Abstract
RNA polymerase (pol) III transcription, responsible for the synthesis of various stable RNAs, including 5 S rRNAs and tRNAs, is regulated by oncogenic proteins and tumor suppressors. Although it is well established that RNA pol III-dependent transcription is deregulated in transformed cells and malignant tumors, it has not been determined whether this represents a cause or consequence of these processes. We show that Rat1a fibroblasts undergoing oncogenic transformation by the TATA-binding protein or c-Myc display enhanced RNA pol III transcription. Decreased expression of the RNA pol III-specific transcription factor Brf1 prevented this increase in RNA pol III transcription. Although the overall proliferation rates of these cells remained unchanged, the ability of cells to grow in an anchorage-independent manner and form tumors in mice was markedly reduced. Although overexpression of Brf1 modestly stimulated RNA pol III transcription, expression of a phosphomimic, Brf1-T145D, more significantly induced transcription. However, these increases in transcription were not sufficient to promote cellular transformation. Together, these results demonstrate that enhanced RNA pol III transcription is essential for anchorage-independent growth and tumorigenesis and that these events can be uncoupled from effects on anchorage-dependent proliferation.
RNA polymerase (pol)2 III synthesizes a variety of small untranslated RNAs, including tRNA, 5 S rRNA, U6 RNA, and 7SL RNA. RNA pol III products play essential roles in protein synthesis, RNA processing, and protein trafficking. rRNA synthesis by RNA pol I and pol III is a limiting step in ribosome accumulation (1), and the overall rate of translation is determined by the number of ribosomes (2). Thus, the rate of RNA pol I and pol III transcription dictates the biosynthetic capacity of cells and may be an important determinant of a cell's capacity to grow. Consistent with this idea, decreasing RNA pol I transcription has been shown to result in reduced cellular growth rates (3). Recent results showed that decreased RNA pol III transcription by the bacterial compound Tagetin prevented hypertropic enlargement of cardiomyocytes (4). Activation of RNA pol III-dependent transcription may thus be needed for increased cell mass.
RNA pol III transcription is regulated by a variety of tumor suppressors and oncogenes. Oncogenic Ras (5), c-Myc (6), and activated phosphatidylinositol 3-kinase (7) all serve to induce transcription, whereas the tumor suppressors Rb (8), p53 (9), and PTEN (7) repress RNA pol III transcription. The RNA pol III-specific transcription factor (TF) IIIB complex, composed of TATA-binding protein (TBP), Brf1, and Bdp1, is the target of these regulatory proteins. Both Rb and p53 interact directly with components of the TFIIIB complex to inhibit its function, whereas c-Myc stimulates RNA pol III transcription by directly associating with the TFIIIB complex and enhancing its recruitment to promoters. In contrast, through its ability to regulate phosphatidylinositol 3-kinase, PTEN indirectly modulates the association between TBP and Brf1, determining the number of functional TFIIIB complexes.
Expression of oncogenic Ras induces expression of TBP in a variety of cell lines (5, 7, 10). Changes in TBP expression differentially affect the transcription of RNA pol II genes depending on the promoter architecture (11, 12). Alterations in cellular TBP expression have profound phenotypic consequences. In mouse embryonic fibroblasts, changes in cellular TBP concentrations alter cellular proliferation rates (13). Heterozygous disruption of TBP in chicken DT cells results in slower growth rates and delayed mitosis (14). In Rat1a fibroblasts, increased cellular TBP expression does not alter proliferation rates but promotes anchorage-independent growth and tumor formation in athymic mice (15). Although it is clear that oncogenic proteins can stimulate RNA pol III transcription, the role of enhanced RNA pol III transcription in their transforming function remains unclear.
In addition to its regulation by oncogenes and tumor suppressors, RNA pol III transcription is deregulated in transformed and tumor cells. A hallmark of cancer cells is their abnormally enlarged nucleoli, which pathologists have long used as a diagnostic criterion for malignancy (for example, see Ref. 16). This increase in nucleolar size is a result of enhanced rRNA synthesis as a consequence of deregulation of both RNA pol I and pol III transcription. In addition, the accumulation of RNA pol III transcripts around the nucleolus is particularly evident in transformed cells (17). Many types of virus-transformed cell lines display enhanced production of RNA pol III transcripts, including those transformed with simian virus 40 (18), papovavirus (19), and viral products such as hepatitis B virus X protein (20) and human T-cell leukemia virus type 1 Tax protein (21). Overproduction of RNA pol III transcripts has been observed in a wide variety of human tumors. Both tRNA and 5 S rRNA are overproduced in ovarian tumors relative to matched normal tissue (22), whereas 7 SL RNA was found to be more abundant in 80 tumor samples compared with matched normal tissue derived from 19 different cancers (23). Furthermore, in situ hybridization of various RNA pol III transcripts revealed that the levels of these transcripts are increased in neoplastic cells compared with the normal surrounding tissue (23, 24). These results argue that enhanced RNA pol III transcription may be essential for tumorigenesis.
Here, we show that anchorage-independent growth and tumorigenicity can be inhibited by selectively preventing TBP- and c-Myc-mediated increases in RNA pol III-dependent transcription. This effect on transformation can be uncoupled from overall cellular proliferation rates, which remain unchanged. These results provide new evidence that increased transcription by RNA pol III is not simply a consequence of the transformation process, but it is actually required to promote cellular transformation and tumorigenesis.
