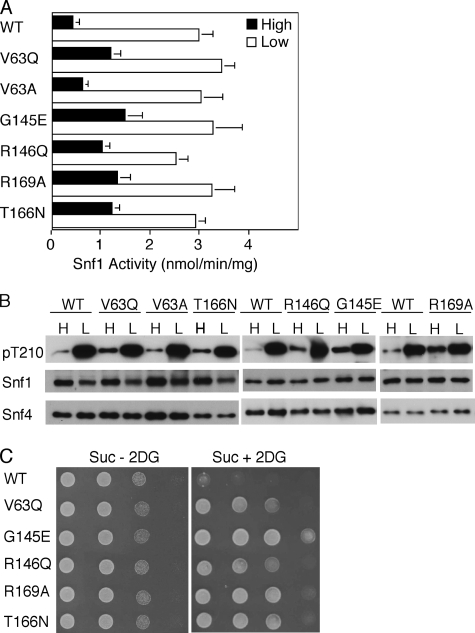
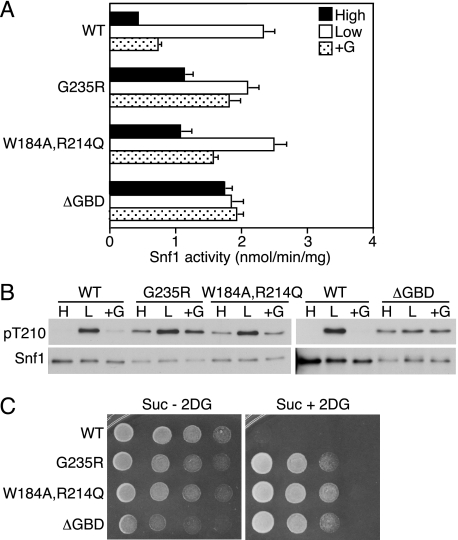
Effects of mutations in the Bateman1 domain on SNF1 catalytic activity, Thr-210 phosphorylation, and resistance to 2-deoxyglucose. Wild-type (WT) and mutant Snf4 proteins were expressed at native levels from plasmids in snf4Δ mutant cells. A and B, cells were grown to mid-log phase in selective SC plus 2% (high, H) glucose, and an aliquot of cells was collected by rapid filtration for assay (filled bars). Another aliquot was collected, resuspended in SC plus 0.05% (low, L) glucose for 10 min, and collected by filtration for assay (open bars). A, partially purified SNF1 was assayed for catalytic activity by phosphorylation of the SAMS peptide, and the values are the averages of results of at least six assays from two independent experiments. B, protein fractions used in A were analyzed by immunoblotting with anti-Thr(P)-172-AMPK antibody to detect phosphorylated Thr-210 (pT210). The blots were reprobed with Snf1 and Snf4 antibodies. C, cells were grown overnight in selective SC plus 2% glucose and spotted with serial 2-fold dilutions on selective SC solid medium containing 2% sucrose, 2-deoxyglucose, and antimycin A(Suc + 2DG) and on control medium lacking 2-deoxyglucose (Suc - 2DG). The plates were incubated at 30 °C and photographed after 36 h (Suc - 2DG) or 60 h (Suc + 2DG).
Together, these assays provided information on mutations causing a range of phenotypes. The assay of catalytic activity provided quantitative data for those mutants with dramatically altered regulation, whereas growth provided the most sensitive assay for the detection of subtle mutant phenotypes, as well as evidence for physiologically relevant effects in vivo.
Mutations in the Bateman1 Domain of Snf4 Corresponding to Human Disease-causing Mutations—We first introduced into Snf4 the substitution V63Q, which is equivalent to R70Q and R302Q in human γ1 and γ2, respectively (Fig. 1). R70Q increases basal phosphorylation and activity of AMPK in cells and reduces allosteric control by AMP in vitro (27). R302Q causes human cardiac disease (22, 25) and affects binding of AMP and ATP to isolated Bateman domains and allosteric activation of AMPK by AMP (19, 28). In pig, the mutation R200Q in γ3 (R226Q in human) increases glycogen levels in skeletal muscle (21). In the structure of the mammalian enzyme, this Arg residue interacts with bound nucleotide (20). In S. cerevisiae, the mutant Snf4V63Q subunit modestly increased SNF1 activity in glucose-grown cells, indicating a loss of glucose inhibition, whereas activity in glucose-depleted cells was normal (Fig. 3A). Phosphorylation of Thr-210 was modestly elevated in glucose-grown cells, as judged by immunoblot analysis of the assayed protein samples (Fig. 3B). Finally, the cells were tested in the growth assay, and the Snf4V63Q subunit conferred resistance to 2-deoxyglucose (Fig. 3C). V63A had little or no effect (Fig. 3 and data not shown).
The mutation H383R in the γ2 subunit (Fig. 1) is associated with human disease (24) and causes defects in binding of AMP and allosteric activation of AMPK (19, 28), and the substitution H151G at the analogous site in γ1 virtually abolishes AMP allosteric control (18). In the mammalian crystal structure, this His residue (His-150) interacts with nucleotide bound to both the site in the Bateman1 domain and the nonexchangeable site in the Bateman2 domain (20). The corresponding Snf4 residue is Gly-145, but the absence of His at this position does not appear to account for the lack of AMP activation of SNF1 because we detected no allosteric effect of AMP (200 μm) on the Snf4G145H mutant kinase (data not shown). The alteration Snf4G145E, however, resulted in a mutant kinase that was highly resistant to glucose inhibition, exhibited marked constitutive phosphorylation of Thr-210, and conferred resistance to 2-deoxyglucose (Fig. 3).
