Abstract
Targeting TRAIL receptors with either recombinant TRAIL or agonistic DR4- or DR5-specific antibodies has been considered a promising treatment for cancer, particularly due to the preferential apoptotic susceptibility of tumor cells over normal cells to TRAIL. However, the realization that many tumors are unresponsive to TRAIL treatment has stimulated interest in identifying apoptotic agents that when used in combination with TRAIL can sensitize tumor cells to TRAIL-mediated apoptosis. Our studies suggest that various apoptosis defects that block TRAIL-mediated cell death at different points along the apoptotic signaling pathway shift the signaling cascade from default apoptosis toward cytoprotective autophagy. We also obtained evidence that inhibition of such a TRAIL-mediated autophagic response by specific knockdown of autophagic genes initiates an effective mitochondrial apoptotic response that is caspase-8-dependent. Currently, the molecular mechanisms linking disabled autophagy to mitochondrial apoptosis are not known. Our analysis of the molecular mechanisms involved in the shift from protective autophagy to apoptosis in response to TRAIL sheds new light on the negative regulation of apoptosis by the autophagic process and by some of its individual components.
Accumulating evidence suggests that autophagy functions as an adaptive cell response, allowing the cell to survive bioenergetic stress via a mechanism associated with clearance of damaged organelles and the degradation of mutant or misfolded proteins (1). Certain therapeutic approaches to cancer, including radiation and cytotoxic drugs that have been known to activate apoptosis, were observed to induce autophagy in certain human cancer cell lines (2). The functional relationship between apoptosis and autophagy and the potential cross-regulation between these two processes are complex and remain to be resolved. The complexity stems partly from the findings that in certain cellular scenarios, autophagy constitutes a stress adaptation response that avoids and suppresses cell death, whereas in other cellular settings, autophagy constitutes an alternative pathway to cellular demise that is called autophagic cell death (type II cell death) (3-5). Thus, the autophagy genes beclin-1 and atg7 are required to induce nonapoptotic cell death in murine fibroblast L929 cells treated with the caspase inhibitor Z-VAD3 (6). In addition, Atg5 and Beclin-1 are required for etoposide- and staurosporin-induced cell death in apoptosis-resistant Bax/Bak double knock-out mouse embryonic fibroblasts (7). Current evidence suggests that the removal or functional inhibition of proteins essential for the apoptotic machinery can switch a cellular stress response from apoptotic default to massive autophagy (4, 6-8). In this regard, dogma-altering studies were reported by Craig Thompson's group, who discovered that when apoptosis-resistant cells are exposed to stress mediated by the decreased availability of growth factor, the ensuing autophagy actually protects cells from death (8). Specifically, they demonstrated that immortalized IL-3-dependent cell lines generated from the bone marrow of Bax-/-Bak-/- mice failed to undergo apoptosis upon IL-3 withdrawal and entered a month-long autophagic process that caused a severe reduction in cell size with the removal of most of the cytoplasm. Inhibition of autophagy by the knockdown of atg5 or atg7 or by the addition of 3-methyladenine (3MA; an inhibitor of Class III phosphatidylinositol 3-kinase) or chloroquine (an inhibitor of lysosomal acidification, which is required for the fusion between autophagosomes and lysosomes) killed Bax-/-Bak-/- cells, indicating that the observed autophagy functions as a survival mechanism. Autophagy in this cellular setting was concluded to be a catabolic process that mobilizes nutrients by macromolecular degradation, thus replenishing the depleted energy reserves of the IL-3-dependent cells and hence preventing a bioenergetic crisis that would otherwise result in an immediate cell death.
Induction of apoptosis due to inhibition of autophagy has mainly been reported for starvation experimental models, where autophagy is the immediate response for the recycling of essential metabolites and the fueling of the bioenergetic machinery. Inhibition of autophagy in starving cells results in accelerated apoptotic cell death, characterized by activation of caspases, mitochondrial permeabilization, and chromatin condensation (9, 10). The applicable nature of targeting autophagy for anticancer therapeutic benefit has recently been indicated by two studies that employed the pharmacological inhibitors of autophagy, chloroquine and 3MA. Thus, inhibition of autophagy by chloroquine was shown to enhance the anticancer activity of the alkylating agent cyclophosphamide in a Myc-driven model of lymphoma (11). In an additional study, both chloroquine and 3MA synergistically augmented the proapoptotic effects and overall anticancer activity of the histone deacetylase inhibitor suberoylanilide hydroxamic acid (12).