EXPERIMENTAL PROCEDURES
Expression Plasmids—The human TBP expression plasmid pLTR-E2TBP and construction of the TBP mutants L185E and K249E were as described previously (15). The human Brf1 expression plasmids pcDNA3-HA-Brf1 and HA-Brf1-T145D were provided by R. J. White (25) and were subcloned into the HindIII and XhoI sites of pCEP4. The Brf1 small interfering RNA targeting sequence 5′-AAG CAC TGC CCC ACT TAT TTG-3′ was used in a small hairpin to make the FG12-U6-Brf1 short hairpin RNA (shRNA) lentiviral expression vector constructed as described (26). FG12-U6 mismatch RNA was described previously (26).
Generation of Lentivirus and Stable Cell Lines—Rat1A cells were grown in Dulbecco's modified Eagle's medium (1 g/liter glucose) with 10% (v/v) fetal calf serum. Stable cell lines expressing TBP or mutant TBPs with or without Brf1 or Brf1 mutants were established as described previously and selected with 600 μg/ml G418 and with or without 80 μg/ml hygromycin (15). Early passage number pooled populations of selected cells were used. Rat1a cells stably expressing c-Myc were described previously (27). Generation of lentiviral stocks and infection of Rat1a cells were described previously (26). Where infection efficiencies were <95%, infected cells were sorted by fluorescence-activated cell sorter analysis and expanded for further use. To re-establish cell lines from tumor tissues, tumors from mice injected with vector, TBP, or TBP-L185E cell lines were surgically removed, minced, and recultured in complete medium for 24 h. After 24 h, the medium was replaced with selection medium containing 600 μg/ml G418 and propagated for 2 weeks. Two independent cell lines were established from tumors from two different mice for each re-established tumor-derived stable cell line. For all experiments, multiple independent stable cell lines were generated for each construct and experimentally tested, and representative results are shown for each.
Immunoblot Analysis—Protein lysates from subconfluent cultures were subjected to SDS-PAGE and immunoblot analysis. Membranes were probed with rabbit polyclonal anti-human TBP or anti-c-Myc (Santa Cruz Biotechnology), rat monoclonal anti-HA (Roche Applied Science), or mouse monoclonal anti-actin (Chemicon) antibody. Anti-Brf1-1 antibody was a gift from R. W. White (28). Primary antibody was visualized with chemiluminescence reagents (Pierce). Densitometry was performed using UN-SCAN-IT software (Silk Scientific).
Quantitative Real-time Reverse Transcription (RT)-PCR—Total RNA was isolated using RNA STAT-60, prepared, and reverse-transcribed for real-time quantitative PCR (qPCR) and analyzed as described previously (26). Primer sets for pre-tRNALeu, 7SL RNA, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (26) and for tRNAMeti (29) were described previously. Additional primer sets were as follows: HA-tagged human TBP, 5′-CCC GAC TAC GCC ACC GGT GAT ATC G-3′ (forward) and 5′-TTG TTG TTG CTG CTG CTG CCT TTG-3′ (reverse); murine TBP, 5′-GCT AGG TTT CTG CGG TCG CGT C-3′ (forward) and 5′-CTG TAC TGA GGC TGC TGC AGT TGC TAC-3′ (reverse); rat Brf1, 5′-CGA CTC ACA GCC TCC AGA GCA C-3′ (forward) and 5′-CCG ATG GCT TGA CAG GCT CA-3′ (reverse); and rat Bdp1, 5′-CTC CGC CCA GGA GGA TAG TCA GAC-3′ (forward) and 5′-CCT GGG TTG CTC AGA CTG CAG ACT-3′ (reverse). Amplification conditions were 94 °C for 30 s, 61 °C for 30 s, and 72 °C for 60 s for all primer sets. Relative transcript amounts were quantified as described (26) using the comparative threshold cycle method with GAPDH serving as the endogenous control reference. Transcript levels were normalized to those of GAPDH, and -fold changes were calculated based on transcript levels in the vector cell line or tumor. Statistics to determine significant differences (p < 0.05) were performed using a one-way analysis of variance followed by Tukey's multiple comparison test.
Cell Accumulation Rate, Anchorage-independent Growth, and Mouse Tumorigenicity Assays—Cell accumulation rates were determined by plating between 10,000 and 50,000 cells/60-mm dish. Cells were harvested daily, and viable cells were counted using a Coulter Counter. Soft agar growth assays were performed as described previously (15). Briefly, 1000 cells (Rat1a-Myc) or 7500 cells (all other Rat1a stable cell lines) were evenly single cell-suspended in 1.5 ml of 0.35% (w/v) low-melting agarose at 42 °C and overlaid onto each well of a 6-well dish containing a 0.7% (w/v) low-melting agarose bed in triplicate. All agarose suspensions were made Dulbecco's modified Eagle's medium (1 g/ml glucose) with 10% (v/v) fetal calf serum (final concentrations). Colonies >100 μm were counted after 21 days. Tumorigenicity assay with athymic nude (nu/nu) mice using Rat1a stable cell lines and infected cell lines (1 × 106 cells/animal) was conducted as described previously (15). Tumor volumes were determined by measuring tumor dimensions (height × width × depth). Statistics were performed as described above.
