Abstract
Visual perception begins with the absorption of a photon by an opsin pigment, inducing isomerization of its 11-cis-retinaldehyde chromophore. After a brief period of activation, the resulting all-trans-retinaldehyde dissociates from the opsin apoprotein rendering it insensitive to light. Restoring light sensitivity to apo-opsin requires thermal re-isomerization of all-trans-retinaldehyde to 11-cis-retinaldehyde via an enzyme pathway called the visual cycle in retinal pigment epithelial (RPE) cells. Vertebrates can see over a 108-fold range of background illumination. This implies that the visual cycle can regenerate a visual chromophore over a similarly broad range. However, nothing is known about how the visual cycle is regulated. Here we show that RPE cells, functionally or physically separated from photoreceptors, respond to light by mobilizing all-trans-retinyl esters. These retinyl esters are substrates for the retinoid isomerase and hence critical for regenerating visual chromophore. We show in knock-out mice and by RNA interference in human RPE cells that this mobilization is mediated by a protein called “RPE-retinal G protein receptor” (RGR) opsin. These data establish that RPE cells are intrinsically sensitive to light. Finally, we show that in the dark, RGR-opsin inhibits lecithin:retinol acyltransferase and all-trans-retinyl ester hydrolase in vitro and that this inhibition is released upon exposure to light. The results of this study suggest that RGR-opsin mediates light-dependent translocation of all-trans-retinyl esters from a storage pool in lipid droplets to an “isomerase pool” in membranes of the endoplasmic reticulum. This translocation permits insoluble all-trans-retinyl esters to be utilized as substrate for the synthesis of a new visual chromophore.
The opsins include a group of light-sensitive G protein-coupled receptors. The ligand for most opsins is 11-cis-retinaldehyde (11-cis-RAL),3 which is covalently coupled to a Lys residue in the opsins as a Schiff base. Absorption of a photon by most photoreceptor opsins isomerizes the 11-cis-RAL to all-trans-retinaldehyde (all-trans-RAL), inducing a conformational change that activates the G protein (transducin) and begins the visual transduction cascade (1). In the case of vertebrate photoreceptor opsins (rhodopsin and the cone opsins), the Schiff base is hydrolyzed and all-trans-RAL dissociates following photoisomerization. Restoration of light sensitivity to the resulting apo-opsin requires thermal re-isomerization of all-trans-RAL to 11-cis-RAL via a multistep enzyme pathway called the visual cycle in cells of the adjacent retinal pigment epithelium (RPE) (2) (Fig. 1). In contrast, all-trans-RAL remains covalently bound to many invertebrate opsins following photoisomerization. Here, the bound all-trans-RAL is re-isomerized in situ back to 11-cis-RAL by absorption of a second photon through a process called photoregeneration (3-5). Besides rhodopsin and the cone opsins, vertebrates contain several “nonvisual” opsins with sequence similarity to the invertebrate opsins. These include melanopsin (Opn4) (6), neuropsin (Opn5) (7), encephalopsin (Opn3) (8), peropsin (9), and RPE-retinal G protein receptor opsin (RGR) (10). Melanopsin is expressed in a subset of retinal ganglion cells and mediates circadian photo-entrainment and the pupillary light response (11, 12). The function of the other vertebrate nonvisual opsins is unknown.
FIGURE 1.
Visual cycle. Absorption of a photon (hv) by a rhodopsin pigment molecule in a rod photoreceptor induces isomerization of 11-cis-RAL to all-trans-RAL, resulting in activated metarhodopsin II. Decay of metarhodopsin II yields apo-opsin and free all-trans-RAL, which is reduced to all-trans-ROL by all-trans-ROL dehydrogenase (all-trans-RDH). The all-trans-ROL is released by photoreceptors and taken up by RPE cells where it is esterified to a fatty acid lecithin:retinol acyltransferase (LRAT). Rpe65 isomerase uses all-trans-REs as substrate to form 11-cis-ROL. 11-cis-ROL is oxidized to 11-cis-RAL by one of several 11-cis-RDHs. 11-cis-RAL is released by the RPE and taken up by the photoreceptor where it combines with apo-opsin to re-form rhodopsin.
RGR-opsin is a 32-kDa protein expressed in internal membranes of RPE and Müller glial cells (10, 13). RGR is 20.4% identical to mammalian rhodopsin and 23.6% identical to retinochrome, an opsin homolog in squid that regenerates visual chromophore by photoisomerization (14). Dark-adapted RGR contains predominantly all-trans-RAL, coupled via a Schiff base to a Lys residue, similar to rhodopsin (15). RGR has absorption maxima (λmax) at 370 and 469 nm from the unprotonated and protonated forms, respectively, of this Schiff base (16). Exposing RGR to light induces photoisomerization of the all-trans-RAL to 11-cis-RAL (15). Unlike rhodopsin, RGR does not readily release its bound chromophore following photoisomerization (15). Instead, the 11-cis-RAL decays thermally back to all-trans-RAL in the dark (15). Mice with a knock-out mutation in the rgr gene show elevated retinyl esters and reduced 11-cis-RAL following exposure to bright light (17). These observations led to the suggestion that RGR functions as a “reverse” photoisomerase to regenerate 11-cis-RAL chromophore, similar to the proposed function of squid retinochrome (14). However, several observations argue against this hypothesis. The retinoid isomerase in RPE cells is Rpe65 (18-20). Mice with a knock-out mutation in the rpe65 gene contain no detectable 11-cis-retinoids (21). RGR-opsin is present in rpe65-/- mice (22). However, exposing rpe65-/- mice to light does not result in synthesis of 11-cis-RAL (22). Also, RGR was shown to enhance rhodopsin regeneration in the dark (23). These observations are inconsistent with the proposed role of RGR as a regenerative photoisomerase. Finally, given the 100-fold lower molar abundance (24) and the nearly 6-fold lower quantum efficiency of RGR versus rhodopsin (15), the primary function of RGR-opsin is unlikely to be synthesis of 11-cis-RAL for regeneration of rhodopsin. Nonetheless, RGR appears to play an important role. Mutations in the RGR gene cause the severe inherited blinding disease, retinitis pigmentosa (25), suggesting that RGR is required for normal functioning of the retina.
In this study, we show that RGR-opsin provides light modulation of all-trans-retinyl ester (all-trans-RE) synthesis and degradation. These all-trans-REs are substrates for Rpe65-isomerase, which catalyze the critical all-trans- to 11-cis-retinoid isomerization reaction (18-20) (Fig. 1). Furthermore, we show that loss of RGR significantly reduces isomerase activity in mice without affecting levels of the Rpe65 protein. These results establish that the RPE is intrinsically light-sensitive and that RGR-opsin plays a regulatory role in the visual cycle.
