Abstract
Mitogen-activated protein kinases (MAPKs) mediate cellular responses to a wide variety of extracellular stimuli. MAPK signal transduction cascades are tightly regulated, and individual MAPKs display exquisite specificity in recognition of their target substrates. All MAPK family members share a common phosphorylation site motif, raising questions as to how substrate specificity is achieved. Here we describe a peptide library screen to identify sequence requirements of the DEF site (docking site for ERK FXF), a docking motif separate from the phosphorylation site. We show that MAPK isoforms recognize DEF sites with unique sequences and identify two key residues on the MAPK that largely dictate sequence specificity. Based on these observations and computational docking studies, we propose a revised model for MAPK interaction with substrates containing DEF sites. Variations in DEF site sequence requirements provide one possible mechanism for encoding complex target specificity among MAPK isoforms.
Mitogen-activated protein kinases (MAPKs)4 lie at the bottom of conserved three-component phosphorylation cascades that integrate cellular responses to a wide variety of extracellular stimuli, including growth factors, cytokines, UV irradiation, and oxidative stress (1). Canonical MAPKs are classified into three major subfamilies, extracellular signal-regulated kinases (ERK), p38 MAPK (p38), and c-Jun N-terminal kinases (JNK), based on sequence homology, shared upstream kinases, and activating stimuli. In addition, the different MAPK subfamilies phosphorylate a distinct set of protein substrates. These substrates act as the critical effectors that enable cells to mount the appropriate responses to varied stimuli.
Analysis of reported MAPK substrate phosphorylation sites suggests a nearly absolute requirement for a Pro residue immediately downstream of the Ser or Thr phosphoacceptor. Extensive peptide library studies on ERK1, ERK2, and p38α have defined a common phosphorylation site motif for these MAPKs. In addition to the requirement for a Pro residue at the +1 position relative to the phosphorylation site, this motif includes a weak preference for Pro and other aliphatic residues at the -2 position (2–4). Mutagenesis experiments with the JNK substrate JunB have also suggested additional specificity for JNK family MAPKs at positions downstream of the phosphorylation site (5). MAPKs have affinities for short peptide substrates that are at least an order of magnitude lower than for full-length protein substrates (4, 6–10). Thus although phosphorylation site motifs likely serve to direct the MAPK to phosphorylate a specific serine or threonine residue within a protein, they are insufficient to fully account for protein substrate recognition. Although mechanisms such as subcellular localization and the use of scaffolding proteins can contribute to kinase substrate targeting in vivo, MAPKs also exhibit a high degree of target specificity in vitro (11–13).
For MAPKs, substrate specificity is ensured through the use of docking interactions occurring between sequence motifs in the substrate distal from the phosphorylation site and regions of the kinase outside of the active site (1, 14, 15). Two common types of docking interactions have been identified between MAPKs and their substrates that serve to enhance affinity and specificity. Both involve interaction of short linear sequence motifs present within substrates with a complementary pocket or groove on the kinase. The best characterized docking site utilized by MAPKs is the D-site (also referred to as the D-domain, δ-domain, or DEJL domain), which consists of two or more basic residues followed by a short linker and a cluster of hydrophobic residues. Interactions between the MAPK and substrate D-sites have been mapped by mutagenesis, hydrogen exchange-mass spectrometry, and x-ray crystallography (16–20). Docking occurs along a groove on the opposite face of the MAPK surface relative to the active site that includes acidic and hydrophobic regions complementary to the D-site. D-sites are present in many ERK, JNK, and p38 substrates and have been implicated in specific targeting by the different subfamilies. In addition, enzymes that regulate MAPKs, including their activating MAPK kinases (MKKs) and inactivating MAPK phosphatases, also contain D-sites that are critical for specific interactions with these kinases (16).
The second major MAPK docking site, known as the DEF site (docking site for ERK FXF, also called the F-site), has been identified in a number of ERK substrates. DEF sites are generally characterized as an FX(F/Y)P sequence, typically located between 6 and 20 amino acids C-terminal to the phosphorylation site. Hydrogen exchange-mass spectrometry experiments with ERK2 in complex with DEF site peptides have allowed the DEF interaction site on ERK to be mapped to a hydrophobic pocket adjacent to the active site cleft (18). Interestingly, accessibility of this pocket to docking sites is coupled to activation of the kinase. MAPKs are activated by dual phosphorylation of closely spaced threonine and tyrosine residues within a conserved region known as the activation loop (15, 21). Phosphorylation triggers a conformational rearrangement of the activation loop, aligning residues important for catalysis, forming the phosphoacceptor binding site, and exposing the DEF site interaction pocket (22).
Most reported substrates bearing DEF sites are transcription factors, including Elk-1, c-Fos, c-Myc, and JunD (23–26). However, DEF sites have also been identified in proteins with diverse functions, such as the ERK scaffolding protein KSR1 (23), the phosphodiesterase PDE4 (27), the proapoptotic Bcl-2 family protein BimEL (28), the GTPase-activating protein CdGAP (29), and the focal adhesion protein vinexin (30). In addition to playing a role in substrate recruitment, DEF sites can also mediate interactions with regulators of ERK, including the MKP-1 phosphatase and nucleoporins that facilitate nuclear import of the kinase (18, 31–33). Although generally described as a docking site for ERK MAPKs, the DEF site within the transcription factor SAP-1 has also been implicated in contributing to efficient phosphorylation by p38α (34).
In contrast to its phosphorylation site motif, which has been extensively characterized using peptide libraries, our understanding of DEF site interactions for ERK are based solely on a relatively small number of sites that have been identified in protein substrates. In the absence of a comprehensive analysis, the essential sequence requirements that constitute an optimal DEF site for ERK are not clear. Furthermore, it is unknown if MAPKs other than ERK can interact with DEF sites in substrates, or whether they recognize distinct DEF site sequence motifs. To address these questions, we adapted peptide library screening methods to elucidate consensus sequences for kinase docking sites. We identified significant differences in the DEF site docking motifs recognized by MAPK family members. Peptide library screening with a series of point mutants allowed us to identify critical determinants of specificity within the hydrophobic pocket of the kinase. The distinct sequence motifs recognized by different MAPKs may contribute to targeting these kinases to their unique substrates.
EXPERIMENTAL PROCEDURES
Peptides—Peptides for library screening were synthesized on a 2-mg scale by Sigma Genosys and used without further purification. All peptides were characterized by matrix-assisted laser desorption ionization time-of-flight mass spectrometry to confirm identity. The P4 Tyr peptide failed synthesis and was omitted from the final peptide library. Peptides were dissolved in DMSO, adjusted to a concentration of 25 mm based on absorbance at 280 nm, and stored at -20 °C. Working 0.5 mm aqueous stocks were prepared by diluting the DMSO stock rapidly into 50 mm HEPES, pH 7.4. Aqueous solutions were aliquoted into 384-well polypropylene stock plates in 40-ml aliquots, sealed with adhesive foil, and stored at -20 °C. Individual peptides were synthesized on a 0.1-mmol scale and high pressure liquid chromatography-purified at the Tufts University Core Facility (Boston). Lyophilized peptides were resuspended in DMSO and quantified by absorbance at 280 nm. Stock solutions were adjusted to 10 mm peptide and stored at -20 °C.
Plasmids—Full-length ERK2, p38α, p38γ, and p38δ MAPK coding sequences (all from rat) were cloned into pGEX4T-1 (GE Healthcare) for bacterial expression as GST fusion proteins. Coding sequences were cloned as BamHI/NotI fragments with the endogenous initiator methionine codon immediately downstream of the BamHI site and the native stop codon intact. DEF site attenuation and specificity swapping mutations were made by PCR-based site-directed mutagenesis in these expression plasmids. Coding sequences for the constitutively active MKK mutants, MKK1ΔN3 (35) and MKK6 S207E/T211E (36), were cloned into pET28a(+) (Novagen) with their initiator methionine codons immediately downstream of the BamHI site. All expression plasmids were confirmed by bidirectional sequence analysis of the full MAPK coding sequence.