RESULTS
Enhanced TBP Expression Induces RNA pol III-dependent Transcription—Our previous study demonstrated that increased expression of TBP in Rat1a fibroblasts induces both anchorage-independent growth and tumor formation in athymic mice (15). The Rat1a system is a sensitive experimental approach that has been extensively used to detect transforming properties of proteins (30, 31). To identify the changes in gene expression required for TBP to promote these processes, we asked whether increased TBP expression induces RNA pol III transcription and, if so, whether this is necessary for TBP-mediated transforming function.
Rat1a cells were stably transfected with expression plasmids encoding HA-tagged TBP, TBP-L185E, or TBP-K249E. These two mutant forms, which differ from wild-type TBP by single amino acid substitutions, were shown previously to be selectively defective for RNA pol III transcription in cultured cells due to decreased association with the RNA pol III-specific factor Brf1 (32). Expression of both mutant and wild-type proteins in these stable lines was 20% above that of the endogenous TBP protein (Fig. 1A).
FIGURE 1.
RNA pol III transcription is increased by enhanced TBP expression. A, stable cell lines expressing increased TBP or mutant TBP proteins. Protein lysates from Rat1a cells stably transfected with empty vector or expression plasmids encoding HA-tagged wild-type or mutant (L185E and K249E) human (H) TBP cDNAs were subjected to immunoblot analysis using antibody against HA, TBP, or β-actin. B, RNA pol III gene activity is increased in cell lines expressing increased TBP but not TBP-L185E or TBP-K249E. RT-qPCR was performed using RNA isolated from stable cell lines in A with primers for pre-tRNALeu, 7SL RNA, and GAPDH. Values are the means ± S.E. (n ≥ 3). C, RNA pol III transcription is enhanced in tumors derived from stable cell lines expressing increased TBP or TBP-L185E. RT-qPCR was performed as described for B using RNA isolated from tumors from mice injected with the vector, TBP, and TBP-L185E cell lines. D, relative levels of TBP, Brf1, and Bdp1 mRNAs in tumors derived from stable cell lines. RT-qPCR using primers for murine TBP, rat Brf1, rat Bdp1, HA-tagged human TBP, and GAPDH was performed with RNA from tumors in C. -Fold changes were calculated based on GAPDH-normalized transcript levels in the tumors derived from the vector cell line (rat TBP, rat Brf1, and rat Bdp1) or TBP cell line (HA-tagged human TBP). Values are the means ± S.E. (n ≥ 4) from tumors from two different mice (C and D). E, tumor cells expressing TBP-L185E recultured in vitro display reduced RNA pol III transcription capacity. Stable cell lines were established from tumors from mice injected with the vector, TBP, and TBP-L185E cell lines. Following propagation in selection medium, total RNA was isolated, and RT-qPCR was performed as described for B. Values are the means ± S.E. (n ≥ 4) from two independently derived cell lines.
To determine the consequence of increased TBP expression on RNA pol III transcription, RNA was isolated from each of the stable cell lines, and RT-qPCR analysis was used to determine the amount of precursor transcripts. Enhanced TBP expression induced increases in both precursor tRNALeu and 7SL RNA transcripts (Fig. 1B). However, expression of TBP-L185E and TBP-K249E did not increase production of these transcripts. These results indicate that increased TBP expression induces RNA pol III transcription in vitro. Furthermore, these results suggest that increases in TBP directly support the RNA pol III transcription process because TBP-Brf1 interactions are necessary to augment transcription.
We next examined whether the stable cell line expressing TBP-L185E would similarly fail to support an increase in transcription in vivo. Cell lines containing empty vector or those expressing increased TBP or TBP-L185E were each subcutaneously injected into athymic mice. RNA was isolated from the resultant tumor cells, and RT-qPCR analysis was conducted. Unlike what was observed in vitro, expression of TBP-L185E displayed an increase in RNA pol III transcripts in the in vivo environment compared with the vector control, comparable with that seen with enhanced expression of TBP (Fig. 1C). To determine whether the stable cell lines maintained expression of the introduced cDNAs, the amounts of exogenous TBP and TBP-L185E and endogenous TBP mRNAs were measured (Fig. 1D). All tumors derived from vector, TBP, and TBP-L185E cell lines expressed comparable amounts of endogenous TBP and the TFIIIB-associated factors Brf1 and Bdp1. Similar levels of ectopically expressed TBP and TBP-L185E were also expressed.
To further examine whether the TBP-L185E-mediated increase in RNA pol III transcription in the tumors is reversible, the tumor cells were recultured in vitro. Although the tumor-derived TBP stable cell line continued to show an increase in RNA pol III transcripts relative to the vector control line, cells expressing TBP-L185E did not support this increase (Fig. 1E), similar to what we observed initially for these stable lines in culture (Fig. 1B). These results indicate that although TBP-L185E does not enhance transcription in vitro, it does so in vivo. This provides us with a tool to examine the effects of transcriptional activity on malignant potential.