TRAIL has been considered a promising candidate for cancer therapeutics because of its selective cytotoxicity against malignancies (13-15). Induction of apoptosis by TRAIL is mediated by its interaction with two death domain-containing receptors, TRAIL-R1/DR4 and TRAIL-R2/DR5. This, in turn, orchestrates the assembly of the death-inducing signaling complex (DISC) that contains FADD, an adaptor that activates initiator caspases, caspase-8 and caspase-10, leading (in Type I cells) to direct activation of effector caspases, such as caspase-3 (16, 17), or to Bid cleavage and subsequent mitochondrial apoptosis (in Type II cells) (18-20). Unfortunately, as with a multitude of other chemotherapeutic compounds, TRAIL-responsive tumors acquire a resistant phenotype, which renders TRAIL therapy ineffective (21, 22). The realization that many tumors are unresponsive to TRAIL treatment has stimulated interest in identifying apoptotic agents that when used in combination with TRAIL can sensitize tumor cells to TRAIL-mediated apoptosis. Indeed, multiple studies have demonstrated that a variety of apoptotic agents and proteins sensitize several classes of tumor cells to TRAIL-induced apoptosis (13, 15).
The current study was designed to investigate the involvement of autophagy in the response to TRAIL treatment of T leukemic and colon carcinoma cells with various apoptotic defects. Our findings suggest that various apoptosis defects that block TRAIL-mediated cell death at different points along the apoptotic signaling pathway shift the signaling cascade toward cell-protective autophagy. We also obtained evidence that inhibition of such a TRAIL-mediated autophagic response initiates an effective TRAIL-dependent apoptotic cascade. We propose that divergent mechanisms of resistance to TRAIL-mediated apoptosis can be reversed by a common approach of targeting for inhibition-specific components of the autophagic process. This novel concept may have significant implications for the development of new strategies to circumvent TRAIL resistance in tumors.
EXPERIMENTAL PROCEDURES
Reagents—Recombinant TRAIL was purchased from Pepro-Tec (Rocky Hill, NJ); Abs against Fas and Cortactin (cortical actin-associated protein) were from Upstate (Lake Placid, NY); Abs specific for LAMP2, PARP, caspase-9, FLIP, Beclin-1, β-tubulin, Histone-1, Bid, AIF, cytochrome c, Bak, Atg7, and Atg5 were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-caspase-3 Abs from StressGen (Victoria, Canada) were utilized for detection of active subunits and from Cell Signaling Technology (Danvers, MA) for prodomain detection; Abs to caspase-7 and -8 were from BD Pharmingen; anti-β-actin Ab, pepstatin A, 3MA, Hoechst, and DAPI were from Sigma; anti-UVRAG Ab was from Abgent (San Diego, CA); anti-MAP-LC3 Abs for immunoblotting were from AnaSpec (San Jose, CA) and from MBL (Woburn, MA); anti-MAP-LC3 Abs for immunostaining were from Santa Cruz; anti-SMAC Ab was from Cell Signaling (Beverly, MA); 5,6-carboxysuccinimidylfluorescein ester, rhodamine phalloidine, fluorescent carbofluorescein-DEVD-fmk and fluorescent carbofluorescein-IETD-fmk, anti-Cox IV Ab, and Alexa Fluor 488 or 647-conjugated anti-rabbit or anti-mouse Ig were from Invitrogen; E64D was from Calbio-chem; TUNEL detection kit was from Roche Applied Science; and Z-VAD-fmk and Z-IETD-fmk were from ICN (Aurora, OH).
RNAi—atg5, atg7, bak, beclin-1, caspase-8, cortactin, and UVRAG siRNAs were obtained as siGENOME SMARTpool reagents from Dharmacon. beclin-1 siRNA was also obtained as ON-TARGET plus SMARTpool siRNA from Dharmacon. Both siGENOME SMARTpool and ON-TARGET plus SMARTpool siRNAs consist of four distinct RNA oligoduplexes per target gene, and both have a guaranteed silencing effectiveness of at least 75% at the mRNA level. To confirm results obtained with beclin-1 siGENOME SMARTpool siRNA, some of the experiments were repeated with beclin-1 ON-TARGET plus SMARTpool siRNA, which is reported to reduce off-target effects up to 90%. bid siRNA was obtained as a duplex in purified and desalted form (Option C) from Dharmacon with the following sense strand sequence: 5′-GAAGACATCATCCGGAATAdTdT-3′. The nontargeting siRNA control used in our RNAi experiments is the siCONTROL nontargeting siRNA pool 2 (Dharmacon), which contains four nontargeting siRNAs. The nontargeting control for beclin-1 ON-TARGET plus SMARTpool siRNA also consists of four nontargeting oligoduplexes. WT Hct116, Hct116-FLIP, or Bax-/- Hct116 cells (2.5 × 105) were plated in a 6-well plate and, following 16 h (at ∼30% confluence), were transfected with 200 nm siRNA in Opti-MEM medium (Invitrogen) without fetal calf serum using Oligofectamine reagent (Invitrogen) according to the manufacturer's transfection protocol. After 4 h, fetal calf serum was added to a final concentration of 10%. At the end of the siRNA treatment period (48-72 h), the medium over the cells was adjusted to 1 ml before the addition of an apoptotic agent.