EXPERIMENTAL PROCEDURES
Mice—To generate rgr-/- mice homozygous for the rpe65 Leu450-allele, we crossed Met450 rgr-/- mice (generously provided by Henry Fong) with wild-type 129/Sv mice (homozygous for the Leu450 allele) and then crossed F1 offspring and determined the genotype of the F2 mice at rgr and rpe65. To distinguish between the normal and disrupted rgr alleles, we used the following primers: 5′-TGCATTTTCCTGTGAGATGG, 5′-GCTCAGTACCAGCAGGTTGC, and 5′-GGGGAACTTCCTGACTAGGG. PCR conditions were 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 60 s, 35 cycles total. The normal rgr allele yielded a 314-bp product and the disrupted allele a 464-bp product. To determine the genotype of rpe65 at codon 450, we used the following primers: 5′-CCTTTGAATTTCCTCAAATCAATTA and 5′-TTCCAGAGCATCTGGTTGAG. PCR conditions were 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 60 s, 35 cycles total. The 300-bp amplification product was then sequenced using the DYNamic™ ET terminator kit (GE Healthcare) under the following conditions: 95 °C for 20 s, 50 °C for 15 s, and 60 °C for 60 s, 28 cycles total. The reaction product was sequenced on an ABI 3100 sequencer. Double knock-out rpe65-/-, rgr-/- mice were generated by crossing single knock-out mice (rpe65-/- mice were generously provided by Michael Redmond), as described above for rgr-/-, Leu450 mice. To distinguish between the normal and disrupted rpe65 allele, we used the following primers: 5′-GATGTGGGCCAGGGCTCTTTGAAG, 5′-CCCAATAGTCTAGTAATCACAGATG, and 5′-GGGAACTTCCTGACTAGGGGAGG. PCR conditions were 94 °C for 60 s, 60 °C for 30 s, and 72 °C for 30 s, 40 cycles total. The normal rpe65 allele yielded a 546-bp amplification product and the disrupted allele a 459-bp product.
Mice were raised in 12-h cyclic light at 20-40 lux. They were studied at 8-12 weeks of age, and all were dark-adapted overnight. As indicated, some mice were light-adapted for 2 h at 400-500 lux before use. Mice were euthanized by cervical dislocation under deep anesthesia with ketamine and xylazine. Following euthanasia, eyeballs were collected and hemisected. Posterior hemispheres containing retina, RPE, choroid, and sclera (eyecups) were frozen in liquid N2 and stored at -80 °C for analysis of retinoid content. For enzyme assays and immunoblot analysis, RPE-containing eyecups were homogenized following removal of the neural retina. All tissue manipulations were done under dim red light (Kodak Wratten 1A).
Retinoid Analysis—Individual eyecups were homogenized in phosphate buffer containing 200 mm hydroxylamine. One ml of ethanol was added, and retinoids were extracted twice with 3 ml of hexane. The samples were centrifuged at 3000 × g for 5 min. The organic phases were collected, dried under a stream of argon gas, and re-dissolved in 100 μl of hexane. Hexane solutions were analyzed by normal phase high performance liquid chromatography (HPLC) using a gradient elution (0.2-10% dioxane in hexane) at a flow rate of 2 ml/min on a silica column (Agilent-Zorbax-Sil 5 μm, 250 × 4.6 mm) in an Agilent model 1100 liquid chromatograph equipped with a photodiode-array detector (Agilent Technologies, Wilmington, DE). The identity of each retinoid (including the syn- and anti-oximes of each retinaldehyde) was confirmed by on-line spectral analysis and co-elution with authentic retinoid standards. We obtained all-trans-ROL, all-trans-RAL, and all-trans-retinyl palmitate from Sigma. 11-cis-ROL and 11-cis-retinyl palmitate were synthesized from 11-cis-RAL (generously provided by Rosalie Crouch) using published methods (26). Retinaldehyde-oxime standards were prepared by reacting 11-cis-RAL or all-trans-RAL with hydroxylamine, as described (27, 28). Quantitation of the retinoids was done by comparing the sample peak areas to calibration curves established for each retinoid standard using the published molar extinction coefficients (29, 30).
Enzyme Activity Analysis—The enzyme sources for the in vitro assays were RPE-containing eyecup homogenates prepared from light- or dark-adapted mice of the indicated genotypes. All the assays were done in 200-μl reaction volumes using 50-100 μg of total protein. The retinoid substrates were added in 2 μl of DMSO. The reaction mixture for the LRAT assay was 10 mm Tris-HCl (pH 7.4), 2 mm CaCl2, 2 mm MgCl2, 1 mm DTT, 1% BSA, and 5 μm all-trans-ROL (Sigma, catalog number 95144). After adding protein, the mixture was incubated for 4 min at 37 °C. The reaction mixture for the all-trans-REH assay was 20 mm HEPES (pH 8.0), 150 mm NaCl, 1 mm DTT, 1% BSA, and 50 μm all-trans-retinyl acetate (Sigma, catalog number R4632). After adding protein, the mixture was incubated for 5 min at 37 °C. The reaction mixture for the isomerase assay was 10 mm Tris-HCl (pH 7.4), 2 mm CaCl2, 2 mm MgCl2, 1 mm DTT, 2.5 mm sodium cholate, 5% BSA and 5 μm all-trans-retinyl palmitate (Sigma, catalog number R3375). After adding protein, the mixture was incubated at 37 °C for 120 min. Finally, the reaction mixture for the 11-cis-ROL dehydrogenase (11-cis-RDH) assay was 50 mm MES (pH 5.5), 1 mm DTT, 2.5% BSA, 500 μm NADH, and 50 μm 11-cis-RAL. Here, 11-cis-RDH was assayed in the reducing direction, as described previously (31). After adding protein, the mixture was incubated at 37 °C for 15 min. All reactions were quenched by adding 400 μl of ice-cold methanol. For the 11-cis-RDH assay, we added hydroxylamine and NaCl to 250 mm final concentrations each followed by a 15-min incubation at room temperature to protect the retinaldehydes by forming oximes (27, 28). SDS was added to all reaction mixtures yielding a final concentration of 0.1%, and the samples were extracted twice with 2 ml of hexane. The extracts were dried under argon, re-dissolved in 100 μl of hexane, and analyzed by HPLC as described above. Protein determinations were done on all homogenates using the Micro BCA protein assay kit (Thermo Scientific). In control experiments we established that the above-described assays were in the linear range of product formation per unit of time. Activities were reported as pmol/min/mg of protein.
Immunoblot Analysis—Five to 10 μg of protein from the indicated eyecup homogenates were heated in Laemmli sample buffer for 10 min at 75 °C, separated by SDS-PAGE in a 10% polyacrylamide gel, and transferred to an Immobilon-P membrane (Millipore). The membrane was incubated in blocking buffer (PBS (pH 7.2) and 5% nonfat milk) for 2 h at 37 °C and then overnight at 4 °C with rabbit polyclonal antisera against the Rpe65 peptide, NFITKINPETLETIK (32), or human truncated LRAT (33). Membranes were washed three times in PBS, incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG, and visualized with the ECL-plus Western blot detection system (Amersham Biosciences).