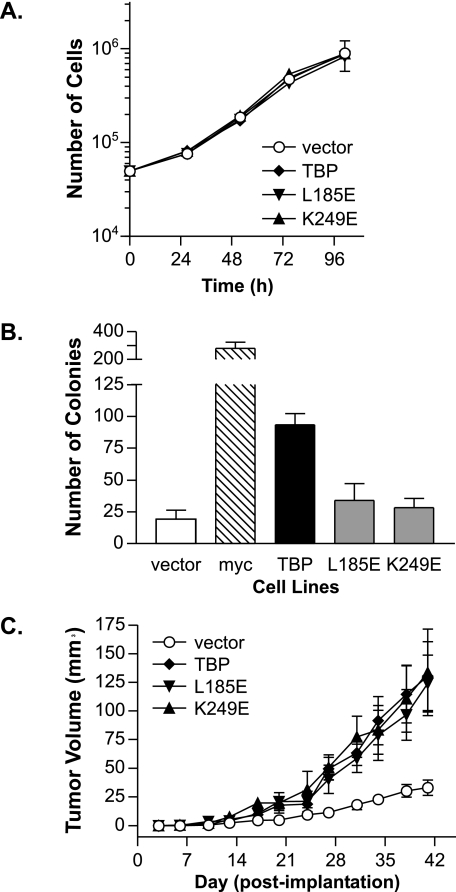
TBP-mediated Induction of RNA pol III Transcription Does Not Alter Cellular Proliferation but Induces Anchorage-independent Growth—We next determined the phenotypic consequences of enhanced TBP expression in the presence and absence of increased RNA pol III transcription. We compared the cellular proliferation rates of the TBP, TBP-L185E, and TBP-K249E stable cell lines. Neither increased TBP expression nor expression of TBP-L185E and TBP-K249E affected proliferation rates (Fig. 2A). However, there were discernible differences in the ability of these cells to form colonies in soft agar. Although enhanced TBP expression supported anchorage-independent growth, expression of TBP-L185E or TBP-K249E did not (Fig. 2B). Thus, enhanced RNA pol III transcription is necessary for TBP-mediated anchorage-independent cellular growth.
FIGURE 2.
Tumor formation is enhanced by increased TBP or mutant TBP expression. A, proliferation rates are not altered by changes in RNA pol III transcription. Stable cell lines on duplicate plates were trypsinized and counted daily. A representative plot of three independent experiments is shown. B, enhanced expression of TBP, but not mutant TBP proteins, promotes anchorage-independent growth. Stable cell lines in A or cells expressing c-Myc were analyzed for growth in soft agar. Values are the means ± S.E. (n ≥ 3). C, tumor formation in mice with cells overexpressing TBP or mutant TBP proteins. Cell lines expressing TBP, TBP-L185E, or TBP-K249E were injected subcutaneously into the groins of nude mice (six mice/group). Representative results from two independent experiments are shown.
We assessed the ability of the stable cell lines expressing TBP-L185E or TBP-K249E to form tumors in mice. In light of our findings that the mutations did not affect transcription in vivo (Fig. 1C), we anticipated that there would be no measurable difference in the tumorigenic potential of the cells expressing increased TBP versus the mutant TBP proteins. Indeed, compared with the vector control cell line, the TBP, TBP-L185E, and TBP-K249E stable lines all displayed similar enhancements in tumor formation (Fig. 2A). Thus, tumorigenic potential is directly influenced by the RNA pol III transcription capacity. These results indicate that enhanced RNA pol III transcription, together with other TBP-mediated changes in gene expression, correlates with tumorigenesis.
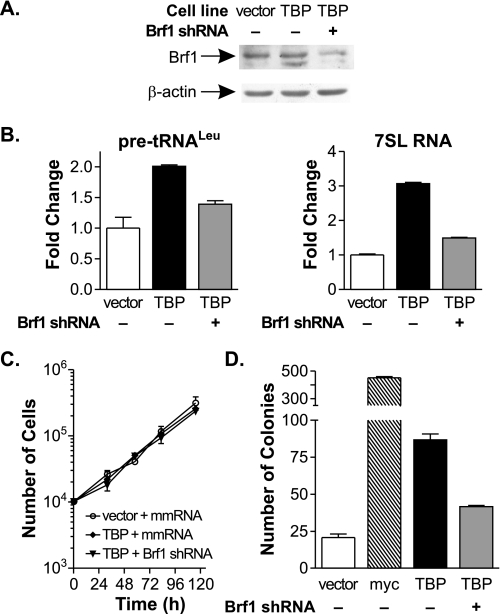
Preventing RNA pol III Transcription Induction Inhibits TBP-mediated Tumorigenesis—We used an additional approach to further confirm that RNA pol III transcription induction is required for TBP-mediated cellular transformation and tumorigenesis. Expression of the RNA pol III-specific TBP-associated factor Brf1 was reduced to prevent TBP-mediated increases in RNA pol III transcription. Stable cell lines expressing increased amounts of TBP were infected with lentiviral constructs containing a mismatch RNA or an shRNA for Brf1. A 2-fold decrease in Brf1 expression was observed in the TBP-Brf1 shRNA stable cell lines (Fig. 3A). This abrogated the TBP-mediated increase in RNA pol III transcription, although the level of transcription was not lower than that observed in the vector control cell line (Fig. 3B).
FIGURE 3.
Decreased Brf1 expression inhibits TBP-mediated increase in RNA pol III transcription and anchorage-independent growth. A, Brf1 levels in stable cell lines expressing Brf1 shRNA. Protein lysates from vector and TBP stable lines expressing mismatch (mmRNA) or Brf1 shRNA were subjected to immunoblot analysis with anti-Brf1 or anti-β-actin antibodies. B, decreased Brf1 expression abrogates TBP-mediated increases in RNA pol III transcription. RT-qPCR was performed on RNA from infected stable cell lines in A. Values are the means ± S.E. (n ≥ 3). C, accumulation rate of stable lines expressing Brf1 shRNA. Infected stable cell lines in A on duplicate plates were trypsinized and counted daily. A representative plot of three independent experiments is shown. D, down-regulating Brf1 expression reduces TBP-mediated anchorage-independent growth. Cells in A or those expressing c-Myc were analyzed for growth in soft agar. Values are the means ± S.E. (n ≥ 3).