Expression and Purification of MAPKs—Active MAPKs were produced as GST fusion proteins by co-expression in bacteria with activating kinases similar to Khokhlatchev et al. (37) with the following modifications. ERK2 was co-expressed with the constitutively active MKK1ΔN3 mutant. Active p38α, p38γ, and p38δ were generated by co-expression with MKK6 S207E/T211E. Bacterially produced p38β (a gift from Stefan Knapp, Oxford University Structural Genomics Consortium) has constitutive activity and was expressed alone. Active His-JNK2 was produced by co-expression in bacteria with MEKK1 and MKK4 and purified as described previously (37). Full procedures for protein expression and purification are provided in the supplemental material.
Peptide Library Screening—Phosphorylation site specificity was determined using a positional scanning peptide library as described previously (38). For the DEF site library, peptides were reacted in 384-well plates in 10-μl reactions with 10 μm peptide in 20 mm HEPES, pH 7.4, 10 mm MgCl2, 1 mm dithiothreitol, 0.1% Tween 20, 100 μm ATP, 0.25 μCi of [γ-33P]ATP, and 20 ng of kinase for 2 h at 30 °C. A strip of 24 slotted capillary pins (VP Scientific) was used to spot 1-μl aliquots from each reaction onto streptavidin-coated filters (SAM2® biotin capture membrane, Promega). Reactions were quenched by immediately submerging the membrane in 0.1% SDS in TBS (10 mm Tris, pH 7.5, 140 mm NaCl). Additional washes were performed for 5 min each to remove unincorporated radiolabel (once in 0.1% SDS/TBS, twice in 2 m NaCl, and once in 1% H3PO4 in 2 m NaCl). After drying, the membrane was exposed to a phosphor storage screen for 1–3 days and scanned on a PhosphorImager (GE Healthcare). Relative phosphorylation rates for each of the amino acid substitutions were quantified by optical densitometry (using ImageQuant 5.0) as the ratio of the individual signal intensity to the average signal intensity for all 19 amino acid substitutions at that DEF site position (P1–P4). Pilot experiments with DEF site peptides were carried out in a similar manner with reactions carried out in microcentrifuge tubes.
Consensus Peptide Kinase Assays—The phosphorylation rates of MAPKs for individual consensus peptides was determined by P81 phosphocellulose filter binding assay essentially as described (39) (the detailed procedure is described in the supplemental material). To normalize docking site-independent activity between individual MAPK preparations, we determined the rate of phosphorylation of 100 μm MAPKtide for each kinase. All p38 preparations (WT and DEF site interaction pocket mutants) were re-assayed against 100 μm MAPKtide during each experiment to ensure proper normalization.
Elk-1 Phosphorylation Assays—Phosphorylation of Elk-1 and its docking site mutants in vitro was studied using a fragment containing the MAPK phosphorylation sites (residues 307–428) expressed in bacteria as a GST fusion protein (7). Reactions contained 20 μg/ml GST-Elk-1 and were carried out under the same conditions as the peptide phosphorylation assays. Reactions were split and subjected to SDS-PAGE followed either by immunoblotting with antibodies against Elk-1 phosphoserine 383 and phospho-ERK (both from Cell Signaling) or by Coomassie staining and autoradiography to detect total protein phosphorylation.
For expression as a GST fusion protein in mammalian cells, the sequence encoding the same fragment of Elk-1 was shuttled into the plasmid pEBG2. NIH3T3 cells were transfected using Lipofectamine PLUS (Invitrogen) as instructed by the manufacturer. At 6 h following transfection, cells were washed once with serum-free media, changed into media containing 0.1% calf serum, and incubated for 18 h. Cells were preincubated with either 5 μm U0126 or DMSO carrier for 30 min prior to stimulation with media containing 10% serum plus U0126 or DMSO as indicated. After 15 min, cells were washed once with ice-cold phosphate-buffered saline and then extracted into lysis buffer (1% Triton X-100 in 20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 2 μg/ml pepstatin A, and 10 μg/ml aprotinin). Lysates were cleared by centrifugation (10 min at 16,000 × g) and subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies.
Docking Studies—The DEF site-MAPK interactions were modeled using the automatic molecular docking program Autodock 4 (40). AutoDockTools, the graphical interface to Autodock 4, was used to set up the docking and subsequently analyze the clustering after completion of docking. Throughout the docking experiment a rigid receptor and flexible ligand were assumed. The published structure of the active form of ERK2 (Protein Data Bank code 2ERK) was used as the receptor, and a capped pentapeptide sequence, acetyl-SFQFP-amide, was used as the ligand. A grid map of 50 × 50 × 50 points with a spacing of 0.375 Å was centered on the previously identified hydrophobic pocket on the ERK2 surface (18). We used the Lamarckian genetic algorithm, the default search method of Autodock 4, which combines the traditional genetic algorithm with a local search capacity. The number of runs was set to 256. The remaining docking parameters were set to the default values in AutoDockTools as follows: the number of individuals in population was set to 150; the maximum number of energy evaluations was 2,500,000; and the maximum number of generations was 27,000. The program automatically grouped potential receptor-ligand complex conformations into clusters based on their r.m.s.d. from one another, using the default threshold (2.0 Å r.m.s.d.).
RESULTS
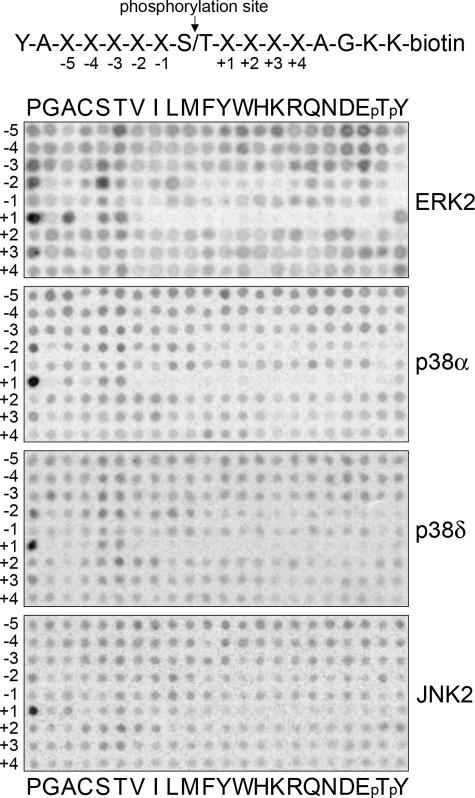
MAPKs Have a Common Phosphorylation Site Motif—All MAPKs are regarded as proline-directed kinases, in that they have a major preference for Pro at the +1 position at their phosphorylation sites. However, not all members of the family have been fully characterized with respect to active site-mediated specificity, and differences in selectivity at other positions near the phosphorylation site could potentially distinguish MAPKs from one another. We therefore first addressed whether targeting of distinct protein substrates by different MAPKs might be due to differences in their respective phosphorylation site motifs. We screened a positional scanning peptide library that systematically substitutes each of the 20 amino acid residues plus phosphothreonine and phosphotyrosine at 9 positions surrounding a central phosphoacceptor (3). Using this approach we found no significant differences among any of four representative MAPKs (ERK2, p38α, p38δ, and JNK2) in their phosphorylation site consensus sequences (Fig. 1). Our results confirm the phosphorylation site consensus motif previously reported for ERK2 and p38α, with a Ser or Thr phosphoacceptor followed by an obligate Pro at the +1 position and a less stringent preference for Pro at -2 (3, 4). Most other residues surrounding the phosphoacceptor are well tolerated with the exception of Pro at the -1 position and aromatic residues at the -2 position. This simple motif is common in eukaryotic proteomes and overlaps with phosphorylation site motifs of more distantly related kinases (e.g. cyclin-dependent kinases) (41). Therefore, it appears unlikely that the phosphorylation site motif contributes to the unique target specificities of the various MAPK isoforms.