Studies of γ1 showed that the substitutions R152Q and R171Q abolished allosteric control by AMP and inhibition by ATP (18). In the mammalian structure, the residue equivalent to Arg-152 interacted with bound ATP in both exchangeable sites (20). The Snf4 analogs, R146Q and R169A (Fig. 1), relieved glucose inhibition of SNF1 activity and Thr-210 phosphorylation and conferred growth in the presence of 2-deoxyglucose (Fig. 3).
The human mutation T400N in γ2 causes glycogen storage disorder, cardiac hypertrophy, and Wolff-Parkinson-White syndrome (25) and affects nucleotide binding and activation of AMPK (19), and this Thr residue was close to nucleotide-binding sites in the structure (20). Previous studies showed that the equivalent yeast mutation, T166N, caused a modest increase in the interaction of Snf4 and Snf1 in the two-hybrid system (25), which could reflect increased nuclear localization of SNF1 resulting from increased activation (55). Consistent with these results, we found that T166N increased SNF1 activity in glucose-grown cells and conferred resistance to 2-deoxyglucose (Fig. 3). T166A had little or no effect (data not shown).
These findings indicate that the regulation of SNF1 is affected, to varying extents, by mutations in the Bateman1 domain of Snf4 corresponding to, or at the same sites as, human mutations that affect regulation of AMPK by AMP and ATP in vitro. Taken together, the assays of catalytic activity, phosphorylation of Thr-210, and cell growth phenotype indicate that these mutations relieve glucose inhibition of SNF1 in vivo. In all cases, activity in glucose-depleted cells was normal.
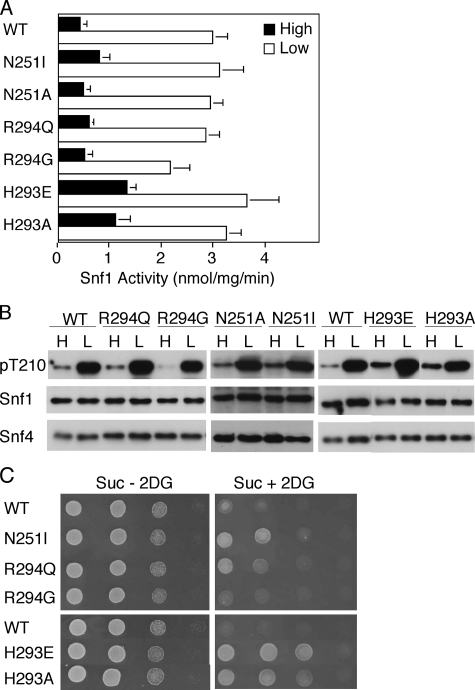
Mutations in the Bateman2 Domain at Sites Corresponding to Disease-causing Mutations—The substitution N488I in γ2 is associated with human cardiac glycogen storage disorder and Wolff-Parkinson-White syndrome (25) and has been reported to cause increased basal activity and loss of nucleotide regulation in mouse heart (69, 70), but this Asn residue is not located close to the nucleotide-binding sites in the crystal structure (20). The corresponding substitution in Snf4, N251I, showed somewhat improved resistance to 2-deoxyglucose, but the effects on kinase activation were marginal; N251A had no effect (Fig. 4 and data not shown).
FIGURE 4.
Effects of mutations in the Bateman2 domain. Wild-type (WT) and mutant Snf4 proteins were expressed in snf4Δ mutant cells. Analysis was as in Fig. 3. A, cells were grown in high glucose or shifted to low glucose, and SNF1 catalytic activity was assayed in two independent experiments. The values are the averages of at least six assays. B, protein fractions used in A were subjected to immunoblot analysis to detect phosphorylated Thr-210 (pT210), Snf1, and Snf4. C, cells were tested for growth on Suc + 2DG and Suc - 2DG medium.
Residue His-293 is located in CBS4 of Snf4 at a position corresponding to that of Gly-145 in CBS2. Similarly to G145E, the substitution H293E, and also H293A, elevated SNF1 activity and phosphorylation of Thr-210 in glucose-grown cells and conferred resistance to 2-deoxyglucose (Fig. 4). The corresponding residue in mammalian AMPK, His-297, interacts with phosphate groups of nucleotides bound to both sites in the Bateman2 domain (20), whereas in the S. pombe structure, His-289 contacts ADP bound to the Bateman1 site (30).
The substitutions R531G/Q in human γ2 cause very strong phenotypes, particularly R531Q, which causes fatal congenital cardiac glycogenosis (23, 26). Both mutations strongly reduce binding of nucleotides and allosteric AMP activation of AMPK and increase basal activity of AMPK in cells (19, 26, 28). The equivalent R298G in rat γ1 also causes defects (16), and Arg-298 interacts with nucleotide bound to the exchangeable site in the Bateman2 domain (20). In the S. pombe structure with one molecule of nucleotide bound, the corresponding residue coordinated nucleotide phosphate (29), although the functional significance of this residue remains to be determined. In contrast to the very substantial effects on mammalian AMPK, the analogous substitutions R294G/Q in Snf4 had little or no effect on SNF1. Catalytic activity was not elevated in glucose-grown cells, nor was phosphorylation of Thr-210, and the very sensitive growth assay showed only slight growth of cells expressing Snf4R294Q (Fig. 4). These findings indicate differences between Snf4 and mammalian γ subunits.