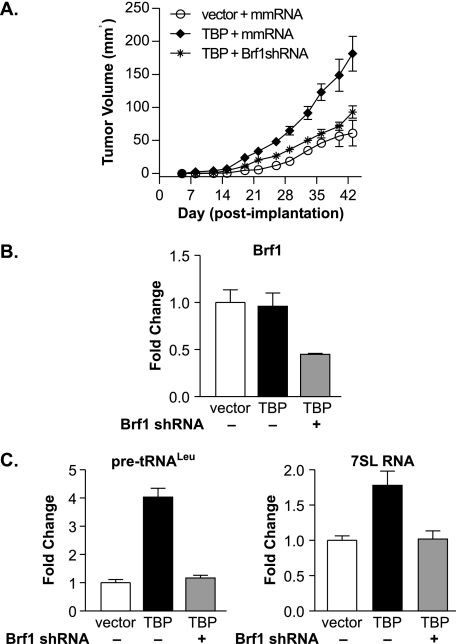
No differences in cellular proliferation rates were observed upon reduction of Brf1 levels (Fig. 3C). However, decreased Brf1 expression significantly inhibited TBP-mediated anchorage-independent growth (Fig. 3D). As expected, when these cell lines were assayed for their ability to form tumors in mice, the stable line expressing an increase in TBP without a change in Brf1 expression displayed a marked increase in tumor formation compared with the vector control line (Fig. 4A). Decreased Brf1 expression resulted in a significant reduction in tumor volume. RNA analysis of the tumor-derived cells confirmed that a reduction in Brf1 expression was maintained in the resultant tumor cells (Fig. 4B). Furthermore, decreased Brf1 expression was sufficient to maintain a reduction in RNA pol III transcription in vivo (Fig. 4C). Thus, selectively preventing induction of RNA pol III transcription through reduced Brf1 expression markedly decreases TBP-mediated anchorage-independent growth and tumorigenesis, independent of any effect on cellular proliferation.
FIGURE 4.
Enhanced RNA pol III transcription is required for TBP-mediated tumorigenesis. A, down-regulating Brf1 expression inhibits TBP-mediated tumorigenesis. Vector and TBP stable cell lines expressing mismatch RNA (mmRNA) or Brf1 shRNA were injected subcutaneously into nude mice (six mice/group), and tumor volumes were determined. B, Brf1 expression remains repressed in tumors from cells expressing Brf1 shRNA. RT-qPCR was performed on RNA from tumors derived from mice injected with stable cell lines using primers for Brf1 or GAPDH. C, RNA pol III transcription remains repressed in tumors from cells expressing Brf1 shRNA. RT-qPCR was performed on RNA from tumors in B using primers for pre-tRNALeu, 7SL RNA, or GAPDH. Values are the means ± S.E. (n ≥ 3) from tumors from two different mice per cell line.
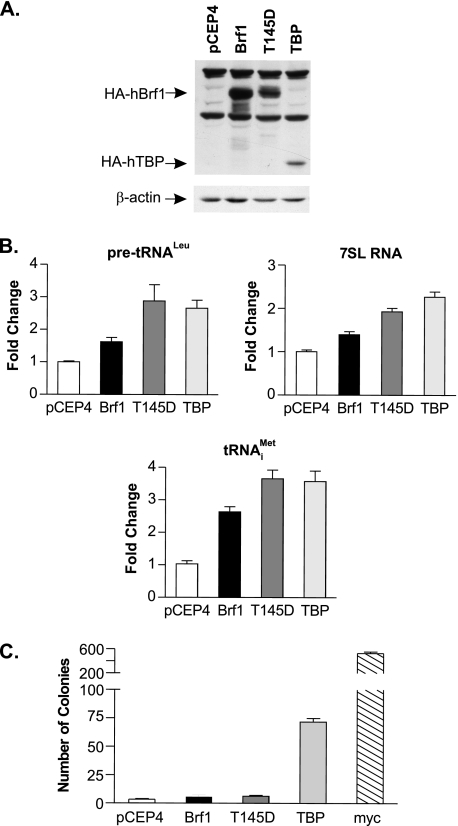
Increased RNA pol III Transcription Is Not Sufficient to Promote Cellular Transformation—As decreased Brf1 expression prevented TBP-mediated RNA pol III transcription induction and limited the transforming properties of TBP, we sought to determine whether increased Brf1 expression is sufficient for altering cellular growth properties. Stable cell lines were generated that expressed either HA-tagged Brf1 or a mutant form of Brf1, Brf1-T145D, which mimics phosphorylation at this position. Phosphorylation of Brf1 at threonine 145 by extracellular signal-regulated kinase (ERK) enhances Brf1 interactions with RNA pol III and TFIIIC (25). HA-Brf1 was consistently expressed at levels ∼30% higher than HA-Brf1-T145D (Fig. 5A). Brf1 overexpression resulted in a modest enhancement of pre-tRNALeu and 7SL RNA and a more pronounced increase in tRNAMeti (Fig. 5B). However, expression of Brf1-T145D resulted in a more significant increase in all of these RNA pol III transcripts, demonstrating the importance of phosphorylation of this site in Brf1 for transcription. RNA pol III transcription induction by Brf1-T145D was comparable with that observed in cells expressing increased TBP. Analyses of the resultant growth properties of these stable cell lines revealed that expression of Brf1 or Brf1-T145D did not alter cellular proliferation rates (data not shown). In addition, expression of these proteins did not confer anchorage-independent growth (Fig. 5C). Together, these results demonstrate that although overexpression of Brf1 or expression of a Brf1 phosphomimic does induce RNA pol III transcription, this is not sufficient to promote transformation of Rat1a fibroblasts.