FIGURE 1.
MAPKs share a common consensus phosphorylation site specificity. Preferred amino acids flanking the Ser or Thr phosphoacceptor were identified for four representative MAPKs by measuring the relative rate of phosphorylation of 198 members within a positional scanning peptide library (sequence shown at top). Kinase reactions were performed in parallel in solution in the presence of [γ-33P]ATP. Biotin-labeled peptides were then captured on a streptavidin membrane, and the relative rate of phosphorylation was assessed by phosphorimaging. ERK2, p38α, p38δ, and JNK2 share a common active site motif with a requirement for Pro at the +1 position and a preference for Pro at -2. pT, phosphothreonine; pY, phosphotyrosine.
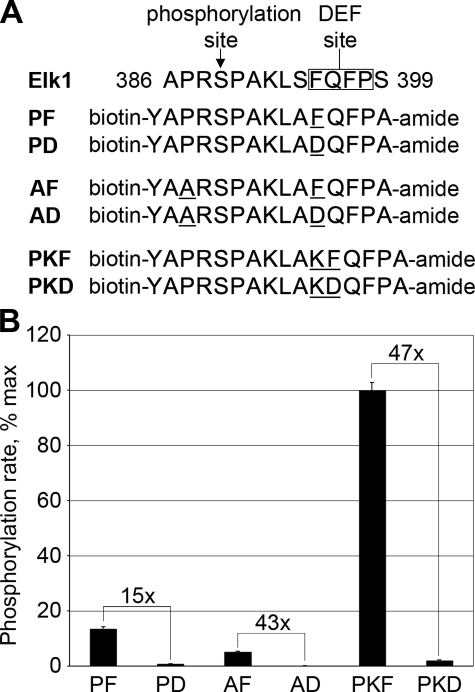
Identification of MAPK Docking Motifs—We next turned our attention to interactions outside of the kinase active site, and asked whether targeting by specific MAPKs might involve recognition of distinct docking site sequences. The close proximity of the DEF site interacting pocket to the active site cleft on ERK2 means that both a MAPK phosphorylation site and a DEF site docking sequence can be encoded in a single extended peptide (7, 23). We therefore developed a substrate optimized for DEF site-dependent phosphorylation using peptides based on amino acids 386–399 of Elk-1 (Fig. 2A). This region encompasses a single ERK phosphorylation site and a functional DEF site (23). In these peptides, we independently varied the sequence surrounding the phosphoacceptor and the spacing between the phosphorylation site and DEF docking site. In each case, we compared the phosphorylation rate between peptides containing an intact DEF site (FQFP) and those with a point substitution within the site (DQFP). To ensure that ERK phosphorylation was directed exclusively to its endogenous target site (Ser-389), other potential phosphoacceptors (Ser-394 and Ser-399) were changed to Ala. Initial rates of phosphorylation of these peptides were measured at subsaturating concentrations (10 μm). Disruption of the DEF site by changing FQFP to DQFP decreased ERK phosphorylation of the peptide closest to the native Elk-1 sequence (peptide PF) by ∼15-fold (Fig. 2B). Substitution of the nonessential Pro residue at the -2 position with Ala slowed the phosphorylation rate by slightly less than 3-fold (compare the phosphorylation rates of PF and AF). However, disruption of the DEF site in the context of this modification further decreased the phosphorylation rate by 43-fold (AF versus AD). Alternatively, inserting an additional residue between the phosphoacceptor Ser and the DEF site both increased the peptide phosphorylation rate by 7-fold (PF versus PKF) and provided the maximal DEF site dependence (a 47-fold difference between PKF and PKD).
FIGURE 2.
Optimization of peptide substrates containing DEF sites. A, pairs of DEF site peptides based on amino acids 386–399 of human Elk-1 that were evaluated for phosphorylation by ERK2. Peptides contain the endogenous ERK phosphorylation site (Ser-389) and either an intact DEF site (FQFP) or a nonfunctional DEF site with charged residue in position P1 (DQFP). B, extent of phosphorylation was measured by radiolabel incorporation from [γ-33P]ATP. The increase in phosphorylation rate afforded by an intact DEF site is potentiated by incorporating a suboptimal phosphorylation site sequence (peptide AF) or by separating the phosphorylation site from the DEF site by one additional residue (peptide PKF). Error bars reflect the standard deviation of three separate experiments.
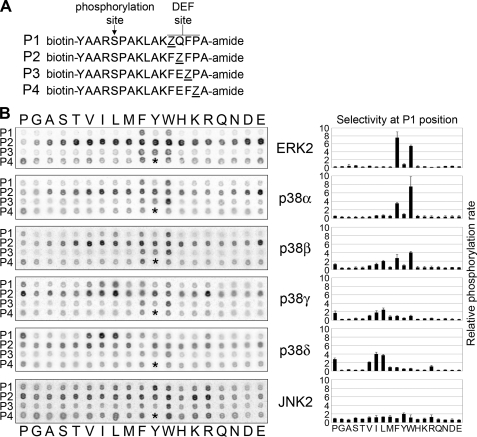
These results suggested that the increased phosphorylation rate afforded by a functional DEF site is enhanced in peptide substrates bearing either a suboptimal phosphorylation site sequence or having a DEF site that begins seven residues downstream of the phosphorylated Ser residue. Both of these features were incorporated into a positional scanning peptide library designed to determine consensus sequences at the DEF site (Fig. 3A). The DEF peptide library consists of 73 biotinylated peptides, in which each of the four positions within the FQFP sequence (designated P1 through P4) is substituted one at a time with the remaining 18 possible amino acids (excluding cysteine). Peptides were arrayed in a 384-well plate and subjected simultaneously to radiolabel kinase assays with a given MAPK. At the end of the reaction, peptides were spotted onto a streptavidin membrane and washed to remove unincorporated radiolabel. Subsequently, the extent of phosphorylation of each peptide was quantified (Fig. 3B).
FIGURE 3.
Identification of MAPK DEF site motifs using a novel positional scanning peptide library. A, peptide library incorporates 19 amino acid substitutions (Z) at each of the four positions within the DEF site (P1–P4). B, peptides were subjected to phosphorylation by the MAPK in the presence of [γ-33P]ATP followed by capture on streptavidin membrane and exposure to a phosphor screen (left panel). p38 MAPK isoforms displayed unique selectivity at the P1 position. The p38α selectivity is similar to ERK2, with a preference for aromatic residues at P1, although p38δ favors aliphatic residues at P1. p38β and p38γ displayed intermediate P1 selectivity with p38β more similar to p38α and p38γ more similar to p38δ. JNK2 did not display a strong sequence preference in the DEF site (<2-fold difference for any substitution). The P4 Tyr peptide was not included because of failed synthesis (space indicated with an asterisk). Histograms depict the quantified radiolabel incorporation for the P1 position. The selectivity value is calculated as the level of phosphorylation of a given peptide divided by the average level of phosphorylation for all 19 peptides. Error bars represent the standard deviation from three separate experiments.