Human RPE Cell Cultures and RNAi—Human fetal RPE cells were grown as confluent monolayers on nitrocellulose in two-chamber Millicell culture dishes (Millipore, catalog number PIHA 01250) in medium that favors RPE cell differentiation (34). These dishes permit independent manipulation of media bathing the basal and apical surface of RPE cells. Only cultures with a measured trans-epithelial resistance of 500-600 Ω/cm2 were used. We first transfected human RPE (hRPE) cells with a fluorescein-labeled, nonsilencing control RNA included in the siRNA starter kit (Qiagen, catalog number 301699) following the manufacturer's recommendations. Greater than 80% of cells were fluorescently labeled (not shown). We designed three synthetic, double-stranded, small-interfering RNAs (siRNAs) against the human RGR mRNA (NM_002921) using the Qiagen on-line design tool: RGR siRNA 1 (5′-AAACACCACTCTGCCAGCAAG), siRNA 2 (5′-AAAATGGTGCCCACGATCAAT), and siRNA 3 (5′-AACTATGCCCTGGGCAATGAG). We transfected each siRNA into hRPE cells at different ratios of RNA to RNAiFect transfection reagent (Qiagen). The greatest reductions in RGR mRNA level were obtained by mixing 1 μg of siRNA (200 nm final concentration), 6 μl of RNAiFect reagent, buffer EC-R (Qiagen) to a final volume of 100 μl, plus 300 μl of hRPE culture medium. This mixture was added to the apical and basal media of the Millicell dishes. Cells were incubated with the transfection media at 37 °C for 3 days. Following transfection, the medium was replaced with fresh minimum Eagle's medium containing 1% BSA and 5 μm all-trans-ROL, and the incubation continued for an additional 3 h. The all-trans-ROL-containing medium was replaced with minimum Eagle's medium plus 1% BSA, and selected plates were incubated at 37 °C in the dark for 2 h, whereas other cells were exposed to UV-filtered fluorescent light (1,500-2,000 lux) for 2 h. For the experiments represented in Fig. 3, C and D, 2 μm holo-retinol-binding protein (RBP) (Sigma, catalog number R9388) was added to the basal medium, and 5 μm all-trans-ROL plus 10 μm interphotoreceptor retinoid-binding protein (IRBP) (final concentrations) were added to the apical medium. IRBP was purified from freshly dissected bovine retinas following published procedures (35) and used to stabilize 11-cis-retinoids released by hRPE cells.
FIGURE 3.
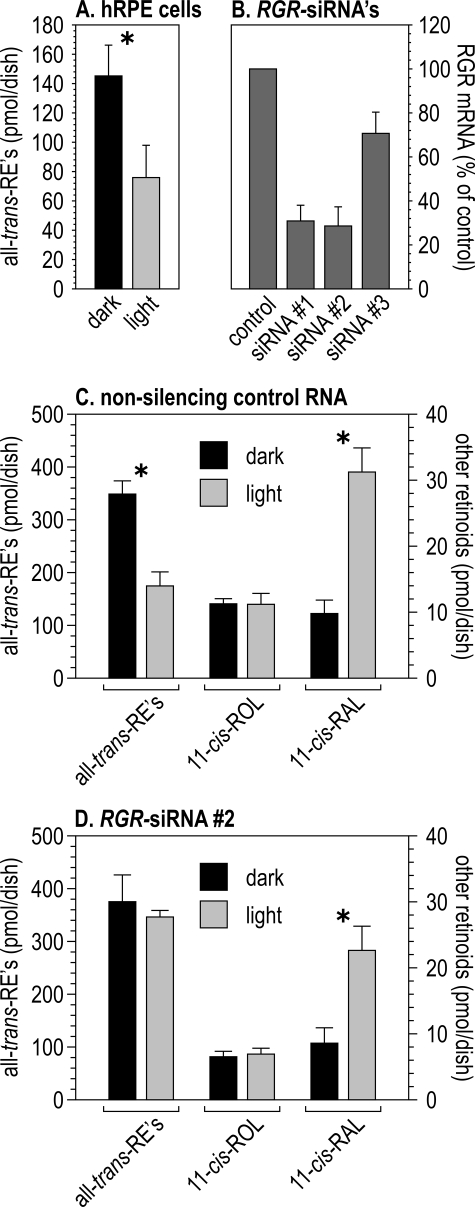
Retinoid levels in cultured human RPE cells. A, levels of all-trans-REs in hRPE cells cultured in the dark or exposed to UV-filtered light for 2 h. Data are expressed as picomoles per dish of confluent cells (n = 4). Asterisk denotes a significant difference between dark and light values (p = 0.032). B, levels of RGR mRNA in hRPE cells from confluent millicell dishes. Cells were transfected with a nonsilencing control RNA (control) or RGR siRNAs 1-3. Quantitation was by qRT-PCR using GAPDH as an internal control. The ΔCt values for each RGR siRNA are expressed as a percent of the nonsilencing control. C, retinoid levels in dark- and light-adapted hRPE cells and media after transfection with the nonsilencing control RNA. The scale on the left is for all-trans-REs (in hRPE cells), and the scale on the right is for 11-cis-ROL and 11-cis-RAL (in the media). Both are expressed as picomoles per confluent dish (n = 4). Asterisks denotes significant differences between indicated values (p = 0.0005 for all-trans-REs and p = 0.002 for 11-cis-RAL). D, retinoid levels in dark- and light-adapted hRPE after transfection with RGR siRNA 2. Quantitation was the same as in C. Asterisk denotes a significant difference between 11-cis-RAL values (p = 0.017).
To determine the levels of the RGR and GAPDH (control) mRNAs, we harvested the hRPE cells, washed them twice with PBS, and then lysed the cells with lysis buffer included in the RNA isolation kit (Stratagene, catalog number 400800). First-strand cDNA was synthesized from 5 μg of total RNA following the manufacturer's procedure (Invitrogen, catalog number 18080-051). We used 1 μl of each cDNA synthesis reaction as a template for quantitative real time PCR (qRT-PCR) using iTaq SYBR Green Supermix with ROX (Bio-Rad, catalog number 170-8551) as the specific primers for RGR and GAPDH. The primer sequences for RGR were 5′-CCCTGACCATCTTCTCTTTC and 5′-GGTGCAGTAGTGGTGATAACG, and for GAPDH were 5′-TGCACCACCAACTGCTTAGC and 5′-GCCTGCTTCACCACCTTCTTG. The qRT-PCRs were done in triplicate for each primer set in an MJR Opticon2 instrument using the following PCR conditions: 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 40 s, 40 cycles total. The predicted amplification products of 294 and 390 bp were obtained for RGR and GAPDH, respectively. We normalized the ΔCt values for RGR (CtRGR - CtGAPDH) from hRPE cells transfected with each RGR siRNA against the ΔCts from hRPE cells transfected with the nonsilencing control RNA.
To determine the content of retinoids in the media and cells, we added 400 μl of media to tubes containing hydroxylamine (100 mm final concentration). The nitrocellulose membranes with adherent hRPE cells were rinsed in PBS and then placed in tubes containing 400 μl of lysis buffer (10 mm HEPES (pH 7.4), 100 mm hydroxylamine, and 0.1% SDS). After vortexing, the nitrocellulose membrane was discarded. We added 200 μl of methanol and then extracted the retinoids twice into 2 ml of hexane. The retinoid extracts were dried under a stream of argon, re-dissolved in 100 μl of hexane, and analyzed by HPLC, as described above.
RESULTS
Light-dependent Mobilization of Retinyl Esters in rpe65-/- Mice—Mice with a knock-out mutation in the rpe65 gene massively accumulate all-trans-REs in the RPE and contain no detectable 11-cis-retinoids (21, 22). Accordingly, these mice are virtually blind by electroretinographic analysis (21). In an exploratory experiment, we dark-adapted 2-month-old wild-type (strain 129) and rpe65-/- mice overnight and then exposed one group of each strain to fluorescent light at 500 lux for 2 h. We measured the content of visual retinoids in the eyes from these dark- and light-adapted mice. As reported previously (36-39), wild-type mice showed a light-dependent increase in levels of all-trans-REs (Fig. 2A). Unexpectedly, all-trans-REs were dramatically lower in the light- versus dark-adapted rpe65-/- mice (Fig. 2C). Representative chromatograms containing retinoid data from wild-type and rpe65-/- mice are shown in Fig. 2, B and D. The magnitude of the light-dependent reduction in all-trans-REs was 300 pmol, almost two-thirds the total retinoid content of a wild-type mouse eye (37). Given the absence of rhodopsin and 11-cis-RAL in rpe65-/- retinas, this mobilization of all-trans-REs is unlikely to be mediated by photoreceptors. Alternatively, RPE cells may be intrinsically sensitive to light and may respond to changes in illumination by regulating activities of the visual cycle.