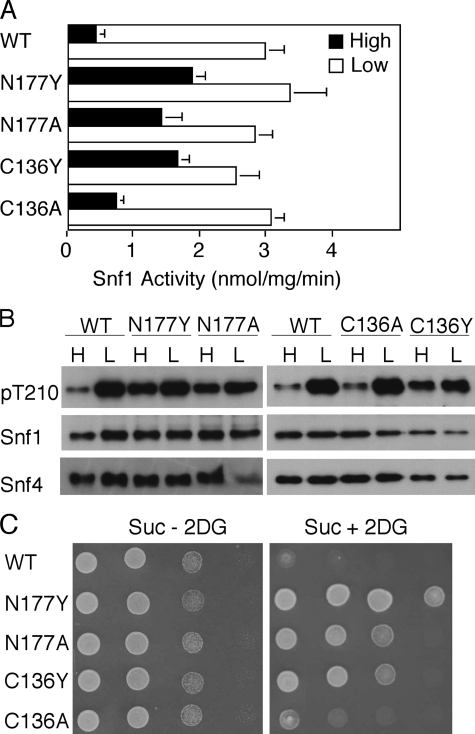
Mutations That Alter Residues Cys-136 and Asn-177—We also examined the effects of two mutations that lack counterparts in mammalian γ subunits and alter residues that are distant from putative nucleotide-binding sites (Fig. 2) but have been shown to affect glucose regulation of transcription. The SNF4-C136Y allele was isolated from a strain carrying a dominant GAL82 allele (71) and was confirmed to affect glucose repression of GAL genes.4 We found that expression of Snf4C136Y caused high level SNF1 activity and strong phosphorylation of Thr-210 in glucose-grown cells (Fig. 5), which was particularly striking because levels of Snf1 and Snf4C136Y proteins were reproducibly low (Fig. 5B). The mutation also conferred resistance to 2-deoxyglucose (Fig. 5C). The substitution C136A had little effect (Fig. 5). We note that a Tyr residue at position 136 is not compatible with the observed crystal structure (58). C136Y, and also R146Q, described above, alter a pseudosubstrate sequence that has been proposed to inhibit the catalytic domain (72); our findings neither support nor refute this model.
FIGURE 5.
Effects of alteration of residues Cys-136 and Asn-177. Wild-type (WT) and mutant Snf4 proteins were expressed in snf4Δ mutant cells. Analysis was as in Fig. 3. A, cells were grown in high glucose, or shifted to low glucose, and SNF1 catalytic activity was assayed. The values are the averages of results of at least six assays from two independent experiments. B, protein fractions used in A were subjected to immunoblot analysis to detect phosphorylated Thr-210 (pT210), Snf1, and Snf4. C, cells were tested for growth on Suc + 2DG and Suc - 2DG medium.
Cys-136 was reported to lie within a consensus elongin C-binding site ((T/S)(L/M)XXX(C/S)XXX(V/L/I)) (73), and the yeast elongin C, called Elc1, binds Snf4 (74). Elc1 is a homolog of Skp1, of the Skp1-Cullin-F-box ubiquitin ligase. Elc1 is a component of nucleotide excision repair factor 4 (75) and has been proposed to be a component of a ubiquitin ligase required for polyubiquitylation of RNA polymerase II in response to DNA damage (76). Snf4 protein levels were modestly reduced in the elc1 mutant (77), but no SNF1-related growth defect was detected (74). We assayed elc1Δ cells and found no defect in glucose regulation of SNF1 activity or Thr-210 phosphorylation (data not shown). These results do not support a regulatory role for Elc1 in the SNF1 pathway, but it is possible that Elc1 affects some as yet unidentified function of Snf4; whereas the molar excess of Snf4 relative to Snf1 is controversial (53, 63, 78), subcellular localization studies suggest that not all Snf4 molecules are associated with Snf1 (53).
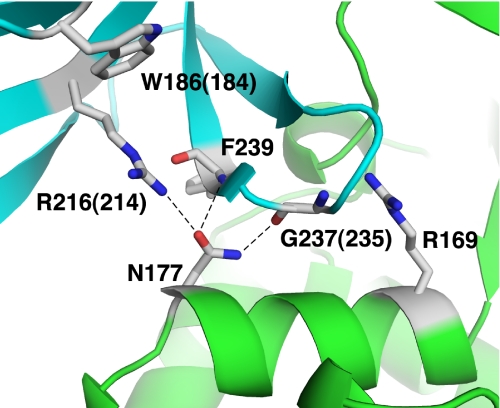
The N177Y allele of SNF4 was recovered as a dominant suppressor that restored INO1 transcription in a TATA-binding protein mutant; it reduced glucose repression of SUC2 and increased the two-hybrid interaction of Snf1 with Snf4 in glucose-grown cells (79). We found that N177Y substantially reduced glucose inhibition of SNF1, conferring high level activity and phosphorylation of Thr-210 in glucose-grown cells, and also allowed strong growth in the presence of 2-deoxyglucose (Fig. 5). Moreover, we found that N177A had nearly the same phenotype (Fig. 5). In the crystal structure of the partial SNF1 heterotrimer (58), Asn-177 is located near the Sip2 GBD and forms hydrogen bonds with Arg-216 and the main chain carbonyl and amide groups of the Gly-237 and Phe-239 residues, respectively (Fig. 6). In addition, we noted that the alteration R169A, described above, would disrupt interactions of Snf4 residue Arg-169 with main chain carbonyl groups of residues 234, 235, and 236 of the GBD. These findings suggested that the interaction of Snf4 and the GBD is important for glucose regulation.