FIGURE 5.
Enhanced RNA pol III transcription is not sufficient for promoting anchorage-independent growth. A, analysis of expression of TBP, Brf1, and Brf1-T145D in stable cell lines. Cells were stably transfected with empty vector or expression plasmids encoding HA-Brf1, HA-Brf1-T145D, or HA-TBP. Protein lysates were subjected to immunoblot analysis with anti-HA or anti-β-actin antibody. h, human. B, expression of Brf1-T145D induces RNA pol III-dependent transcription. RT-qPCR was performed using RNA isolated from each cell line in A and primers for pre-tRNALeu, 7SL RNA, tRNAMeti, and GAPDH. Transcript levels were normalized to GAPDH levels, and -fold change was calculated based on levels in the pCEP4 vector cell line. Values are the means ± S.E. (n ≥ 3). C, enhanced Brf1 or Brf1-T145D expression does not increase anchorage-independent growth. Stable cell lines in A or those expressing c-Myc were analyzed for growth in soft agar. Values are the means ± S.E. (n ≥ 3).
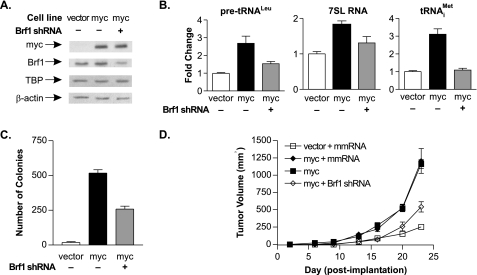
Cellular Transformation by c-Myc Requires Enhanced RNA pol III Transcription—The above studies demonstrated that an increase in RNA pol III transcription is required to drive TBP-mediated cellular transformation and tumorigenesis. To examine whether increased transcription is necessary to drive the transforming function of other oncogenes, we determined whether enhanced RNA pol III transcription is also required to support the transforming function of c-Myc. Stable cell lines expressing c-Myc were infected with either the lentiviral control mismatch RNA or Brf1 shRNA to repress Brf1 expression (Fig. 6A). Expression of Brf1 was effectively reduced by 50% without altering TBP expression. Expression of c-Myc produced an increase in precursor tRNALeu, tRNAMeti, and 7SL RNA expression (Fig. 6B). Reducing Brf1 expression inhibited the increase in all of these RNAs. Moreover, c-Myc-mediated growth in soft agar was repressed by the decrease in Brf1 and RNA pol III transcription (Fig. 6C). This reduction in RNA pol III transcription also significantly reduced c-Myc-mediated tumorigenesis (Fig. 6D). Thus, enhanced RNA pol III transcription is necessary to drive c-Myc-mediated transformation and tumor formation.
FIGURE 6.
Enhanced RNA pol III transcription is required for c-Myc-mediated transformation and tumorigenesis. A, Brf1 and TBP levels in lines expressing c-Myc and Brf1 shRNA. Vector and c-Myc stable cell lines were infected with lentivirus expressing mismatch RNA (mmRNA; –) or Brf1 shRNA (+). Protein lysates were subjected to immunoblot analysis. B, decreased Brf1 expression abrogates c-Myc-induced RNA pol III transcription. RT-qPCR was performed on RNA isolated from each cell line in A with primers for pre-tRNALeu, 7SL RNA, tRNAMeti, and GAPDH. Values are the means ± S.E. (n ≥ 3). C, decreased Brf1 expression inhibits c-Myc-mediated anchorage-independent growth. Cell lines in A were analyzed for growth in soft agar. Values are the means ± S.E. (n ≥ 3). D, decreased Brf1 expression inhibits c-Myc-mediated tumorigenesis. Stable cell lines in A were injected subcutaneously into mice (six mice/group). Tumor volumes were determined by measuring the dimensions (height × width × depth) of tumors.
DISCUSSION
Increased RNA pol III transcription and enhanced production of tRNAs and 5 S rRNAs are well known hallmarks of the malignant phenotype. These findings support the idea that enhanced RNA pol III transcription may not only contribute to the proliferative potential of cells, but could also alter the oncogenic properties of cells. Our study clearly demonstrates that increased RNA pol III transcription is not merely a consequence of the transformation process but is indeed essential for the transformed phenotype. Both TBP- and c-Myc-mediated increases in RNA pol III transcription allow fibroblasts to acquire anchorage-independent growth properties and to form tumors in an athymic mouse model. Consistent with these studies, recent results have also demonstrated that enhanced RNA pol III transcription is required to mediate focus formation of an HCT116/p53 null colon carcinoma cell line (29).