We first screened the DEF peptide library with ERK2, which has the best characterized DEF motif (Fig. 3B). The peptide library yielded a consensus sequence for the ERK2 DEF site characterized by a strong preference for aromatic residues at positions P1 (Phe and Trp) and P3 (Phe, Tyr, and Trp). This motif closely matches functional DEF sites in ERK protein substrates, and confirms previous reports of the importance of the aromatic residues within the Elk-1 DEF site (7, 23). Although Pro does appear to be the optimal residue at the P4 position, substitution of other residues at this position has a relatively modest effect on the rate of phosphorylation. This observation is consistent with the presence of other residues at this position in DEF sites found in PDE4 and BimEL (27, 28).
Although the results with ERK2 were consistent with its known protein substrates, we were interested in determining whether divergent DEF site sequences might be recognized by other MAPKs. We thus screened the DEF peptide library with a panel of MAPKs, including all four p38 isoforms and JNK2 (Fig. 3B). We found that p38α has a DEF site motif similar to ERK2, being highly selective for aromatic residues at the P1 and P3 positions, albeit with a particular preference for Trp at both sites. In contrast, p38δ strikingly preferred aliphatic residues (Ile, Leu, Pro, and Val) over aromatic residues at the P1 position, while having a similar aromatic preference at P3. We were surprised to find such a dramatic difference between members of a single subfamily of MAPKs. However, the p38 subfamily can be further subdivided based on sequence homology into two groups, composed of p38α/β and p38γ/δ. Accordingly, we found that p38β DEF site specificity most closely resembled p38α, whereas p38γ specificity resembled p38δ, although these other p38 isoforms are more tolerant of both aliphatic and aromatic residues at the P1 position. JNK2 displayed no selectivity and phosphorylated all of the peptides in the library with roughly equal efficiency, suggesting that it does not use this particular docking site. Preferred residues at all four positions are summarized in Table 1 (histograms of the full library screening results are shown in supplemental Fig. 1).
TABLE 1.
DEF site motifs for MAPKs Residues positively selected at each position within the DEF site library are shown for each MAPK. Selectivity values in parentheses were calculated as described in the legend to Fig. 3. Residues with selectivity values greater than 1.5 are shown, and positions where no residues were selected to such an extent are indicated with an X. Residues with values greater than 3.0 are shown in boldface.
|
Kinase
|
Position
|
|||||||
|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | P4 | |||||
| ERK2 | Phe | (7.6) | Trp | (1.7) | Phe | (4.7) | Pro | (2.4) |
| Trp | (5.5) | Glu | (1.6) | Trp | (4.5) | Phe | (2.2) | |
| Val | (1.5) | Tyr | (4.3) | Trp | (1.7) | |||
| Glu | (1.6) | |||||||
| p38α | Trp | (7.5) | Glu | (2.1) | Trp | (8.8) | Pro | (2.8) |
| Phe | (3.5) | Val | (1.6) | Tyr | (2.6) | Trp | (1.8) | |
| Phe | (2.1) | Phe | (1.6) | |||||
| p38β | Trp | (4.0) | Ile | (1.8) | Trp | (5.3) | Phe | (2.3) |
| Phe | (2.7) | Glu | (1.8) | Phe | (2.8) | Trp | (1.8) | |
| Leu | (2.0) | Val | (1.8) | Tyr | (2.2) | |||
| Phe | (1.6) | |||||||
| p38γ | Leu | (2.5) | Leu | (1.7) | Trp | (3.5) | X | |
| Ile | (1.8) | Phe | (1.9) | |||||
| Pro | (1.7) | |||||||
| p38δ | Ile | (4.0) | Val | (1.7) | Trp | (3.7) | Pro | (1.9) |
| Leu | (3.7) | Phe | (2.6) | Trp | (1.8) | |||
| Pro | (2.8) | Leu | (1.6) | Phe | (1.7) | |||
| Val | (2.1) | |||||||
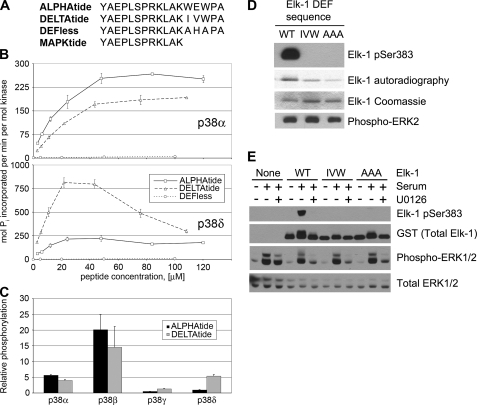
Consensus DEF Site Peptide Substrates Display Selectivity for Specific MAPKs—Short peptide substrates for MAPKs tend to have very high Km values and do not distinguish between different members of the family. To generate efficient and specific MAPK substrates, we evaluated individual consensus peptides for p38α and p38δ, referred to here as ALPHAtide and DELTAtide, respectively (Fig. 4A). These substrates share a common 12-residue N-terminal sequence with an optimized MAPK phosphorylation site (based on the data from Fig. 1), followed by a DEF site sequence predicted to be optimal for its respective kinase. At subsaturating concentrations (below 50 μm) each of these peptides was phosphorylated by its respective kinase over an order of magnitude more efficiently than either a truncated peptide lacking a DEF site (MAPKtide, data not shown) or a peptide with alanine substitutions at P1 and P3 of the DEF site (DEFless, Fig. 4B). At all concentrations tested, p38α and p38δ phosphorylated their respective consensus peptides most efficiently. At higher concentrations (40–100 μm), reaction rates began to decrease, suggestive of substrate inhibition, and we were unable to establish Vmax and Km values for p38δ. We were able to determine these values for p38α phosphorylation of ALPHAtide (Km = 24 μm; Vmax = 366 pmol of Pi/s/nmol kinase), DELTAtide (Km = 49 μm; Vmax = 354 pmol Pi/s/nmol kinase), and DEFless (Km = 1740 μm; Vmax = 45 pmol Pi/s/nmol kinase). This comparison suggests that the enhancement of phosphorylation by p38α afforded by an intact DEF site is because of favorable changes in both Vmax and Km values. Rates of phosphorylation of ALPHAtide and DELTAtide were measured for all p38 isoforms at a single subsaturating concentration (5 μm). For each isoform, the relative rates of phosphorylation of the two peptides reflect their P1 specificities observed in the DEF peptide library (Fig. 4C).
FIGURE 4.
DEF site-dependent phosphorylation of consensus peptide and protein substrates. A, sequences of consensus peptides used in this study. B, phosphorylation rates of consensus peptides by p38α and p38δ were assayed at concentrations ranging from 2.5 to 120 μm. The two p38 isoforms phosphorylate their respective consensus peptides at a faster rate at all concentrations tested. A peptide with Ala substitutions in positions P1 and P3 in the DEF site (DEFless) was phosphorylated inefficiently by both kinases. C, rate of phosphorylation of ALPHAtide or DELTAtide at 5 μm concentration by all four p38 isoforms is shown (relative to their phosphorylation rate of MAPKtide at 100 μm). Selectivity for the two consensus peptides reflects their specificity at the P1 position observed in the DEF peptide library, with p38β and p38γ having intermediate selectivity between p38α and p38δ (cf. Fig. 3B). Error bars reflect the standard deviation of three separate experiments. D, fragment of human Elk-1 containing its MAPK phosphorylation and docking sites (residues 307–428, WT), and the same fragment with its DEF sequence mutated from FQF to either AAA or IVW as indicated, was phosphorylated in vitro with ERK2. Site-specific phosphorylation was determined by immunoblotting with a phosphospecific antibody against phospho-Ser-383, and total phosphorylation was determined by autoradiography. E, NIH3T3 cells were transfected with either an empty vector or plasmids encoding the same fragment of Elk-1 used in D and the indicated mutants. Cells were serum-starved overnight and either harvested without further treatment or treated with media containing 10% calf serum to stimulate ERK activity. Cells were pretreated where indicated with the MEK1/2 inhibitor U0126 (5 μm) to block ERK activation. Shown are immunoblots of cell lysates using antibodies to detect phosphorylated and total Elk-1 and ERK1/2.