FIGURE 6.
Locations of altered Snf4 and Gal83 GBD residues relative to the structure of SNF1. A schematic representation of the interacting regions of Snf4 (green) and the GBD of Sip2 (cyan) within the SNF1 partial heterotrimer has been adapted from that of Amodeo et al. (58). Relevant Gal83 and Snf4 residues are indicated in single-letter code. The numbers correspond to Sip2 residues, with equivalent Gal83 residues indicated in parentheses. Arg-169 forms hydrogen bonds with main chain carbonyl groups of GBD residues 234, 235, and 236 (not shown).
Mutation of the Gal83 GBD Relieves Glucose Inhibition of SNF1—The GBD sequence is conserved in both Gal83 (residues 161–243) and Sip2 (residues 163–245). Gal83 binds glycogen strongly in vitro, whereas Sip2 binds weakly (56). The substitutions W184A,R214Q (and W186A,R216Q in Sip2) and G235R in Gal83 abolished binding (56), and equivalent residues in AMPK β-GBD affected glycogen binding (80) and interaction with β-cyclodextrin in a crystal structure (the equivalent G147R is predicted to affect conformation) (81). Gal83 is the major isoform during growth on glucose (53) and is thus important for SNF1 activation in response to glucose withdrawal (55). Gal83W184A,R214Q and Gal83G235R enhanced various SNF1-dependent processes, including transcriptional activation by Sip4 and activation of the carbon source-responsive element after a shift from glucose to nonfermentable carbon source, glycogen accumulation, and haploid invasive growth (56). SNF1 activity was not elevated in cells after a shift to nonfermentable carbon source, but activity in glucose-grown cells was not tested.
To determine whether these alterations relieve glucose inhibition of SNF1, we expressed Gal83W184A,R214Q and Gal83G235R in the gal83Δ sip1Δ sip2Δ triple mutant, which has the W303 genetic background. In control experiments, Snf4N177Y, Snf4G145E, and Snf4R169A behaved similarly in the W303 background as in the S288C background used above (data not shown). Both mutant Gal83 proteins not only restored SNF1 activation in the triple mutant but also relieved glucose inhibition of activity and Thr-210 phosphorylation; they also prevented re-establishment of inhibition when cells that had been subjected to glucose depletion were returned to glucose-replete medium (Fig. 7, A and B). These alterations also conferred resistance to 2-deoxyglucose (Fig. 7C). Previous findings that Gal83W184A,R214Q and Gal83G235R affect SNF1-dependent functions as assayed following a shift from glucose to nonfermentable carbon source (56) most likely reflect the increased SNF1 activity in cells prior to the shift.
FIGURE 7.
Mutations in the Gal83 GBD alter regulation of SNF1. A and B, wild-type (WT) and mutant Gal83 proteins (fused to green fluorescent protein) were expressed in gal83Δ sip1Δ sip2Δ mutant cells (MCY4099). Analysis was as in Fig. 3, except that after cells were resuspended in SC plus 0.05% glucose for 10 min, an aliquot was also incubated in SC plus 2% glucose (+G) for 10 min and then harvested. A, SNF1 catalytic activity was assayed in three independent experiments. The values are the averages of six assays. B, protein fractions used in A were separated by 8% SDS-PAGE and subjected to immunoblot analysis; separate blots were probed with anti-histidine antibody to detect Snf1 and with antibody specific for phosphorylated Thr-210 (pT210). C, Gal83 proteins were expressed in gal83Δ sip1Δ sip2Δ snf4Δ::hphMX4 cells (derivative of MCY4099) that coexpressed SNF4. The cells were tested for growth on Suc - 2DG and Suc + 2DG medium; for the latter, the plates were photographed after 84 h.
We then deleted codons 153–243 of Gal83 to remove the entire GBD. Expression of Gal83ΔGBD in the triple mutant similarly relieved glucose inhibition of SNF1 activity and Thr-210 phosphorylation (Fig. 7, A and B) and conferred 2-deoxyglucose resistance (Fig. 7C). None of the three mutant Gal83 proteins conferred SNF1 function in the absence of Snf4, as judged by growth assays (data not shown). These findings indicate that the interaction of the GBD and Snf4 is essential for glucose inhibition of SNF1.