FIGURE 2.
Retinyl esters in wild-type and rpe65-/- mice. A, levels of all-trans-REs in light-adapted (LA) and dark-adapted (DA) eyes from wild-type mice. Expressed as picomoles per eye, ± S.D. (n = 4). Asterisk denotes a significant difference between DA and LA values (p = 0.046). B, representative chromatogram of retinoids extracted from dark-adapted, wild-type mouse eyecups in milliabsorbance units (mAU) at 325 nm. Identified peaks are as follows: peak 1, 11-cis-retinyl palmitate; peak 2, all-trans-retinyl palmitate; peak 3, syn-11-cis-RAL-oxime; peak 4, syn-all-trans-RAL-oxime; peak 5, 11-cis-ROL; peak 6, anti-11-cis-RAL-oxime; peak 7, all-trans-ROL; peak 8, anti-all-trans-RAL-oxime. C, levels of all-trans-REs in LA and DA eyes from rpe65-/- mice. Expressed as picomoles per eye, ± S.D. (n = 6). Asterisk denotes a significant difference between DA and LA values (p = 0.038). D, representative chromatogram of retinoids extracted from dark-adapted, rpe65-/- mouse eyecups in milliabsorbance units (mAU) at 325 nm. Identified peaks: peak 2, all-trans-retinyl palmitate; peak 7, all-trans-ROL.
Light-dependent Mobilization of Retinyl Esters in Cultured Human RPE Cells—To test this alternate possibility, we looked for changes in the retinoid content of primary cultured hRPE cells following light exposure. Cells in these monolayer cultures were shown to have normal RPE ultrastructure, to express all known proteins of the visual cycle, and to synthesize 11-cis-retinoids from exogenous all-trans-ROL (35, 40). Photoreceptors are absent from these cultures. Without addition of all-trans-ROL to the medium, retinoids are undetectable in these cells (not shown). We added all-trans-ROL to the media of cultured hRPE cells grown in darkness. After incubating for an additional 6 h in the dark, we exposed one set of dishes to UV-filtered fluorescent light for 2 h, whereas the other set was kept in darkness. We then collected and homogenized the cells, extracted the retinoids, and analyzed by HPLC. The levels of all-trans-REs were 1.9-fold lower in the light-exposed cells (Fig. 3A). These data are consistent with the results from rpe65-/- mice (Fig. 2C) and suggest that RPE cells are intrinsically light-sensitive.
RGR-Opsin Mediates Light Sensitivity in RPE Cells—RGR is present in RPE cells and an excellent candidate protein to mediate the observed light-dependent mobilization of all-trans-REs. To test this possibility, we “knocked down” RGR expression in RPE cells by RNAi. We generated three synthetic, double-stranded siRNAs that correspond to the human RGR mRNA. We transfected these, plus a nonsilencing control double-stranded RNA, into cultured hRPE cells, and we then measured the levels of the RGR mRNA in total RNA extracted from these cells by qRT-PCR. RGR-siRNAs 1 and 2 reduced the RGR mRNA by ∼75%, whereas RGR-siRNA 3 only reduced expression by ∼30% (Fig. 3B). To explore the role of RGR on light-dependent mobilization of all-trans-REs, we transfected hRPE cells with RGR-siRNA 2 or the nonsilencing control RNA. After 3 days of incubation in the dark, we added all-trans-ROL to the media, incubated for an additional 6 h, exposed one set of dishes to UV-filtered light, and then analyzed for retinoid content as described. Cells transfected with the control RNA showed significant mobilization of all-trans-REs after exposure to light (Fig. 3C). In contrast, all-trans-REs were unchanged after light exposure in hRPE cells transfected with RGR-siRNA 2 (Fig. 3D). These data suggest that RGR-opsin mediates light-dependent mobilization of all-trans-REs by RPE cells. Levels of 11-cis-RAL were higher in light-exposed cells, suggesting partial conversion of all-trans-REs to 11-cis-RAL by Rpe65 (Fig. 3, C and D). The higher all-trans-REs observed in untransfected (Fig. 3A) versus control RNA-transfected hRPE cells (Fig. 3C) probably results from our use of RBP and IRBP as carrier proteins to deliver all-trans-ROL in the RNA transfection experiments, instead of BSA. The total retinoid content of the hRPE cells plus media were similar in dark- and light-adapted dishes.
Light-dependent Mobilization of Retinyl Esters Is Lost in Mice Lacking Rpe65 and RGR—If RGR is responsible for the light-stimulated depletion of all-trans-REs in rpe65-/- mice, this effect should disappear in mice lacking both Rpe65 and RGR. To test this prediction, we generated mice homozygous for null mutations in rpe65 and rgr. We dark-adapted wild-type, rpe65-/-, rgr-/-, and rpe65-/-, rgr-/- double knock-out mice overnight, and we exposed one group to fluorescent light for 2 h while the other group was kept in darkness, and we then quantitated visual retinoids in the eyes of mice from both groups. As before, light exposure caused modest accumulation of all-trans-REs in wild type and significant depletion of all-trans-REs from rpe65-/- eyes (Fig. 4, A and C). Single knockout rgr-/- mice showed a larger fractional increase in all-trans-REs following light exposure than wild-type mice (Fig. 4, A and B). Importantly, the light-dependent depletion of all-trans-REs seen in rpe65-/- mice was abolished in rpe65-/-, rgr-/- mice (Fig. 4D). These data corroborate the hypothesis that RGR-opsin is required for light-dependent mobilization of all-trans-REs. Wild-type mice exposed to light could maintain levels of 11-cis-ROL and 11-cis-RAL at nearly dark-adapted levels under these experimental conditions (Fig. 4A). In contrast, 11-cis-ROL and 11-cis-RAL were significantly higher in dark-adapted versus light-exposed rgr-/- mice (Fig. 4B). This effect was seen despite much higher levels of all-trans-REs in light-exposed rgr-/- versus wild-type mice (Fig. 4, A and B). These observations suggest lower isomerase activity in rgr-/- mice.
FIGURE 4.
Retinoid levels in light- and dark-adapted mice. A, levels of retinoids in wild-type mouse eyecups. The scale on the right is for 11-cis-RAL, and the scale on the left is for all other retinoids. The asterisk denotes a significant difference between all-trans-ROL levels in the dark and light (p = 0.0024). B, levels of retinoids in rgr-/- mouse eyecups. The scale on the right is for 11-cis-RAL, and the scale on the left is for all other retinoids. Asterisks denote significant differences between the indicated values for dark and light (p = 0.008 for all-trans-ROL; p = 0.02 for 11-cis-ROL; and p = 0.005 for 11-cis-RAL). C, levels of retinoids in rpe65-/- mouse eyecups. The scale on the right is for all-trans-RE, and the scale on the left is for all other retinoids. Asterisk denotes a significant difference between all-trans-RE values (p = 0.026). D, levels of retinoids in rgr-/-, rpe65-/- double knockout eyecups. The scale on the right is for all-trans-REs, and the scale on the left is for all other retinoids. All values are shown as picomoles per eye (n = 4).