Our previous studies have shown that small changes in the levels of the central eukaryotic transcription initiation factor, TBP, alter cellular growth properties (13, 15). Because TBP is used by all three nuclear RNA polymerases, it is important to identify the genes regulated by changes in TBP concentrations and those that are necessary to support these TBP-mediated phenotypic changes. Our current studies reveal that TBP is a limiting factor for RNA pol III transcription in Rat1a fibroblasts and that increased expression of TBP is sufficient to induce transcription. Increased expression of the RNA pol III-specific TBP-associated factor Brf1 leads to a relatively modest induction of RNA pol III transcription. Expression of the phosphomimic form of Brf1 leads to a more robust increase in transcription. However, these increases in RNA pol III transcription fail to promote cellular transformation. This is consistent with our previous study showing that expression of RNA pol II-defective TBP mutants, which still are able to function in RNA pol I and pol III transcription, abrogates TBP-mediated cellular transformation (15). Together, these results demonstrate that although enhanced RNA pol III transcription is required for cellular transformation, it is not sufficient.
A recent study by Marshall et al. (29) shows that overexpression of Brf1 supports an increase in RNA pol III transcription in mouse embryo fibroblasts and an ovarian epithelial cell line. In addition, enhanced RNA pol III transcription also promotes cell cycle progression and transformation. These phenotypic effects could be mimicked by overexpression of tRNAMeti. However, our studies show that modulating RNA pol III transcription alone does not alter proliferation rates or the transforming properties of Rat1a cells. Although we also found that overexpression of Brf1 or Brf1-T145D increases tRNAMeti levels, this does not appear to be sufficient to induce transformation. Whether the differences observed between our studies and that of Marshall et al. (29) reflect cell type-specific effects or the conditions used to modulate Brf1 expression is not clear. Previous work in cardiomyocytes showed that an increase in Brf1 expression enhances RNA pol III transcription and that this is required for inducing hypertrophic growth (33). Collectively, these results suggest that the mechanisms that drive an increase in RNA pol III transcription to promote cellular growth or proliferation may be distinct from that used to promote transformation.
Expression of mutant TBP proteins did not induce RNA pol III transcription and anchorage-independent growth in vitro, whereas introduction of these cells into mice resulted in stimulation of RNA pol III transcription and tumor formation. Although it is clear that enhanced RNA pol III-dependent transcription is associated with cellular transformation, it is unclear why the mutant TBP proteins are defective for RNA pol III-dependent transcription in vitro but not in vivo. This is likely to be a result of the in vivo environment, where these cells interact with components of the extracellular matrix, which are not present in vitro. These components could alter cellular signaling pathways and impinge on TBP-mediated changes in RNA pol II-dependent transcription that ultimately affect RNA pol III-dependent transcription. We considered that TBP-mediated changes in RNA pol II-dependent transcription might selectively affect the expression of the RNA pol III transcription components in vivo. Because the TBP-containing TFIIIB complex has been shown to be a target of various oncogenic proteins and tumor suppressors (5–7, 9, 34–36), we examined whether the mutant TBP proteins increase expression of the TFIIIB subunits in vivo. However, no apparent changes in the amounts of endogenous TBP, Brf1, or Bdp1 were found in the tumor-derived cells. Because both Brf1 and Bdp1 are phosphorylated at multiple sites (7, 25, 37, 38), it is possible that, in context with the in vivo environment, changes in their phosphorylation state and/or other modifications of these proteins may selectively enhance their function in vivo. Alternatively, recent findings support the idea that certain coactivators of RNA pol II transcription may also be used to drive transcription of RNA pol III genes (39). Selective enhancement of the function of these coactivators in vivo could potentially be used to drive an increase in RNA pol III transcription.
Our study demonstrates that enhanced RNA pol III transcription is necessary for both TBP- and c-Myc-mediated transformation. Although many oncogenic factors and proteins induce RNA pol III transcription, it is likely that this increase is generally necessary to promote and maintain a transformed phenotype. As the link between metabolic activity and cancer has long been known, our findings provide a new molecular level of understanding for this connection. The deregulation of ribosome biogenesis and protein synthesis in cancer has prompted the investigation of potential cancer therapeutic agents that target the translation machinery (40). Our results support the idea that targeting the RNA pol III transcription machinery may represent a novel strategy for controlling tumorigenesis and cancer progression.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Postdoctoral Training Grant 2 T32 CA009659 (to S. A. S. J.) and National Institutes of Health Grants CA74138 and CA108614 (to D. L. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: pol, polymerase; TF, transcription factor; TBP, TATA-binding protein; HA, hemagglutinin; shRNA, short hairpin RNA; RT, reverse transcription; qPCR, quantitative PCR; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Zetterberg, A., and Killander, D. (1965) Exp. Cell Res. 40 1–11 [DOI] [PubMed] [Google Scholar]