An Optimal DEF Site Sequence Is Essential for Efficient Phosphorylation of Elk-1 by ERK2—We also tested the requirement for specific DEF site motifs for efficient phosphorylation of the Elk-1 transcription factor by ERK2. As shown previously, mutation of the P1–P3 DEF site motif residues in Elk-1 from FQF to AAA specifically impairs phosphorylation at Ser-383 but has only a modest effect on the overall level of phosphorylation (Fig. 4D) (7, 18). We found that substitution with the p38δ consensus sequence (IVW) also inhibits phosphorylation by ERK2 specifically at Ser-383 in vitro. Furthermore, when expressed in NIH3T3 cells, the optimal DEF site sequence was required for ERK-dependent phosphorylation of Elk-1 at Ser-383, as neither the AAA nor the IVW mutant was detectably phosphorylated at that site. These results suggest that specific DEF site sequences are required for phosphorylation of MAPK targets both in vitro and in cultured cells.
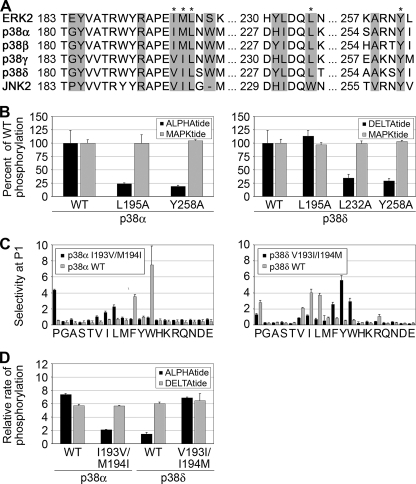
Structural Determinants of MAPK DEF Site Specificity—Sequence alignments of ERK2 and the p38 isoforms show a high degree of conservation of residues within the hydrophobic DEF interacting pocket defined by Lee et al. (18) (Fig. 5A). Alanine substitution of residues lining the hydrophobic pocket selectively impairs phosphorylation of substrates with DEF sites by ERK2 in vitro and in vivo (18, 42). We generated point mutants of a subset of these residues in p38α and p38δ and characterized their effects on phosphorylation of DEF site containing peptides. Alanine substitutions at Leu-195 or Tyr-258 in p38α significantly attenuated phosphorylation of ALPHAtide relative to MAPKtide (Fig. 5B). For p38δ, mutation of Leu-232 or Tyr-258 had a similar attenuating effect, although the L195A mutant displayed a phosphorylation rate equivalent to the wild-type (WT) kinase. We further characterized the effects of these mutations by screening the full DEF peptide library (results are shown in supplemental Fig. 2). Mutation of Leu-195 in p38α led to a loss of selectivity at the P1 position but had little effect at P3. The p38δ L195A mutant displayed a slightly increased tolerance for aromatic groups at P1 but appeared similar to the WT kinase overall. Conversely, Ala substitution at Leu-232 in p38δ abolished selectivity at the P3 position while maintaining wild-type selectivity for aliphatic side chains at P1. The Y258A mutation in both p38α and p38δ displayed reduced selectivity at P1 and P3, suggesting an overall disruption of the hydrophobic pocket.
FIGURE 5.
Mutagenesis of the DEF site interaction pocket. A, primary sequence alignment of the residues surrounding the DEF site interaction pocket (highlighted residues) of ERK2, p38 isoforms, and JNK2 reveals a high degree of sequence conservation. Amino acids that were mutated in this study are indicated with an asterisk. B, single amino acid substitutions to Ala were made in p38α at Leu-195 and Tyr-258 and in p38δ at Leu-195, Leu-232, and Tyr-258. With the exception of p38δ L195A, the phosphorylation rates of DEF site consensus peptides were attenuated ∼4-fold relative to the WT kinase. Results from screening the DEF peptide library with these mutants are shown in supplemental Fig. 2. C, specificity transposition mutants (p38α I193V/M194I and p38δ V193I/I194M) were screened against the DEF peptide library. Mutation of these residues interchanges the selectivity for aromatic versus aliphatic side chains at the P1 position between p38α and p38δ. D, relative rates of phosphorylation of ALPHAtide and DELTAtide consensus peptides by the transposition mutants mirror those for the WT p38 isoforms. Error bars reflect the standard deviation of three separate experiments.
Interestingly, the residues corresponding to Ile-196 and Met-197 of ERK2 are conserved in p38α and p38β but are substituted with Val and Ile, respectively, in both p38γ and p38δ. We hypothesized that these surface-exposed hydrophobic residues might be responsible for the differences observed in P1 selectivity between p38α/β and p38γ/δ. To test this hypothesis, we mutated Ile-193 and Met-194 in p38α to Val and Ile, respectively, and made the converse V193I and I194M mutations in p38δ. Analysis with the DEF peptide library revealed that p38α I193V/M194I displayed p38δ-like selectivity at P1, with a preference for aliphatic side chains, including Pro, whereas aromatic residues were no longer tolerated (Fig. 5C). Similarly, p38δ V193I/I194M exhibited a p38α-like profile, a preference for aromatic residues, and a decreased tolerance of aliphatic amino acids at P1. The transposed specificity of the two double mutants was affirmed by measuring phosphorylation rates of the consensus ALPHAtide and DELTAtide substrates (Fig. 5D). Although wild-type p38α efficiently phosphorylates both peptides, with a slight preference for ALPHAtide, the I193V/M194I mutant is highly specific for DELTAtide. Likewise, the V193I/I194M mutation confers upon p38δ the ability to phosphorylate ALPHAtide.
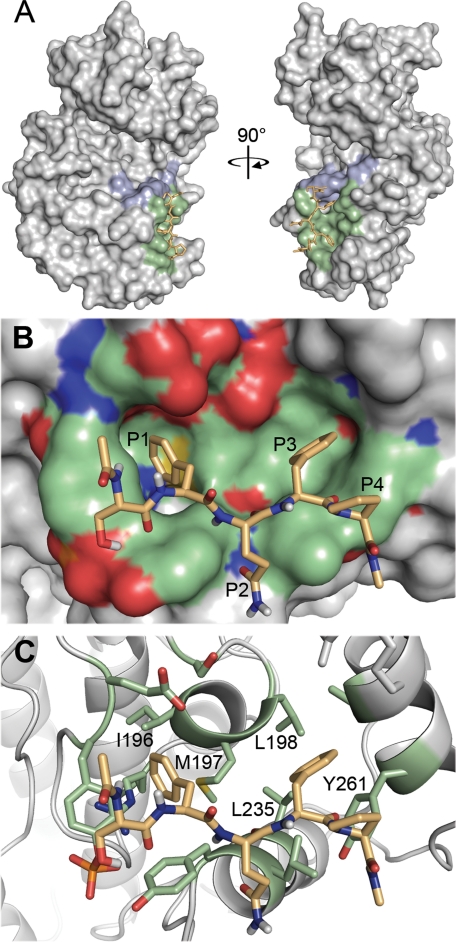
Collectively these mutagenesis experiments support a mode of binding in which the P1 residue contacts residues analogous to Ile-196, Met-197, and Leu-198 of ERK2, whereas the P3 residue makes contact with Leu-235. Because structures of active p38α and p38δ have not been solved, we modeled docking interactions of a capped pentapeptide DEF site ligand (SFQFP) onto the DEF interaction site of the published structure of diphosphorylated ERK2 (22). Of 256 docking runs with random initial conditions, over one-half (139) of the models converged on the lowest energy cluster, with root mean squared deviation (r.m.s.d.) values of less than 2 Å relative to each other. In contrast to the docking model proposed by Lee et al. (18), which seeks to accommodate all four DEF site residues into the hydrophobic pocket, this revised model shows the P1 and P3 Phe residues buried deeply within the pocket, whereas the P4 Pro is more exposed to solvent (Fig. 6). This mode of interaction is consistent with our peptide screening data, which indicates strong selectivity for the P1 and P3 residues with a less stringent preference for the Pro at the P4 position. Based on the structural model, we can rationalize the distinct DEF site specificities observed for the different p38 isoforms. Within the hydrophobic pocket, ERK2 and p38α differ by only two residues, Tyr-231 and Leu-232 of ERK2, which are present at the walls of the pocket and correspond to His-228 and Ile-229 of p38α. As neither residue projects deeply into the pocket, p38α would be anticipated to have DEF site specificity similar to ERK2. By contrast, compared with Met-197 of ERK2, the branched side chain of Ile-194 in p38γ/δ is likely to restrict the depth of the pocket in the region that interacts with the P1 residue. This substitution thus reduces the capacity of the pocket to accommodate large aromatic side chains, while still allowing favorable interaction with smaller aliphatic residues.