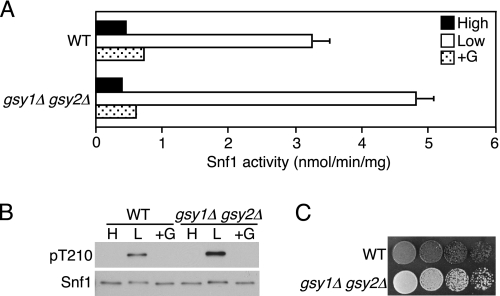
To determine whether glycogen binding by the GBD is important for inhibition of SNF1 activity, we assayed gsy1Δ gsy2Δ mutant cells, which lack glycogen synthase and cannot synthesize glycogen. Glucose inhibition of SNF1 activity and Thr-210 phosphorylation were normal in these cells (Fig. 8, A and B), and the absence of glycogen was confirmed by iodine staining (Fig. 8C). We previously showed that Gal83W184A,R214Q affects Sip4 transcriptional activator function and activation of the carbon source-responsive element in gsy1Δ gsy2Δ cells (56). These results support the view that the GBD interacts with Snf4 to affect glucose inhibition of SNF1 by a mechanism that is independent of glycogen binding. The possibility that glycogen binding has other regulatory effects is not excluded.
FIGURE 8.
Cells lacking glycogen exhibit normal regulation of SNF1. A and B, gsy1Δ gsy2Δ cells (MCY4101, MATa gsy1Δ::kanMX4 gsy2Δ::kanMX4 his3 leu2 lys2 ura3) (56) and wild-type (WT) cells of the same genetic background (BY4741) were grown to mid-log phase in YEP plus 2% glucose. Analysis was as in Fig. 7 except that rich medium was used. A, cells were grown in high glucose, shifted to low glucose, and incubated again in high glucose. SNF1 catalytic activity was assayed in three independent experiments. The values are the averages of six assays. B, protein fractions used in A were subjected to immunoblot analysis to detect phosphorylated Thr-210 (pT210) and Snf1. C, cells were spotted with serial 5-fold dilutions on YEP solid medium containing 2% glucose. The cells were exposed to iodine vapor for 3 min to stain glycogen and were photographed.
DISCUSSION
We present evidence for regulatory roles of Snf4 and the GBD of Gal83 in glucose inhibition of SNF1. Previous studies of mutant cells lacking Snf4 indicated that Snf4 counteracts autoinhibition of Snf1 by C-terminal regulatory sequences but is not required for glucose-regulated phosphorylation of Thr-210 (46, 50, 52, 61–63), implying that at least one glucose regulatory mechanism functions independently of Snf4. Here, studies of cells containing heterotrimeric SNF1 with various mutant Snf4 subunits have revealed that Snf4 also has regulatory roles in glucose inhibition of Thr-210 phosphorylation and SNF1 activity.
We have shown that regulation of SNF1 is affected by alterations of Snf4 at sites corresponding to those of human mutations that affect nucleotide binding by AMPK. In S. cerevisiae cells, these mutations relieved glucose inhibition of SNF1 activity, as judged by a combination of assays of catalytic activity, Thr-210 phosphorylation, and growth. These findings are consistent with the view that Snf4 binds nucleotide and that binding affects function; moreover, a crystal of a partial SNF1 heterotrimer with AMP bound at one site has been reported (58). Our genetic findings, however, are equally compatible with the possibility that, in vivo, Snf4 binds another adenosine derivative, as is the case for other Bateman domains (19). Analysis of Snf4R294G/Q indicates that the regulation of SNF1 is distinct from that of mammalian AMPK in another respect besides the lack of AMP allosteric activation (45, 46, 51). Whereas R531G/Q in γ2 and R298G in g1 cause severe defects (16, 19, 23, 26, 28) and Arg-298 interacts with nucleotide (20), the analogous R294G/Q had little effect, suggesting that Arg-294 does not contribute to ligand binding or that binding to the cognate site has no functional consequence. Although the question of ligand binding remains unresolved, these findings nonetheless establish that Snf4 not only counteracts autoinhibition but also has a regulatory role in glucose inhibition of Thr-210 phosphorylation.
The increased phosphorylation of Thr-210 in glucose-grown cells carrying these mutant Snf4 subunits could result from increased phosphorylation, decreased dephosphorylation, or both. For AMPK, it has been reported that AMP binding promotes phosphorylation of the activation loop Thr-172 by LKB1 (4) and also that it attenuates dephosphorylation of Thr-172 by protein phosphatase 2Cα (13, 15, 16). Previous studies suggested that Reg1-Glc7 protein phosphatase 1 is active toward wild-type SNF1 in glucose-grown cells but not in glucose-limited cells (8) and that access of phosphorylated Thr-210 to Reg1-Glc7 is glucose-regulated (82).
We further found that alteration of Asn-177, which is located distant from putative ligand-binding sites (Fig. 2), strongly relieved glucose inhibition of SNF1 activity. In the crystal structure of the SNF1 partial heterotrimer (58), Asn-177 is located near the Sip2 GBD and forms hydrogen bonds with several residues. Arg-169 also interacts with the GBD, and the R169A substitution relieved glucose inhibition, although less markedly. These findings suggested that N177Y, N177A, and R169A affect glucose inhibition by altering the interaction of Snf4 with the GBD of the β subunit. We then showed that alterations of Gal83 GBD residues that are involved in such interactions, as determined from the structure, similarly relieved glucose inhibition of SNF1 activity and Thr-210 phosphorylation, as did simple deletion of the GBD. Analysis of SNF1 regulation in gsy1Δ gsy2Δ cells lacking glycogen indicated that the GBD affects glucose inhibition of SNF1 by a mechanism that is independent of glycogen binding. These findings reveal a new regulatory role for the GBD and indicate that the interaction of the GBD and Snf4 is essential for glucose inhibition of SNF1.