Stable Transfection—Hct116 cells were washed in cold phosphate-buffered saline and resuspended in electroporation buffer (AMAXA) at a final concentration of 3 × 107 cells/ml. Five μg of linearized FLIP or linearized GFP-LC3 plasmids and linearized pCR3.1 vector (Invitrogen) were mixed with 0.1 ml of cell suspension, transferred to a 2.0-mm electroporation cuvette, and nucleofected with an Amaxa Nucleofector apparatus, utilizing the appropriate program according to the manufacturer's directions. Geneticin-resistant cell lines were grown in the presence of G418 (1500 μg/ml). Geneticin-resistant clonal cell lines harboring either the FLIP-expressing vector or the empty vector were generated by dakocytomation (1 cell/well) utilizing a MOFLO high speed cell sorter and Summit Software.
Cell Microscopy—Cytospins were made from Jurkat cells on noncoated slides. Hct116 cells were grown in culture chamber slides, and cytospins were made only when treatment resulted in floating apoptotic cells. Confocal microscopy was performed with an OlympusFluoview 1000 confocal microscope and the companion software FV10-ASW1.6. For electron microscopy, cells were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.1 m phosphate buffer (pH 7.0), followed by 1% OsO4. After dehydration, thin sections were stained with uranyl acetate and lead citrate for observation under a JEM 1011CX electron microscope (JEOL, Peabody, MA). Images were acquired digitally.
Cell Lines, Cell Lysates, and Cell Extracts—Previously described (23) Jurkat T leukemic cells stably transfected with empty vector or with FLIP-L were grown in RPMI 1640 medium containing 10% fetal calf serum, 20 mm HEPES, 2 mm l-glutamine, and 100 units/ml each of penicillin and streptomycin and 5 μg/ml puromycin. Hct116 cells were grown in Dulbecco's modified Eagle's medium containing 15% fetal calf serum, 20 mm l-glutamine, and 100 units/ml each of penicillin and streptomycin. The cells were treated with various siRNAs for 48-60 h and then with TRAIL (50 or 100 ng/ml) for 6 h. The length of the TRAIL treatment is indicated where it was longer (16 or 24 h). Cell lysates were prepared with 1% Nonidet P-40, 20 mm Tris-HCl, pH 7.4, 137 mm NaCl, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. To prepare cell extracts, cultured cells were washed twice with phosphate-buffered saline and then resuspended in ice-cold buffer (20 mm HEPES, pH 7.0, 10 mm KCl, 1.5 mm MgCl2, 1 mm sodium EDTA, 1 mm sodium EGTA, 1 mm dithiothreitol, 250 mm sucrose, and protease inhibitors). After incubation on ice for 20 min, cells (2.5 × 106/0.5 ml) were disrupted by Dounce homogenization. Nuclei were removed by centrifugation at 650 × g for 10 min at 4 °C. Cellular extracts were obtained as the supernatants resulting from centrifugation at 14,000 × g at 4 °C for 30 min.
Subcellular Fractionation—To obtain an enriched mitochondrial fraction, Hct116 or Jurkat cells were suspended in mitochondrial buffer composed of 0.3 m sucrose, 10 mm MOPS, 1 mm EDTA, and 4 mm KH2PO4, pH 7.4, and lysed by Dounce homogenization. The purified mitochondrial fraction was obtained as previously described (24). The mitochondrial purity was assessed at 95%, with ∼5% or less contamination from the microsomal fraction (24-27).
Western Blot Analysis—Proteins in cell lysates, cell extracts, and the various subcellular fractions were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes, as previously described (24, 27). Following probing with a specific primary Ab and horseradish peroxidase-conjugated secondary Ab, the protein bands were detected by enhanced chemiluminescence (Pierce).