Effects of RGR-Opsin on Retinoid-processing Enzymes of the RPE—Light-dependent mobilization of retinyl esters in cultured hRPE cells and rpe65-/- mice was only observed in the presence of RGR (Figs. 3C and 4C). Retinyl ester levels are determined by the balance of synthase and hydrolase activities. The dominant all-trans-RE synthase in RPE is LRAT (41-43) (Fig. 1). Hydrolysis of all-trans-REs is carried out in the RPE by all-trans-retinyl ester hydrolase (all-trans-REH), which catalyzes the hydrolysis of an all-trans-RE to all-trans-ROL plus a free fatty acid. This enzyme has been characterized kinetically (44-46) but not yet identified. Another enzyme that hydrolyzes all-trans-REs is Rpe65 isomerase, here yielding 11-cis-ROL plus a free fatty acid (18-20, 47). Finally, it has been suggested that LRAT may possess “reverse synthase” or ester hydrolase activity (48). We measured the catalytic activities of these enzymes in vitro, to understand the role of RGR on light-dependent mobilization of retinyl esters.
First, we determined the activity of LRAT in RPE homogenates from dark- and light-adapted wild-type and rgr-/- mice. This experiment yielded three significant observations (Fig. 5A). (i) LRAT activity was 5.6-fold higher in light- versus dark-adapted homogenates from wild-type mice. (ii) LRAT activity was only 1.5-fold higher in light- versus dark-adapted homogenates from rgr-/- mice. (iii) LRAT activity was 10.6-fold higher in rgr-/- versus wild-type homogenates from dark-adapted and 2.7-fold higher in homogenates from light-adapted mice. These results explain the higher retinyl esters in light-adapted rgr-/- (Fig. 4B) versus wild-type (Fig. 4A) eyecups, as reported previously (49).
FIGURE 5.
Specific activities of retinoid-processing enzymes in RPE homogenates from dark- and light-adapted mice. A, LRAT activities in wild-type and rgr-/- RPE homogenates using all-trans-ROL as substrate. Expressed as picomoles of all-trans-RE synthesized per mg of protein per min. Asterisks denote significant differences between LRAT activities in the dark and light (p = 0.008 for wild-type and 0.022 for rgr-/- mice). B, all-trans-REH activities in RPE homogenates from wild-type, rgr-/-, rpe65-/-, and rgr-/-, rpe65-/- double knock-out mice. The substrate was all-trans-retinyl acetate, and activities are expressed as all-trans-ROL synthesized per mg of protein per min. Asterisks denote significant differences between all-trans-REH activities in the dark and light (p = 0.0009 for wild-type and p = 0.0001 for rpe65-/- mice). C, isomerase activities in wild-type and rgr-/- RPE-homogenates using all-trans-retinyl palmitate as substrate. Expressed as picomoles of 11-cis-ROL synthesized per mg of protein per min. Asterisks denote significant differences between isomerase activities in the dark and light (p = 0.024 for wild-type and p = 0.003 for rgr-/- mice). D, 11-cis-RDH activities in wild-type and rgr-/- RPE homogenates using 11-cis-RAL and NADH as substrates. Expressed as picomoles of 11-cis-ROL per mg of protein per min. Asterisk denotes a significant difference between 11-cis-RDH activities in the dark and light for rgr-/- mice (p = 0.003).
Next, we measured all-trans-REH activity in RPE homogenates from dark- and light-adapted mice of several genotypes. Hydrolase activity was 2.2-fold higher in light- versus dark-adapted RPE homogenates from wild-type mice (Fig. 5B). Hydrolase activities in light- and dark-adapted rgr-/- homogenates were similar to the activity in light-adapted wild-type homogenates (Fig. 5B). The hydrolase activities in homogenates from light- and dark-adapted rpe65-/- mice were higher than the activities in the cognate homogenates from wild-type mice (Fig. 5B). Nonetheless, rpe65-/- homogenates retained the light-dependent stimulation of all-trans-REH activity. Similar to rgr-/- single knock-out mice, this light dependence was lost in homogenates from rgr-/-, rpe65-/- double knock-out mice (Fig. 5B). These results suggest that the light-dependent mobilization of retinyl esters in RPE cells is mediated by the effects of RGR on all-trans-REH activity.
We assayed isomerase activity in RPE homogenates from wild-type and rgr-/- mice. Both showed modest (1.3-1.6-fold) light-dependent stimulation of isomerase activity (Fig. 5C). Isomerase activity was ∼3-fold lower in the rgr-/- versus wild-type homogenates (Fig. 5C). This observation explains the light-independent slowing of rhodopsin regeneration previously reported in rgr-/- mice (23).
RGR-opsin was shown to co-precipitate with NADH-dependent 11-cis-RDH in RPE cells (24). We measured 11-cis-RDH activity in RPE homogenates from light- and dark-adapted wild-type and rgr-/- mice. We observed slightly higher 11-cis-RDH activity in the light- versus dark-adapted homogenates but no difference between wild-type and rgr-/- mice (Fig. 5D). Thus, RGR-opsin does not appear to modulate the activity of 11-cis-RDH.
Increased LRAT and Reduced Isomerase Activities in rgr-/- Mice Are Not Caused by Changes in Protein Expression—A possible explanation for the increased LRAT and reduced Rpe65 activity in rgr-/- mice is altered expression of these proteins. We tested this possibility by immunoblot analysis using antisera against LRAT (33) and Rpe65 (32). In repeated immunoblot assays, LRAT immunoreactivities were similar in dark- and light-adapted wild-type and rgr-/- eyecups (representative blot in Fig. 6A). Rpe65 immunoreactivities were also similar in dark- and light-adapted wild-type versus rgr-/- eyecups (representative blot in Fig. 6B). These data suggest that the 2.7-fold higher LRAT and 2.6-fold lower isomerase activities in light-adapted wild-type versus rgr-/- mice (Fig. 5, A and C) are not caused by significant changes in levels of the LRAT or Rpe65 proteins.
FIGURE 6.
Immunoblots of mouse eyecup homogenates. A, LRAT immunoreactivity in homogenates from dark- and light-adapted wild-type and rgr-/- mice. The immunoreactive bands migrated with an apparent mass of 25 kDa compared with protein size standards. B, Rpe65 immunoreactivity in homogenates from dark- and light-adapted wild-type and rgr-/- mice. The immunoreactive bands migrated with an apparent mass of 63 kDa compared with protein size standards.
DISCUSSION
A surprising result of this study is that RPE cells are intrinsically sensitive to light. This sensitivity was revealed in two experimental systems where RPE cells were functionally or physically separated from photoreceptors. The first was the rpe65-/- mouse. These mice lack Rpe65 isomerase and cannot synthesize the 11-cis-RAL chromophore. RPE cells in rpe65-/- mice greatly accumulate all-trans-REs. When rpe65-/- mice were exposed to light, we observed large-scale mobilization of these all-trans-REs (Figs. 2C and 4C). This effect was blocked in rpe65-/-, rgr-/- mice lacking both Rpe65 and RGR (Fig. 4D). The second system was a primary culture of hRPE cells. These cells possess isomerase activity, evidenced by the synthesis of 11-cis-ROL and 11-cis-RAL from all-trans-ROL added to the medium (Fig. 3, C and D). Despite the absence of photoreceptors, hRPE cells responded to light exposure by significant depletion of all-trans-REs and synthesis of 11-cis-RAL (Fig. 3C). Knockdown of RGR expression by RNAi resulted in the loss of this effect (Fig. 3D). These results suggest that the intrinsic light sensitivity of RPE cells is mediated by RGR-opsin to modulate the synthesis and/or degradation of retinyl esters.