FIGURE 6.
Proposed model for DEF site-MAPK interactions. Docking of a capped pentapeptide ligand representing an aromatic-X-aromatic DEF sequence (acetyl-SFQFP-amide) to the active form of ERK2 (Protein Data Bank code 2ERK) was modeled using AutoDock. A, docking of the peptide ligand (orange) was restricted to the previously identified hydrophobic pocket on the ERK2 surface (green). Residues constituting the P +1 selectivity pocket are highlighted in blue. B, close up view of the docking model (rotated 90° counterclockwise from A). Consistent with peptide screening data showing strongest selectivity at the P1 and P3 positions, the two Phe residues extend deeply into the ERK2 hydrophobic pocket, whereas the Pro residue at P4 makes less intimate contact. C, removal of the surface detail from B. Residues lining the hydrophobic pocket are shown in stick representation. The P1 Phe residue is in closest proximity to Met-197, consistent with the change in specificity observed when the analogous residue in p38α is mutated to Ile.
DISCUSSION
Protein kinase specificity is determined in part through recognition of consensus sequences immediately flanking the site of phosphorylation by the kinase active site. Accordingly, much effort has been directed toward identifying these phosphorylation site sequence motifs by peptide library screening (3, 41, 43–46). However, short amino acid sequence motifs can occur at high frequency in proteomes. Furthermore, as we have shown here for MAPKs, related kinases often utilize similar motifs. Active site specificity alone is therefore generally insufficient to direct a particular kinase to a specific protein substrate.
Protein kinase substrate affinity and specificity is enhanced by interactions that occur outside of the active site. Some protein kinases possess relatively high affinity protein interaction domains, such as Src homology 2 and 3, C2, and polobox domains, among others (47). These domains can play a role in both subcellular localization of the kinase and substrate recruitment through direct interactions. The strong affinities of these protein interaction domains for peptide ligands (KD < 1 μm) make them amenable to characterization by peptide library analysis of direct binding interactions (48–51). The relatively weak binding affinities of docking interactions within the kinase catalytic domain (KD > 10 μm), however, preclude the use of such an approach for analyzing docking specificity. Alternatively, methods that characterize the phosphorylation site specificity of protein kinases typically involve analysis of low affinity peptide substrates. By using peptide substrates with a phosphoacceptor and a tethered docking site, we have expanded the repertoire of peptide screening methods to include relatively weak docking interactions within the kinase catalytic domain. Efficient peptide substrates for MAPKs have also been generated by incorporating D-site peptides (10), suggesting that this approach will be generally useful for identifying docking sequence motifs utilized by these kinases.
We have shown that most MAPK family members can interact with DEF sites and that they exhibit distinct sequence preferences for DEF docking sites. To date, DEF sites have been identified almost exclusively in ERK substrates, with a single report suggesting that p38α also can use DEF sites. The idea that DEF site interactions are restricted to these two MAPKs likely arises from the fact that the other p38 isoforms, in particular p38γ and p38δ, are not specific for optimal FXF sequences. These isoforms constitute a separate group within the p38 subfamily, with distinct inhibitor sensitivity, unique protein substrates, and discrete biological functions (52). It is likely that cryptic DEF sites exist within specific substrates for these isoforms that contribute to their selective phosphorylation. In contrast to the p38 family members, we observed an almost complete lack of specificity by JNK2 for peptides within the DEF library. This could mean that JNK2 does not use this particular docking site, and instead relies on other docking interactions to achieve substrate specificity. Alternatively, it is possible that JNK2 has a DEF site sequence specificity that is so divergent from the sequence found in Elk-1 that it does not interact appreciably with any of the peptides within the library that we used. Interestingly, the residue corresponding to Leu-235 in the DEF interacting pocket of ERK2 is conserved across all p38 isoforms but is a Trp residue in JNK isoforms (see Fig. 5A). Substitution of this much larger residue could occlude the pocket in JNK, or simply perturb the overall geometry of the pocket so that it can no longer accommodate the aromatic residues in the DEF library peptides. It is also possible that this region of JNKs binds to docking sites on substrates using an alternative binding mode. For example, the analogous region of glycogen synthase kinase 3β (GSK3β), a related kinase, interacts with peptides that are in a helical rather than extended conformation (53, 54).
A general function of docking sites is to enhance phosphorylation through substrate recruitment, and the affinity of MAPKs for their substrates appears to derive entirely from docking rather than phosphorylation site interactions (8, 9). Accordingly, DEF site interactions have been reported to lower the Km value for phosphorylation of peptides by ERK and effect modest changes in Vmax values (7, 23). We show here for p38α that the presence of a DEF site dramatically lowers the Km value and increases Vmax values. This discrepancy could mean that engagement of the DEF site has different mechanistic consequences for ERK and p38α or, alternatively, that effects on Vmax depend on the substrate used. The hydrophobic pocket that interacts with DEF sites is located very near the kinase activation loop, and it is in fact occluded by the activation loop in the unphosphorylated, inactive form of ERK2. Substrate docking to the hydrophobic pocket would be expected to stabilize the activation loop in its active conformation, potentially increasing Vmax values. Alternatively, DEF site engagement could help to align the substrate such that its phosphorylation site is oriented in a manner that promotes phosphorylation. Other docking interactions have also been implicated in allosteric control of MAPK activity but by distinct mechanisms. D-site binding to ERK, JNK, and p38α influences their activation loop conformations through long range communication, potentially affecting accessibility to MKKs and MAPK phosphatases (17–20, 55). Binding of the Ste5 scaffold to the yeast MAPK Fus3 occurs through two regions of the kinase, including the D-site interacting groove, and induces partial autophosphorylation and activation of the kinase (56). Impacting function through mechanisms beyond substrate recruitment is a recurring theme for MAPK docking interactions and may turn out to be a more general feature of interactions with kinase catalytic domains.