Acknowledgments
We thank Penny Becker and Mark Johnston for communication of results regarding C136Y and Gabriele Amodeo for assistance with Fig. 2.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34095 (to M. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AMPK, AMP-activated protein kinase; CBS, cystathionine β-synthase; GBD, glycogen-binding domain; SC, synthetic complete; Suc, sucrose; 2DG, 2-deoxyglucose.
P. Becker and M. Johnston, personal communication.
References
- 1.Hardie, D. G., Carling, D., and Carlson, M. (1998) Annu. Rev. Biochem. 67 821-855 [DOI] [PubMed] [Google Scholar]
- 2.Kahn, B. B., Alquier, T., Carling, D., and Hardie, D. G. (2005) Cell Metab. 1 15-25 [DOI] [PubMed] [Google Scholar]
- 3.Hong, S.-P., Leiper, F. C., Woods, A., Carling, D., and Carlson, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8839-8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawley, S. A., Boudeau, J., Reid, J. L., Mustard, K. J., Udd, L., Makela, T. P., Alessi, D. R., and Hardie, D. G. (2003) J. Biol. 2 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woods, A., Johnstone, S. R., Dickerson, K., Leiper, F. C., Fryer, L. G., Neumann, D., Schlattner, U., Wallimann, T., Carlson, M., and Carling, D. (2003) Curr. Biol. 13 2004-2008 [DOI] [PubMed] [Google Scholar]
- 6.Shaw, R. J., Kosmatka, M., Bardeesy, N., Hurley, R. L., Witters, L. A., DePinho, R. A., and Cantley, L. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3329-3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawley, S. A., Pan, D. A., Mustard, K. J., Ross, L., Bain, J., Edelman, A. M., Frenguelli, B. G., and Hardie, D. G. (2005) Cell Metab. 2 9-19 [DOI] [PubMed] [Google Scholar]
- 8.Hong, S. P., Momcilovic, M., and Carlson, M. (2005) J. Biol. Chem. 280 21804-21809 [DOI] [PubMed] [Google Scholar]
- 9.Hurley, R. L., Anderson, K. A., Franzone, J. M., Kemp, B. E., Means, A. R., and Witters, L. A. (2005) J. Biol. Chem. 280 29060-29066 [DOI] [PubMed] [Google Scholar]
- 10.Woods, A., Dickerson, K., Heath, R., Hong, S. P., Momcilovic, M., Johnstone, S. R., Carlson, M., and Carling, D. (2005) Cell Metab. 2 21-33 [DOI] [PubMed] [Google Scholar]
- 11.Momcilovic, M., Hong, S. P., and Carlson, M. (2006) J. Biol. Chem. 281 25336-25343 [DOI] [PubMed] [Google Scholar]
- 12.Hawley, S. A., Selbert, M. A., Goldstein, E. G., Edelman, A. M., Carling, D., and Hardie, D. G. (1995) J. Biol. Chem. 270 27186-27191 [DOI] [PubMed] [Google Scholar]
- 13.Davies, S. P., Helps, N. R., Cohen, P. T., and Hardie, D. G. (1995) FEBS Lett. 377 421-425 [DOI] [PubMed] [Google Scholar]
- 14.Hawley, S. A., Davison, M., Woods, A., Davies, S. P., Beri, R. K., Carling, D., and Hardie, D. G. (1996) J. Biol. Chem. 271 27879-27887 [DOI] [PubMed] [Google Scholar]
- 15.Suter, M., Riek, U., Tuerk, R., Schlattner, U., Wallimann, T., and Neumann, D. (2006) J. Biol. Chem. 281 32207-32216 [DOI] [PubMed] [Google Scholar]
- 16.Sanders, M. J., Grondin, P. O., Hegarty, B. D., Snowden, M. A., and Carling, D. (2007) Biochem. J. 403 139-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman, A. (1997) Trends Biochem. Sci 22 12-13 [DOI] [PubMed] [Google Scholar]
- 18.Adams, J., Chen, Z. P., Van Denderen, B. J., Morton, C. J., Parker, M. W., Witters, L. A., Stapleton, D., and Kemp, B. E. (2004) Protein Sci 13 155-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott, J. W., Hawley, S. A., Green, K. A., Anis, M., Stewart, G., Scullion, G. A., Norman, D. G., and Hardie, D. G. (2004) J. Clin. Investig. 113 274-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao, B., Heath, R., Saiu, P., Leiper, F. C., Leone, P., Jing, C., Walker, P. A., Haire, L., Eccleston, J. F., Davis, C. T., Martin, S. R., Carling, D., and Gamblin, S. J. (2007) Nature 449 496-500 [DOI] [PubMed] [Google Scholar]
- 21.Milan, D., Jeon, J.-T., Looft, C., Amarger, V., Robic, A., Thelander, M., Rogel-Gaillard, C., Paul, S., Iannuccelli, N., Rask, L., Ronne, H., Lundstrom, K., Reinsch, N., Gellin, J., Kalm, E., Le Roy, P., Chardon, P., and Andersson, L. (2000) Science 288 1248-1251 [DOI] [PubMed] [Google Scholar]
- 22.Gollob, M. H., Green, M. S., Tang, A. S., Gollob, T., Karibe, A., Ali Hassan, A. S., Ahmad, F., Lozado, R., Shah, G., Fananapazir, L., Bachinski, L. L., and Roberts, R. (2001) N. Engl. J. Med. 344 1823-1831 [DOI] [PubMed] [Google Scholar]