Light exposure induces isomerization of 11-cis-RAL in rhodopsin, resulting in the eventual release of all-trans-ROL by photoreceptors (Fig. 1). This all-trans-ROL is taken up by RPE cells and esterified by LRAT. In a normal retina, light exposure induces net synthesis of all-trans-REs in the RPE (37-39). This synthesis is driven by the availability of all-trans-ROL from bleached photoreceptors (Fig. 1). Formation of all-trans-REs in the light makes teleological sense because these are substrates for the isomerase and hence required for synthesis of the new visual chromophore. However, the results discussed above suggest that RGR-opsin stimulates light-dependent degradation of all-trans-REs. Why at a time of great need for chromophore synthesis should substrates for the isomerase be wastefully degraded? To answer this question we must consider the sites of all-trans-RE storage inside RPE cells.
One compartment for the storage of all-trans-REs is within the lipid bilayers of the endoplasmic reticulum (ER). Several retinoid-processing proteins are associated with the ER, including LRAT (50), 11-cis-RDH (51), RGR-opsin (13), and Rpe65 (52). Another compartment in RPE cells for storage of all-trans-REs is lipid droplets (53). Lipid droplets are cytoplasmic structures containing a core of neutral lipids, including triglycerides, cholesterol and retinyl esters, surrounded by a phospholipid monolayer “skin” (54, 55). Lipid droplets are expandable three-dimensional structures of potentially far greater capacity for retinyl esters than ER membranes. As an example, most of the excess all-trans-REs in rpe65-/- mice are present within lipid droplets (21, 39). Several proteins associate with lipid droplets, including the perilipins and caveolins. Importantly, Rpe65 does not co-localize with lipid droplets in RPE cells (39). Accordingly, lipid droplets may serve as a “spill-over” site for retinyl ester storage after ER membranes have been filled to capacity. Because of their water insolubility (56), all-trans-REs in lipid droplets must first be hydrolyzed by all-trans-REH, transported to the ER as all-trans-ROL, and secondarily re-esterified by LRAT into ER membranes before they can be used as substrate by the isomerase. We propose that the translocation of all-trans-REs from lipid droplets to ER membranes is regulated by light through the RGR-opsin, as depicted in Fig. 7.
FIGURE 7.
Model showing regulation of all-trans-REH and LRAT by RGR. After release by photoreceptors, all-trans-ROL is taken up by RPE cells and esterified by LRAT to form all-trans-REs in two pools. Lipid droplets include the larger “storage pool” not associated with Rpe65. ER membranes include the smaller isomerase pool associated with Rpe65. The “dark-adapted” (all-trans-RAL-bound) form of RGR-opsin strongly inhibits all-trans-REH and partially inhibits LRAT, promoting storage of all-trans-REs in the dark until the supply of all-trans-ROL is consumed by LRAT. RGR-opsin absorbs a photon (hv), which induces isomerization of its bound all-trans-RAL to 11-cis-RAL. RGR-opsin is thus converted to its “light-adapted” form that no longer inhibits all-trans-REH and LRAT. This accelerates hydrolysis of all-trans-REs in lipid droplets and re-esterification of the released all-trans-ROL to all-trans-REs in ER membranes. The all-trans-REs are used by Rpe65 on the ER to generate 11-cis-ROL, which is subsequently oxidized by 11-cis-RDH to yield 11-cis-RAL for the photoreceptor opsins.
Consistent with this model, we observed light-dependent stimulation of LRAT and all-trans-REH activities in RPE homogenates from wild-type mice (Fig. 5, A and B). The activity of all-trans-REH was similar in homogenates from light-adapted wild-type and dark- and light-adapted rgr-/- mice (Fig. 5, A and B). This suggests that the dark-adapted form of RGR inhibits but the light-adapted form has no effect on all-trans-REH activity. LRAT activity was 5.6-fold higher in RPE homogenates from light- versus dark-adapted wild-type mice (Fig. 5A). Under both light conditions, LRAT activity was significantly higher in homogenates from rgr-/- versus wild-type mice (Fig. 5A). In contrast to all-trans-REH activity, where light modulation was completely lost in rgr-/- mice (Fig. 5B), LRAT retained partial light responsiveness (Fig. 5A). These results suggest that RGR inhibits LRAT in both darkness and light but more strongly in the dark. The 1.5-fold stimulation of LRAT seen in light- versus dark-adapted rgr-/- homogenates (Fig. 5A) may result from the influence of another receptor in RPE cells.
The observed effects of RGR on all-trans-REH and LRAT-specific activities were not due to light-dependent changes in substrate availability, because these enzymes were assayed under saturating substrate concentrations. The specific activities of all-trans-REH were 30-70-fold higher than those of LRAT in the identical homogenates from light- or dark-adapted wild-type mice (Fig. 5, A and B). Accordingly, the inhibitory effect of RGR on the hydrolysis of all-trans-REs is probably greater than its effect on all-trans-RE-synthesis. This would explain the net mobilization of all-trans-REs mediated by RGR in rpe65-/- eyecups (Figs. 2C and 4C) and hRPE cells (Fig. 3, A and C). The unbalanced increase of LRAT with unchanged all-trans-REH activities in light-adapted rgr-/- mice explains the higher all-trans-REs in these animals (Fig. 4, A and B). The increased all-trans-REs following light exposure in wild-type RPE (37-39) reflects the increased all-trans-ROL substrate for LRAT from bleached photoreceptors (Fig. 1). This source of substrate is unavailable in rpe65-/- eyes and cultured hRPE cells, which explains the net depletion following light exposure of all-trans-REs in these systems.
LRAT has also been shown to be active in the reverse direction, effectively functioning as a retinyl ester hydrolase (48). An alternative explanation for the results presented here is that RGR-opsin modulates this reverse activity of LRAT. Specifically, the dark-adapted form of RGR-opsin may inhibit both the synthase and hydrolase activities of LRAT. Light “activation” of RGR would release both activities of LRAT from inhibition and cause increased “churning” of all-trans-REs. Whether mediated by the reverse activity of LRAT, or a separate all-trans-REH as proposed above, release of inhibition by light-adapted RGR should cause a net flux of all-trans-REs from lipid droplets to ER membranes for subsequent isomerization (Fig. 7).
Isomerase catalytic activity was ∼3-fold lower in RPE homogenates from rgr-/- versus wild-type mice (Fig. 5C). This reduced activity explains the slower regeneration of rhodopsin seen in rgr-/- versus wild-type mice (17, 23). We observed modest (∼1.3-fold) light stimulation of isomerase activity, but the effect was present in both wild-type and rgr-/- homogenates (Fig. 5C) suggesting that it is mediated by another receptor. The increased LRAT and reduced isomerase activities cannot be explained by changes in the levels of these proteins, because the immunoblot signal intensities of LRAT and Rpe65 were similar in wild-type and rgr-/- homogenates (Fig. 6, A and B).