Another important function of docking sites is to augment the specificity of kinase-substrate interactions, and understanding sequence requirements for interaction with docking sites can help to identify new kinase substrates. Approximately 30% of human proteins in the Swiss-Prot data base have at least one occurrence of the optimal MAPK phosphorylation site motif (PX(S/T)P), and by itself the phosphorylation site motif is insufficient to direct specific targeting. Furthermore, searching protein data bases for MAPK consensus sequences has failed to uncover new substrates or even significantly enrich for known substrates (4). The presence of docking sites can facilitate substrate targeting by specific kinases, and the characterization of docking site motifs has predictive value to identify novel substrates by computational scanning. This principle was demonstrated recently for the yeast polo-like kinase Cdc5, where a data base search for both the optimal phosphorylation site and docking site sequences led to the identification of several novel substrates (57). As a relatively small number (306) of human proteins have an ERK/p38α consensus DEF sequence ((F/W)X(F/Y/W)) within 20 residues downstream of the optimal MAPK phosphorylation site, this approach is anticipated to be fruitful for identifying new MAPK substrates as well.
Therapeutic targeting of MAPKs is currently of great clinical interest. Inhibitors of the ERK, p38, and JNK pathways have all reached clinical trials for diverse indications, including cancer, inflammation, and neurodegenerative disease (52, 58, 59). Efficient kinase-specific peptide substrates such as those described here are adaptable to high throughput assays and are thus valuable tools for drug discovery. Incorporation of these peptides into fluorescent biosensors could also provide a means for cell-based screening (60–62). In addition, docking grooves offer an alternative to the ATP-binding site as a strategy for targeting with small molecule inhibitors (63). The identification of differences in docking specificity between the various MAPKs not only illuminates fundamental mechanisms of substrate discrimination but also suggests an avenue for designing isoform-specific inhibitors of potential clinical value.
Supplementary Material
Acknowledgments
We thank Jeff Peterson and Anton Bennett for providing helpful comments on the manuscript. We gratefully acknowledge technical assistance from Ved Desai. We thank Stefan Knapp for providing a sample of p38β protein and Natalie Ahn for sharing coordinates of the ERK2-DEF site peptide model and plasmids carrying the constitutively active MKK1 mutant, WT Elk-1, and mutant Elk-1.
This work was supported, in whole or in part, by National Institutes of Health Grants GM079498 (to B. E. T.) and GM59802 (to K. N. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods and Figs. 1 and 2.
Footnotes
The abbreviations used are: MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; WT, wild type; GST, glutathione S-transferase; r.m.s.d., root mean square deviation; MKK, MAPK kinase.
References
- 1.Raman, M., Chen, W., and Cobb, M. H. (2007) Oncogene 26 3100-3112 [DOI] [PubMed] [Google Scholar]
- 2.Songyang, Z., Lu, K. P., Kwon, Y. T., Tsai, L. H., Filhol, O., Cochet, C., Brickey, D. A., Soderling, T. R., Bartleson, C., Graves, D. J., DeMaggio, A. J., Hoekstra, M. F., Blenis, J., Hunter, T., and Cantley, L. C. (1996) Mol. Cell. Biol. 16 6486-6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutti, J. E., Jarrell, E. T., Chang, J. D., Abbott, D. W., Storz, P., Toker, A., Cantley, L. C., and Turk, B. E. (2004) Nat. Meth. 1 27-29 [DOI] [PubMed] [Google Scholar]
- 4.Manke, I. A., Nguyen, A., Lim, D., Stewart, M. Q., Elia, A. E., and Yaffe, M. B. (2005) Mol. Cell 17 37-48 [DOI] [PubMed] [Google Scholar]
- 5.Kallunki, T., Deng, T., Hibi, M., and Karin, M. (1996) Cell 87 929-939 [DOI] [PubMed] [Google Scholar]
- 6.Hawkins, J., Zheng, S., Frantz, B., and LoGrasso, P. (2000) Arch. Biochem. Biophys. 382 310-313 [DOI] [PubMed] [Google Scholar]
- 7.Fantz, D. A., Jacobs, D., Glossip, D., and Kornfeld, K. (2001) J. Biol. Chem. 276 27256-27265 [DOI] [PubMed] [Google Scholar]
- 8.Rainey, M. A., Callaway, K., Barnes, R., Wilson, B., and Dalby, K. N. (2005) J. Am. Chem. Soc. 127 10494-10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callaway, K. A., Rainey, M. A., Riggs, A. F., Abramczyk, O., and Dalby, K. N. (2006) Biochemistry 45 13719-13733 [DOI] [PubMed] [Google Scholar]
- 10.Fernandes, N., Bailey, D. E., Vanvranken, D. L., and Allbritton, N. L. (2007) ACS Chem. Biol. 2 665-673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knebel, A., Morrice, N., and Cohen, P. (2001) EMBO J. 20 4360-4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, S. H., Galanis, A., and Sharrocks, A. D. (1999) Mol. Cell. Biol. 19 4028-4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao, M., New, L., Kravchenko, V. V., Kato, Y., Gram, H., di Padova, F., Olson, E. N., Ulevitch, R. J., and Han, J. (1999) Mol. Cell. Biol. 19 21-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biondi, R. M., and Nebreda, A. R. (2003) Biochem. J. 372 1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsmith, E. J., Akella, R., Min, X., Zhou, T., and Humphreys, J. M. (2007) Chem. Rev. 107 5065-5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanoue, T., Adachi, M., Moriguchi, T., and Nishida, E. (2000) Nat. Cell Biol. 2 110-116 [DOI] [PubMed] [Google Scholar]
- 17.Chang, C. I., Xu, B. E., Akella, R., Cobb, M. H., and Goldsmith, E. J. (2002) Mol. Cell 9 1241-1249 [DOI] [PubMed] [Google Scholar]
- 18.Lee, T., Hoofnagle, A. N., Kabuyama, Y., Stroud, J., Min, X., Goldsmith, E. J., Chen, L., Resing, K. A., and Ahn, N. G. (2004) Mol. Cell 14 43-55 [DOI] [PubMed] [Google Scholar]
- 19.Zhou, T., Sun, L., Humphreys, J., and Goldsmith, E. J. (2006) Structure (Lond.) 14 1011-1019 [DOI] [PubMed] [Google Scholar]
- 20.Liu, S., Sun, J. P., Zhou, B., and Zhang, Z. Y. (2006) Proc. Natl. Acad. Sci. U. S. A. 281 38834-38844 [Google Scholar]
- 21.Nolen, B., Taylor, S., and Ghosh, G. (2004) Mol. Cell 15 661-675 [DOI] [PubMed] [Google Scholar]
- 22.Canagarajah, B. J., Khokhlatchev, A., Cobb, M. H., and Goldsmith, E. J. (1997) Cell 90 859-869 [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, D., Glossip, D., Xing, H., Muslin, A. J., and Kornfeld, K. (1999) Genes Dev. 13 163-175 [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy, L. O., Smith, S., Chen, R. H., Fingar, D. C., and Blenis, J. (2002) Nat. Cell Biol. 4 556-564 [DOI] [PubMed] [Google Scholar]