- 23.Gollob, M. H., Seger, J. J., Gollob, T. N., Tapscott, T., Gonzales, O., Bachinski, L., and Roberts, R. (2001) Circulation 104 3030-3033 [DOI] [PubMed] [Google Scholar]
- 24.Blair, E., Redwood, C., Ashrafian, H., Oliveira, M., Broxholme, J., Kerr, B., Salmon, A., Ostman-Smith, I., and Watkins, H. (2001) Hum. Mol. Genet. 10 1215-1220 [DOI] [PubMed] [Google Scholar]
- 25.Arad, M., Benson, D. W., Perez-Atayde, A. R., McKenna, W. J., Sparks, E. A., Kanter, R. J., McGarry, K., Seidman, J. G., and Seidman, C. E. (2002) J. Clin. Investig. 109 357-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burwinkel, B., Scott, J. W., Buhrer, C., van Landeghem, F. K., Cox, G. F., Wilson, C. J., Hardie, D. G., and Kilimann, M. W. (2005) Am. J. Hum. Genet. 76 1034-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton, S. R., Stapleton, D., O'Donnell, J. B., Jr., Kung, J. T., Dalal, S. R., Kemp, B. E., and Witters, L. A. (2001) FEBS Lett. 500 163-168 [DOI] [PubMed] [Google Scholar]
- 28.Daniel, T., and Carling, D. (2002) J. Biol. Chem. 277 51017-51024 [DOI] [PubMed] [Google Scholar]
- 29.Townley, R., and Shapiro, L. (2007) Science 315 1726-1729 [DOI] [PubMed] [Google Scholar]
- 30.Jin, X., Townley, R., and Shapiro, L. (2007) Structure 15 1285-1295 [DOI] [PubMed] [Google Scholar]
- 31.Hedbacker, K., and Carlson, M. (2008) Front. Biosci. 13 2408-2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celenza, J. L., and Carlson, M. (1986) Science 233 1175-1180 [DOI] [PubMed] [Google Scholar]
- 33.Thompson-Jaeger, S., Francois, J., Gaughran, J. P., and Tatchell, K. (1991) Genetics 129 697-706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchin, S., Vyas, V. K., and Carlson, M. (2002) Mol. Cell. Biol. 22 3994-4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlova, M., Kanter, E., Krakovich, D., and Kuchin, S. (2006) Eukaryot. Cell 5 1831-1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alepuz, P. M., Cunningham, K. W., and Estruch, F. (1997) Mol. Microbiol. 26 91-98 [DOI] [PubMed] [Google Scholar]
- 37.Portillo, F., Mulet, J. M., and Serrano, R. (2005) FEBS Lett. 579 512-516 [DOI] [PubMed] [Google Scholar]
- 38.Platara, M., Ruiz, A., Serrano, R., Palomino, A., Moreno, F., and Arino, J. (2006) J. Biol. Chem. 281 36632-36642 [DOI] [PubMed] [Google Scholar]
- 39.Dubacq, C., Chevalier, A., and Mann, C. (2004) Mol. Cell. Biol. 24 2560-2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong, S. P., and Carlson, M. (2007) J. Biol. Chem. 282 16838-16845 [DOI] [PubMed] [Google Scholar]
- 41.Carlson, M., Osmond, B. C., and Botstein, D. (1981) Genetics 98 25-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlson, M., and Botstein, D. (1982) Cell 28 145-154 [DOI] [PubMed] [Google Scholar]
- 43.Young, E. T., Dombek, K. M., Tachibana, C., and Ideker, T. (2003) J. Biol. Chem. 278 26146-26158 [DOI] [PubMed] [Google Scholar]
- 44.Hardy, T. A., Huang, D., and Roach, P. J. (1994) J. Biol. Chem. 269 27907-27913 [PubMed] [Google Scholar]
- 45.Mitchelhill, K. I., Stapleton, D., Gao, G., House, C., Michell, B., Katsis, F., Witters, L. A., and Kemp, B. E. (1994) J. Biol. Chem. 269 2361-2364 [PubMed] [Google Scholar]
- 46.Woods, A., Munday, M. R., Scott, J., Yang, X., Carlson, M., and Carling, D. (1994) J. Biol. Chem. 269 19509-19516 [PubMed] [Google Scholar]
- 47.Sutherland, C. M., Hawley, S. A., McCartney, R. R., Leech, A., Stark, M. J., Schmidt, M. C., and Hardie, D. G. (2003) Curr. Biol. 13 1299-1305 [DOI] [PubMed] [Google Scholar]
- 48.Nath, N., McCartney, R. R., and Schmidt, M. C. (2003) Mol. Cell. Biol. 23 3909-3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanz, P., Alms, G. R., Haystead, T. A. J., and Carlson, M. (2000) Mol. Cell. Biol. 20 1321-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCartney, R. R., and Schmidt, M. C. (2001) J. Biol. Chem. 276 36460-36466 [DOI] [PubMed] [Google Scholar]
- 51.Wilson, W. A., Hawley, S. A., and Hardie, D. G. (1996) Curr. Biol. 6 1426-1434 [DOI] [PubMed] [Google Scholar]
- 52.Jiang, R., and Carlson, M. (1996) Genes Dev. 10 3105-3115 [DOI] [PubMed] [Google Scholar]