The results presented above suggest that RGR-opsin regulates the activities of all-trans-REH and LRAT in a light-dependent fashion. The mechanism of this regulation is not clear. It may involve direct modulation of catalytic activities or changes in the stability of a protein complex through a second messenger system. The motifs “D(E)RY” and “NPXXY” are critical for the interaction of a G protein-coupled receptor with a G protein α-subunit (57). RGR contains the variant motifs “GRY” and “NAXXY.” RGR-opsin may interact with a G protein via these variant motifs or may modulate LRAT and all-trans-REH through another signaling mechanism. Several G proteins have been identified in human RPE cells, including Gsα, Gi-1α, Gi-2α, Gi-3α, and Gzα (58). Additional studies will be required to define the signaling pathway for RGR-opsin. Unlike the photoreceptor opsins, RGR does not require Rpe65 for synthesis of its chromophore because RGR functions as a photoisomerase (15).
In a previous study, 9-cis-retinaldehyde (9-cis-RAL) was administered to rpe65-/- mice in a successful attempt to correct the visual phenotype (22). Similar to 11-cis-RAL, 9-cis-RAL combines with apo-opsin to form a functional visual pigment (iso-rhodopsin). Unexpectedly, rpe65-/- mice treated with 9-cis-RAL showed a dramatic reduction in the level of all-trans-REs. This observation was difficult to understand, because supplemental 9-cis-RAL does nothing to correct the inherited isomerase deficiency that induces all-trans-RE accumulation. The results presented here suggest a possible explanation for the reduced all-trans-REs after administration of 9-cis-RAL to rpe65-/- mice. Purified RGR is sensitive to hydroxylamine and binds exogenous all-trans-RAL chromophore (30, 59), indicating that RGR is available for chromophore exchange. In the above-described experiment with rpe65-/- mice, the exogenous 9-cis-RAL may have replaced the bound all-trans-RAL, converting RGR to its light-adapted form. As we have shown (Fig. 5B), light adaptation of RGR frees all-trans-REH from inhibition, causing mobilization of all-trans-REs. This effect is analogous to the all-trans-RE-mobilization seen in rpe65-/- mice (Figs. 2C and 4C) and hRPE cells (Fig. 3, A and C) following exposure to light.
In summary, we have identified a new mechanism for regulation of the visual cycle in RPE cells. This mechanism involves light-dependent redistribution of all-trans-REs, mediated by RGR-opsin. The likely purpose of this redistribution is to move all-trans-REs from the lipid droplet storage pool to the smaller ER “isomerase pool” where the all-trans-REs can be utilized for synthesis of visual chromophore. Biochemically, this regulatory mechanism involves inhibition of all-trans-REH and LRAT by RGR-opsin in the dark, with release of inhibition following photoisomerization of the RGR chromophore to 11-cis-RAL. Further characterization of this regulatory mechanism will require identification of the all-trans-REH protein associated with lipid droplets.
Acknowledgments
We gratefully acknowledge Minshan Hu for excellent technical assistance; Henry Fong for insightful comments and the gift of rgr-/- knock-out mice; Mike Redmond for the gift of the rpe65-/- knock-out mice, and Rosalie Crouch for the gift of 11-cis-RAL.
This work was supported, in whole or in part, by National Institutes of Health Grant NEI R01-EY01584. This work was also supported by the Foundation Fighting Blindness. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: 11-cis-RAL, 11-cis-retinaldehyde; 11-cis-ROL, 11-cis-retinol; 11-cis-RDH, 11-cis-retinol dehydrogenase; all-trans-RAL, all-trans-retinaldehyde; all-trans-ROL, all-trans-retinol; all-trans-RE, all-trans-retinyl ester; all-trans-REH, all-trans-retinyl ester hydrolase; DA, dark-adapted; ER, endoplasmic reticulum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HPLC, high performance liquid chromatography; hRPE, human RPE; IRBP, interphotoreceptor retinol-binding protein; LA, light-adapted; LRAT, lecithin:retinol acyltransferase; PBS, phosphate-buffered saline; RBP, retinol-binding protein; RGR, RPE-retinal G protein receptor; RNAi, RNA interference; RPE, retinal pigment epithelium; siRNA, small interfering RNA; MES, 4-morpholineethanesulfonic acid; qRT, quantitative real time.
References
- 1.Arshavsky, V. Y., Lamb, T. D., and Pugh, E. N. (2002) Annu. Rev. Physiol. 64 153-187 [DOI] [PubMed] [Google Scholar]
- 2.Travis, G. H., Golczak, M., Moise, A. R., and Palczewski, K. (2007) Annu. Rev. Pharmacol. Toxicol. 47 469-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard, R., and St George, R. C. C. (1958) J. Gen. Physiol. 41 501-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamsdorf, K. (1979) in Handbook of Sensory Physiology (Autrum, H., ed) Vol. VII, part 6A, pp. 45-224, Springer-Verlag, Berlin [Google Scholar]
- 5.Schwemer, J. (1985) in The Molecular Mechanism of Photoreception (Sieve, H., ed) p. 326, Springer-Verlag, Berlin
- 6.Provencio, I., Jiang, G., De Grip, W. J., Hayes, W. P., and Rollag, M. D. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 340-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarttelin, E. E., Bellingham, J., Hankins, M. W., Foster, R. G., and Lucas, R. J. (2003) FEBS Lett. 554 410-416 [DOI] [PubMed] [Google Scholar]
- 8.Blackshaw, S., and Snyder, S. H. (1999) J. Neurosci. 19 3681-3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun, H., Gilbert, D. J., Copeland, N. G., Jenkins, N. A., and Nathans, J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 9893-9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, M., Pandey, S., and Fong, H. K. (1993) Invest. Ophthalmol. Vis. Sci. 34 3669-3678 [PubMed] [Google Scholar]
- 11.Fu, Y., Liao, H. W., Do, M. T., and Yau, K. W. (2005) Curr. Opin. Neurobiol. 15 415-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nayak, S. K., Jegla, T., and Panda, S. (2007) Cell Mol. Life Sci. 64 144-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey, S., Blanks, J. C., Spee, C., Jiang, M., and Fong, H. K. (1994) Exp. Eye Res. 58 605-613 [DOI] [PubMed] [Google Scholar]
- 14.Hara-Nishimura, I., Matsumoto, T., Mori, H., Nishimura, M., Hara, R., and Hara, T. (1990) FEBS Lett. 271 106-110 [DOI] [PubMed] [Google Scholar]