- 25.Murphy, L. O., MacKeigan, J. P., and Blenis, J. (2004) Mol. Cell. Biol. 24 144-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vinciguerra, M., Vivacqua, A., Fasanella, G., Gallo, A., Cuozzo, C., Morano, A., Maggiolini, M., and Musti, A. M. (2004) J. Biol. Chem. 279 9634-9641 [DOI] [PubMed] [Google Scholar]
- 27.MacKenzie, S. J., Baillie, G. S., McPhee, I., Bolger, G. B., and Houslay, M. D. (2000) J. Biol. Chem. 275 16609-16617 [DOI] [PubMed] [Google Scholar]
- 28.Ley, R., Hadfield, K., Howes, E., and Cook, S. J. (2005) J. Biol. Chem. 280 17657-17663 [DOI] [PubMed] [Google Scholar]
- 29.Tcherkezian, J., Danek, E. I., Jenna, S., Triki, I., and Lamarche-Vane, N. (2005) Mol. Cell. Biol. 25 6314-6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsushima, M., Suwa, A., Amachi, T., Ueda, K., and Kioka, N. (2004) J. Biol. Chem. 279 34570-34577 [DOI] [PubMed] [Google Scholar]
- 31.Lin, Y. W., and Yang, J. L. (2006) J. Biol. Chem. 281 915-926 [DOI] [PubMed] [Google Scholar]
- 32.Matsubayashi, Y., Fukuda, M., and Nishida, E. (2001) J. Biol. Chem. 276 41755-41760 [DOI] [PubMed] [Google Scholar]
- 33.Whitehurst, A. W., Wilsbacher, J. L., You, Y., Luby-Phelps, K., Moore, M. S., and Cobb, M. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 7496-7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galanis, A., Yang, S. H., and Sharrocks, A. D. (2001) J. Biol. Chem. 276 965-973 [DOI] [PubMed] [Google Scholar]
- 35.Mansour, S. J., Matten, W. T., Hermann, A. S., Candia, J. M., Rong, S., Fukasawa, K., Vande Woude, G. F., and Ahn, N. G. (1994) Science 265 966-970 [DOI] [PubMed] [Google Scholar]
- 36.Raingeaud, J., Whitmarsh, A. J., Barrett, T., Derijard, B., and Davis, R. J. (1996) Mol. Cell. Biol. 16 1247-1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khokhlatchev, A., Xu, S., English, J., Wu, P., Schaefer, E., and Cobb, M. H. (1997) J. Biol. Chem. 272 11057-11062 [DOI] [PubMed] [Google Scholar]
- 38.Turk, B. E., Hutti, J. E., and Cantley, L. C. (2006) Nat. Protoc. 1 375-379 [DOI] [PubMed] [Google Scholar]
- 39.Hastie, C. J., McLauchlan, H. J., and Cohen, P. (2006) Nat. Protoc. 1 968-971 [DOI] [PubMed] [Google Scholar]
- 40.Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., and Olson, A. J. (1998) J. Comput. Chem. 19 1639-1662 [Google Scholar]
- 41.Songyang, Z., Blechner, S., Hoagland, N., Hoekstra, M. F., Piwnica-Worms, H., and Cantley, L. C. (1994) Curr. Biol. 4 973-982 [DOI] [PubMed] [Google Scholar]
- 42.Dimitri, C. A., Dowdle, W., MacKeigan, J. P., Blenis, J., and Murphy, L. O. (2005) Curr. Biol. 15 1319-1324 [DOI] [PubMed] [Google Scholar]
- 43.Schutkowski, M., Reimer, U., Panse, S., Dong, L., Lizcano, J. M., Alessi, D. R., and Schneider-Mergener, J. (2004) Angew. Chem. Int. Ed. Engl. 43 2671-2674 [DOI] [PubMed] [Google Scholar]
- 44.Fujii, K., Zhu, G., Liu, Y., Hallam, J., Chen, L., Herrero, J., and Shaw, S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13744-13749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim, Y. G., Shin, D. S., Kim, E. M., Park, H. Y., Lee, C. S., Kim, J. H., Lee, B. S., Lee, Y. S., and Kim, B. G. (2007) Angew. Chem. Int. Ed. Engl. 46 5408-5411 [DOI] [PubMed] [Google Scholar]
- 46.Pouchain, D., Diaz-Mochon, J. J., Bialy, L., and Bradley, M. (2007) ACS Chem. Biol. 2 810-818 [DOI] [PubMed] [Google Scholar]
- 47.Seet, B. T., Dikic, I., Zhou, M. M., and Pawson, T. (2006) Nat. Rev. Mol. Cell Biol. 7 473-483 [DOI] [PubMed] [Google Scholar]
- 48.Songyang, Z., Shoelson, S. E., Chaudhuri, M., Gish, G., Pawson, T., Haser, W. G., King, F., Roberts, T., Ratnofsky, S., Lechleider, R. J., Neel, B. G., Birge, R. B., Fajardo, J. E., Chou, M. M., Hanafusa, H., Schaffhausen, B., and Cantley, L. C. (1993) Cell 72 767-778 [DOI] [PubMed] [Google Scholar]
- 49.Yu, H., Chen, J. K., Feng, S., Dalgarno, D. C., Brauer, A. W., and Schreiber, S. L. (1994) Cell 76 933-945 [DOI] [PubMed] [Google Scholar]
- 50.Elia, A. E., Rellos, P., Haire, L. F., Chao, J. W., Ivins, F. J., Hoepker, K., Mohammad, D., Cantley, L. C., Smerdon, S. J., and Yaffe, M. B. (2003) Cell 115 83-95 [DOI] [PubMed] [Google Scholar]
- 51.Benes, C. H., Wu, N., Elia, A. E., Dharia, T., Cantley, L. C., and Soltoff, S. P. (2005) Cell 121 271-280 [DOI] [PubMed] [Google Scholar]
- 52.Cuenda, A., and Rousseau, S. (2007) Biochim. Biophys. Acta 1773 1358-1375 [DOI] [PubMed] [Google Scholar]
- 53.Bax, B., Carter, P. S., Lewis, C., Guy, A. R., Bridges, A., Tanner, R., Pettman, G., Mannix, C., Culbert, A. A., Brown, M. J., Smith, D. G., and Reith, A. D. (2001) Structure (Lond.) 9 1143-1152 [DOI] [PubMed] [Google Scholar]
- 54.Dajani, R., Fraser, E., Roe, S. M., Yeo, M., Good, V. M., Thompson, V., Dale, T. C., and Pearl, L. H. (2003) EMBO J. 22 494-501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heo, Y. S., Kim, S. K., Seo, C. I., Kim, Y. K., Sung, B. J., Lee, H. S., Lee, J. I., Park, S. Y., Kim, J. H., Hwang, K. Y., Hyun, Y. L., Jeon, Y. H., Ro, S., Cho, J. M., Lee, T. G., and Yang, C. H. (2004) EMBO J. 23 2185-2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattacharyya, R. P., Remenyi, A., Good, M. C., Bashor, C. J., Falick, A. M., and Lim, W. A. (2006) Science 311 822-826 [DOI] [PubMed] [Google Scholar]
- 57.Snead, J. L., Sullivan, M., Lowery, D. M., Cohen, M. S., Zhang, C., Randle, D. H., Taunton, J., Yaffe, M. B., Morgan, D. O., and Shokat, K. M. (2007) Chem. Biol. 14 1261-1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fecher, L. A., Cummings, S. D., Keefe, M. J., and Alani, R. M. (2007) J. Clin. Oncol. 25 1606-1620 [DOI] [PubMed] [Google Scholar]
- 59.Zhang, G. Y., and Zhang, Q. G. (2005) Exp. Opin. Investig. Drugs 14 1373-1383 [DOI] [PubMed] [Google Scholar]
- 60.Allen, M. D., DiPilato, L. M., Rahdar, M., Ren, Y. R., Chong, C., Liu, J. O., and Zhang, J. (2006) ACS Chem. Biol. 1 371-376 [DOI] [PubMed] [Google Scholar]
- 61.Green, H. M., and Alberola-Ila, J. (2005) BMC Chem. Biol. 5 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato, M., Kawai, Y., and Umezawa, Y. (2007) Anal. Chem. 79 2570-2575 [DOI] [PubMed] [Google Scholar]
- 63.Hancock, C. N., Macias, A., Lee, E. K., Yu, S. Y., Mackerell, A. D., Jr., and Shapiro, P. (2005) J. Med. Chem. 48 4586-4595 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.