- 53.Vincent, O., Townley, R., Kuchin, S., and Carlson, M. (2001) Genes Dev. 15 1104-1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vincent, O., and Carlson, M. (1999) EMBO J. 18 6672-6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hedbacker, K., Hong, S. P., and Carlson, M. (2004) Mol. Cell. Biol. 24 8255-8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiatrowski, H. A., van Denderen, B. J. W., Berkey, C. D., Kemp, B. E., Stapleton, D., and Carlson, M. (2004) Mol. Cell. Biol. 24 352-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang, R., and Carlson, M. (1997) Mol. Cell. Biol. 17 2099-2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amodeo, G. A., Rudolph, M. J., and Tong, L. (2007) Nature 449 492-495 [DOI] [PubMed] [Google Scholar]
- 59.Neigeborn, L., and Carlson, M. (1984) Genetics 108 845-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Celenza, J. L., Eng, F. J., and Carlson, M. (1989) Mol. Cell. Biol. 9 5045-5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Celenza, J. L., and Carlson, M. (1989) Mol. Cell. Biol. 9 5034-5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leech, A., Nath, N., McCartney, R. R., and Schmidt, M. C. (2003) Eukaryot. Cell 2 265-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elbing, K., Rubenstein, E. M., McCartney, R. R., and Schmidt, M. C. (2006) J. Biol. Chem. 281 26170-26180 [DOI] [PubMed] [Google Scholar]
- 64.Rudolph, M. J., Amodeo, G. A., Iram, S. H., Hong, S. P., Pirino, G., Carlson, M., and Tong, L. (2007) Structure 15 65-74 [DOI] [PubMed] [Google Scholar]
- 65.Davies, S. P., Carling, D., and Hardie, D. G. (1989) Eur. J. Biochem. 186 123-128 [DOI] [PubMed] [Google Scholar]
- 66.Estruch, F., Treitel, M. A., Yang, X., and Carlson, M. (1992) Genetics 132 639-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neigeborn, L., and Carlson, M. (1987) Genetics 115 247-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hedbacker, K., and Carlson, M. (2006) Eukaryot. Cell 5 1950-1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arad, M., Moskowitz, I. P., Patel, V. V., Ahmad, F., Perez-Atayde, A. R., Sawyer, D. B., Walter, M., Li, G. H., Burgon, P. G., Maguire, C. T., Stapleton, D., Schmitt, J. P., Guo, X. X., Pizard, A., Kupershmidt, S., Roden, D. M., Berul, C. I., Seidman, C. E., and Seidman, J. G. (2003) Circulation 107 2850-2856 [DOI] [PubMed] [Google Scholar]
- 70.Zou, L., Shen, M., Arad, M., He, H., Lofgren, B., Ingwall, J. S., Seidman, C. E., Seidman, J. G., and Tian, R. (2005) Circ. Res. 97 323-328 [DOI] [PubMed] [Google Scholar]
- 71.Matsumoto, K., Toh-e, A., and Oshima, Y. (1981) Mol. Cell. Biol. 1 83-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scott, J. W., Ross, F. A., Liu, J. K., and Hardie, D. G. (2007) EMBO J. 26 806-815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kamura, T., Sato, S., Haque, D., Liu, L., Kaelin, W. G., Jr., Conaway, R. C., and Conaway, J. W. (1998) Genes Dev. 12 3872-3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jackson, T., Kwon, E., Chachulska, A. M., and Hyman, L. E. (2000) Biochim. Biophys. Acta 1491 161-176 [DOI] [PubMed] [Google Scholar]
- 75.Ramsey, K. L., Smith, J. J., Dasgupta, A., Maqani, N., Grant, P., and Auble, D. T. (2004) Mol. Cell. Biol. 24 6362-6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ribar, B., Prakash, L., and Prakash, S. (2006) Mol. Cell. Biol. 26 3999-4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hyman, L. E., Kwon, E., Ghosh, S., McGee, J., Chachulska, A. M., Jackson, T., and Baricos, W. H. (2002) J. Biol. Chem. 277 15586-15591 [DOI] [PubMed] [Google Scholar]
- 78.Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., and Weissman, J. S. (2003) Nature 425 737-741 [DOI] [PubMed] [Google Scholar]
- 79.Shirra, M. K., and Arndt, K. M. (1999) Genetics 152 73-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polekhina, G., Gupta, A., Michell, B. J., van Denderen, B., Murthy, S., Feil, S. C., Jennings, I. G., Campbell, D. J., Witters, L. A., Parker, M. W., Kemp, B. E., and Stapleton, D. (2003) Curr. Biol. 13 867-871 [DOI] [PubMed] [Google Scholar]
- 81.Polekhina, G., Gupta, A., van Denderen, B. J., Feil, S. C., Kemp, B. E., Stapleton, D., and Parker, M. W. (2005) Structure 13 1453-1462 [DOI] [PubMed] [Google Scholar]
- 82.Rubenstein, E. M., McCartney, R. R., Zhang, C., Shokat, K. M., Shirra, M. K., Arndt, K. M., and Schmidt, M. C. (2008) J. Biol. Chem. 283 222-230 [DOI] [PMC free article] [PubMed] [Google Scholar]