- 15.Hao, W. S., and Fong, H. K. W. (1999) J. Biol. Chem. 274 6085-6090 [DOI] [PubMed] [Google Scholar]
- 16.Shen, D., Jiang, M., Hao, W., Tao, L., Salazar, M., and Fong, H. K. (1994) Biochemistry 33 13117-13125 [DOI] [PubMed] [Google Scholar]
- 17.Chen, P., Hao, W. S., Rife, L., Wang, X. P., Shen, D. W., Chen, J., Ogden, T., Van Boemel, G. B., Wu, L. Y., Yang, M., and Fong, H. K. W. (2001) Nat. Genet. 28 256-260 [DOI] [PubMed] [Google Scholar]
- 18.Jin, M., Li, S., Moghrabi, W. N., Sun, H., and Travis, G. H. (2005) Cell 122 449-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moiseyev, G., Chen, Y., Takahashi, Y., Wu, B. X., and Ma, J. X. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 12413-12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redmond, T. M., Poliakov, E., Yu, S., Tsai, J. Y., Lu, Z., and Gentleman, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13658-13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Redmond, T. M., Yu, S., Lee, E., Bok, D., Hamasaki, D., Chen, N., Goletz, P., Ma, J. X., Crouch, R. K., and Pfeifer, K. (1998) Nat. Genet. 20 344-351 [DOI] [PubMed] [Google Scholar]
- 22.Van Hooser, J. P., Liang, Y., Maeda, T., Kuksa, V., Jang, G. F., He, Y. G., Rieke, F., Fong, H. K. W., Detwiler, P. B., and Palczewski, K. (2002) J. Biol. Chem. 277 19173-19182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wenzel, A., Oberhauser, V., Pugh, E. N., Jr., Lamb, T. D., Grimm, C., Samardzija, M., Fahl, E., Seeliger, M. W., Reme, C. E., and von Lintig, J. (2005) J. Biol. Chem. 280 29874-29884 [DOI] [PubMed] [Google Scholar]
- 24.Chen, P., Lee, T. D., and Fong, H. K. W. (2001) J. Biol. Chem. 276 21098-21104 [DOI] [PubMed] [Google Scholar]
- 25.Morimura, H., Saindelle-Ribeaudeau, F., Berson, E. L., and Dryja, T. P. (1999) Nat. Genet. 23 393-394 [DOI] [PubMed] [Google Scholar]
- 26.Bridges, C. D., and Alvarez, R. A. (1982) Methods Enzymol. 81 463-485 [DOI] [PubMed] [Google Scholar]
- 27.Groenendijk, G. W., De Grip, W. J., and Daemen, F. J. (1980) Biochim. Biophys. Acta 617 430-438 [DOI] [PubMed] [Google Scholar]
- 28.Ozaki, K., Terakita, A., Hara, R., and Hara, T. (1986) Vision Res. 26 691-705 [DOI] [PubMed] [Google Scholar]
- 29.Garwin, G. G., and Saari, J. C. (2000) Methods Enzymol. 316 313-324 [DOI] [PubMed] [Google Scholar]
- 30.Yang, M., and Fong, H. K. W. (2002) J. Biol. Chem. 277 3318-3324 [DOI] [PubMed] [Google Scholar]
- 31.Jang, G. F., Van Hooser, J. P., Kuksa, V., McBee, J. K., He, Y. G., Janssen, J. J. M., Driessen, C., and Palczewski, K. (2001) J. Biol. Chem. 276 32456-32465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mata, N. L., Moghrabi, W. N., Lee, J. S., Bui, T. V., Radu, R. A., Horwitz, J., and Travis, G. H. (2004) J. Biol. Chem. 279 635-643 [DOI] [PubMed] [Google Scholar]
- 33.Bok, D., Ruiz, A., Yaron, O., Jahng, W. J., Ray, A., Xue, L., and Rando, R. R. (2003) Biochemistry 42 6090-6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu, J., and Bok, D. (2001) Mol. Vis. 7 14-19 [PubMed] [Google Scholar]
- 35.Carlson, A., and Bok, D. (1992) Biochemistry 31 9056-9062 [DOI] [PubMed] [Google Scholar]
- 36.Hubbard, R., and Dowling, J. E. (1962) Nature 193 341-343 [DOI] [PubMed] [Google Scholar]
- 37.Weng, J., Mata, N. L., Azarian, S. M., Tzekov, R. T., Birch, D. G., and Travis, G. H. (1999) Cell 98 13-23 [DOI] [PubMed] [Google Scholar]
- 38.Saari, J. C., Nawrot, M., Kennedy, B. N., Garwin, G. G., Hurley, J. B., Huang, J., Possin, D. E., and Crabb, J. W. (2001) Neuron 29 739-748 [DOI] [PubMed] [Google Scholar]
- 39.Imanishi, Y., Batten, M. L., Piston, D. W., Baehr, W., and Palczewski, K. (2004) J. Cell Biol. 164 373-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bok, D., O. Day, W., and Rodriguez-Boulan, E. (1992) Exp. Eye Res. 55 853-860 [DOI] [PubMed] [Google Scholar]
- 41.Saari, J. C., and Bredberg, D. L. (1988) J. Biol. Chem. 263 8084-8090 [PubMed] [Google Scholar]
- 42.MacDonald, P. N., and Ong, D. E. (1988) J. Biol. Chem. 263 12478-12482 [PubMed] [Google Scholar]
- 43.Batten, M. L., Imanishi, Y., Maeda, T., Tu, D. C., Moise, A. R., Bronson, D., Possin, D., Van Gelder, R. N., Baehr, W., and Palczewski, K. (2004) J. Biol. Chem. 279 10422-10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mata, N. L., Tsin, A. T., and Chambers, J. P. (1992) J. Biol. Chem. 267 9794-9799 [PubMed] [Google Scholar]
- 45.Mata, N. L., Mata, J. R., and Tsin, A. T. (1996) J. Lipid Res. 37 1947-1952 [PubMed] [Google Scholar]
- 46.Mata, J. R., Mata, N. L., and Tsin, A. T. (1998) J. Lipid Res. 39 604-612 [PubMed] [Google Scholar]
- 47.Rando, R. R. (1991) Biochemistry 30 595-602 [DOI] [PubMed] [Google Scholar]
- 48.Saari, J. C., Bredberg, D. L., and Farrell, D. F. (1993) Biochem. J. 291 697-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maeda, T., Van Hooser, J. P., Driessen, C. A., Filipek, S., Janssen, J. J., and Palczewski, K. (2003) J. Neurochem. 85 944-956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moise, A. R., Golczak, M., Imanishi, Y., and Palczewski, K. (2007) J. Biol. Chem. 282 2081-2090 [DOI] [PubMed] [Google Scholar]
- 51.Simon, A., Romert, A., Gustafson, A. L., McCaffery, J. M., and Eriksson, U. (1999) J. Cell Sci. 112 549-558 [DOI] [PubMed] [Google Scholar]
- 52.Hamel, C. P., Tsilou, E., Harris, E., Pfeffer, B. A., Hooks, J. J., Detrick, B., and Redmond, T. M. (1993) J. Neurosci. Res. 34 414-425 [DOI] [PubMed] [Google Scholar]
- 53.Tsuiki, E., Fujita, A., Ohsaki, Y., Cheng, J., Irie, T., Yoshikawa, K., Senoo, H., Mishima, K., Kitaoka, T., and Fujimoto, T. (2007) Invest. Ophthalmol. Vis. Sci. 48 2858-2867 [DOI] [PubMed] [Google Scholar]
- 54.Brown, D. A. (2001) Curr. Biol. 11 R446-R449 [DOI] [PubMed] [Google Scholar]
- 55.Brasaemle, D. L. (2007) J. Lipid Res. 48 2547-2559 [DOI] [PubMed] [Google Scholar]
- 56.Ho, M. T., Pownall, H. J., and Hollyfield, J. G. (1989) J. Biol. Chem. 264 17759-17763 [PubMed] [Google Scholar]
- 57.Mirzadegan, T., Benko, G., Filipek, S., and Palczewski, K. (2003) Biochemistry 42 2759-2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang, M., Pandey, S., Tran, V. T., and Fong, H. K. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 3907-3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hao, W., and Fong, H. K. (1996) Biochemistry 35 6251-6256 [DOI] [PubMed] [Google Scholar]