- 2.Liebhaber, S. A., Wolf, S., and Schlessinger, D. (1978) Cell 13 121–127 [DOI] [PubMed] [Google Scholar]
- 3.White, R. J. (2005) Nat. Rev. Mol. Cell Biol. 6 69–78 [DOI] [PubMed] [Google Scholar]
- 4.Goodfellow, S. J., and White, R. J. (2007) Cell Cycle 6 2323–2326 [DOI] [PubMed] [Google Scholar]
- 5.Wang, H. D., Trivedi, A., and Johnson, D. L. (1997) Mol. Cell. Biol. 17 6838–6846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Roman, N., Grandori, C., Eisenman, R. N., and White, R. J. (2003) Nature 421 290–294 [DOI] [PubMed] [Google Scholar]
- 7.Woiwode, A., Johnson, S. A., Zhong, S., Zhang, C., Roeder, R. G., Teichmann, M., and Johnson, D. L. (2008) Mol. Cell. Biol., in press [DOI] [PMC free article] [PubMed]
- 8.White, R. J., Trouche, D., Martin, K., Jackson, S. P., and Kouzarides, T. (1996) Nature 382 88–90 [DOI] [PubMed] [Google Scholar]
- 9.Crighton, D., Woiwode, A., Zhang, C., Mandavia, N., Morton, J. P., Warnock, L. J., Milner, J., White, R. J., and Johnson, D. L. (2003) EMBO J. 22 2810–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, S. A., Mandavia, N., Wang, H. D., and Johnson, D. L. (2000) Mol. Cell. Biol. 20 5000–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colgan, J., and Manley, J. L. (1992) Genes Dev. 6 304–315 [DOI] [PubMed] [Google Scholar]
- 12.Majello, B., Napolitano, G., and Lania, L. (1998) AIDS 12 1957–1964 [DOI] [PubMed] [Google Scholar]
- 13.Zhong, S., Fromm, J., and Johnson, D. L. (2007) Mol. Cell. Biol. 27 54–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Um, M., Yamauchi, J., Kato, S., and Manley, J. L. (2001) Mol. Cell. Biol. 21 2435–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, S. A., Dubeau, L., Kawalek, M., Dervan, A., Schonthal, A. H., Dang, C. V., and Johnson, D. L. (2003) Mol. Cell. Biol. 23 3043–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rzymowska, J. (1997) Tumori 83 938–942 [DOI] [PubMed] [Google Scholar]
- 17.Wang, C., Politz, J. C., Pederson, T., and Huang, S. (2003) Mol. Biol. Cell 14 2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White, R. J., Stott, D., and Rigby, P. W. (1990) EMBO J. 9 3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felton-Edkins, Z. A., and White, R. J. (2002) J. Biol. Chem. 277 48182–48191 [DOI] [PubMed] [Google Scholar]
- 20.Wang, H. D., Yuh, C. H., Dang, C. V., and Johnson, D. L. (1995) Mol. Cell. Biol. 15 6720–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottesfeld, J. M., Johnson, D. L., and Nyborg, J. K. (1996) Mol. Cell. Biol. 16 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter, A. G., Sourvinos, G., Allison, S. J., Tosh, K., Scott, P. H., Spandidos, D. A., and White, R. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12619–12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, W., Bocker, W., Brosius, J., and Tiedge, H. (1997) J. Pathol. 183 345–351 [DOI] [PubMed] [Google Scholar]
- 24.Chen, W., Heierhorst, J., Brosius, J., and Tiedge, H. (1997) Eur. J. Cancer 33 288–292 [DOI] [PubMed] [Google Scholar]
- 25.Felton-Edkins, Z. A., Fairley, J. A., Graham, E. L., Johnston, I. M., White, R. J., and Scott, P. H. (2003) EMBO J. 22 2422–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, S. S., Zhang, C., Fromm, J., Willis, I. M., and Johnson, D. L. (2007) Mol. Cell 26 367–379 [DOI] [PubMed] [Google Scholar]
- 27.Hoang, A. T., Cohen, K. J., Barrett, J. F., Bergstrom, D. A., and Dang, C. V. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6875–6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alzuherri, H. M., and White, R. J. (1998) J. Biol. Chem. 273 17166–17171 [DOI] [PubMed] [Google Scholar]
- 29.Marshall, L., Kenneth, N. S., and White, R. J. (2008) Cell 133 78–89 [DOI] [PubMed] [Google Scholar]
- 30.Stone, J., de Lange, T., Ramsay, G., Jakobovits, E., Bishop, J. M., Varmus, H., and Lee, W. (1987) Mol. Cell. Biol. 7 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Small, M. B., Hay, N., Schwab, M., and Bishop, J. M. (1987) Mol. Cell. Biol. 7 1638–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen, Y., Kassavetis, G. A., Bryant, G. O., and Berk, A. J. (1998) Mol. Cell. Biol. 18 1692–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodfellow, S. J., Innes, F., Derblay, L. E., MacLellan, W. R., Scott, P. H., and White, R. J. (2006) EMBO J. 25 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirsch, H. A., Jawdekar, G. W., Lee, K. A., Gu, L., and Henry, R. W. (2004) Mol. Cell. Biol. 24 5989–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutcliffe, J. E., Brown, T. R., Allison, S. J., Scott, P. H., and White, R. J. (2000) Mol. Cell. Biol. 20 9192–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong, S., Zhang, C., and Johnson, D. L. (2004) Mol. Cell. Biol. 24 5119–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fairley, J. A., Scott, P. H., and White, R. J. (2003) EMBO J. 22 5841–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottesfeld, J. M., Wolf, V. J., Dang, T., Forbes, D. J., and Hartl, P. (1994) Science 263 81–84 [DOI] [PubMed] [Google Scholar]
- 39.Kenneth, N. S., Ramsbottom, B. A., Gomez-Roman, N., Marshall, L., Cole, P. A., and White, R. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14917–14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruggero, D., and Pandolfi, P. P. (2003) Nat. Rev. Cancer 3 179–192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.