Cell Survival Assays—Cell viability based on the ATP levels present in live cells was tested with the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer's protocol. Clonogenic assays were performed with methylcellulose-based semisolid medium (MethoCult H4230; StemCell Technologies) according to the manufacturer's protocol. In brief, after treatment, the cells were washed, suspended in MethoCult medium, and cultured in triplicates (300 cells/3 ml) in 35-mm Petri dishes. The cultures were maintained at 37 °C in 5% CO2 for 14 days, and colonies were counted using an inverted microscope and gridded scoring dishes. Cytofluorometric analyses of apoptosis were performed by co-staining with propidium iodide and fluorescein isothiocyanate-Annexin V conjugates (BD Biosciences).
RESULTS
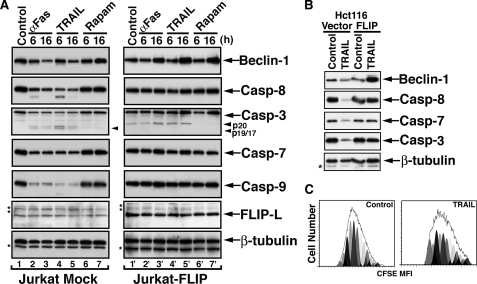
TRAIL Induces a Protective Autophagic Response in Apoptosis-defective Tumor Cells—Recent studies suggest that an autophagic response is developed in apoptotic defective cells subjected to various forms of stress (8, 28). FLIP is a potent inhibitor of death receptor-initiated apoptotic cascades that functions at the DISC to block the development of downstream apoptotic events. It has been well established that the apoptotic response to either TRAIL or Fas ligand is blocked by FLIP, but the possibility that such an apoptotic blockade gives rise to an autophagic process has not yet been explored. Therefore, we revisited the response of FLIP-overexpressing cells to TRAIL. As previously reported, the ligation of either Fas or TRAIL-Rs by their agonistic ligands in cells overexpressing FLIP does not result in productive apoptosis. Thus, no caspase activity is detected in either Jurkat or Hct116 cells stably transfected with FLIP (Fig. 1, A and B), and the cells maintain their pretreatment proliferation rate (Fig. 1C). However, we observed an unexpected change in the expression levels of Beclin-1, whereas under apoptotic conditions in vector control cells, the Beclin-1 expression level was reduced, in FLIP-overexpressing cells, agonistic ligation of either Fas or TRAIL-Rs resulted in increased Beclin-1 (Fig. 1, A and B). Likewise, treatment with rapamycin, an inhibitor of mTOR that induces autophagy, also resulted in significant increased expression of Beclin-1. These findings indicate that despite the apoptotic block mediated by FLIP overexpression, the ligated death receptors transmit a signal that leads at least to an increase in the expression level of Beclin-1.
FIGURE 1.
TRAIL-mediated up-regulation of Beclin-1 in FLIP-overexpressing cells. A, induction of Beclin-1 in Jurkat-FLIP cells treated with anti-Fas Ab or TRAIL. Clonal Jurkat cell lines stably transfected with an empty vector (mock) or with FLIP-L were treated with agonistic anti-Fas Ab (CH11; 100 ng/ml), TRAIL (100 ng/ml), or rapamycin (2 mm) for the indicated time periods. The cells were then lysed and assessed by immunoblotting and successive probing of the same membranes for the indicated proteins. The asterisks indicate unidentified protein bands. B, induction of Beclin-1 in TRAIL-treated Hct116-FLIP cells. Hct116 cells stably transfected with empty vector or with FLIP were treated with TRAIL (100 ng/ml, 16 h), and the cell lysates were assessed by immunoblotting for the expression of the indicated proteins. C, TRAIL treatment of Jurkat-FLIP cells does not interfere with cell proliferation. 5,6-carboxysuccinimidylfluorescein ester (CFSE)-labeled Jurkat-FLIP cells were treated with TRAIL (100 ng/ml, 6 h) or with vehicle control. The cells were cultured for 5 days, and each day a sample was assessed by flow cytometry for the dilution effect of the fluorescent dye as a result of cell proliferation. A similar pattern of proliferation was observed for each of the 5 days, and the results obtained for day five are shown. The different peaks represent different daughter generations. The presence of 13-15 daughter generations indicates robust proliferation of each of the cultures. A representative experiment of at least three performed with similar results is shown for each panel.
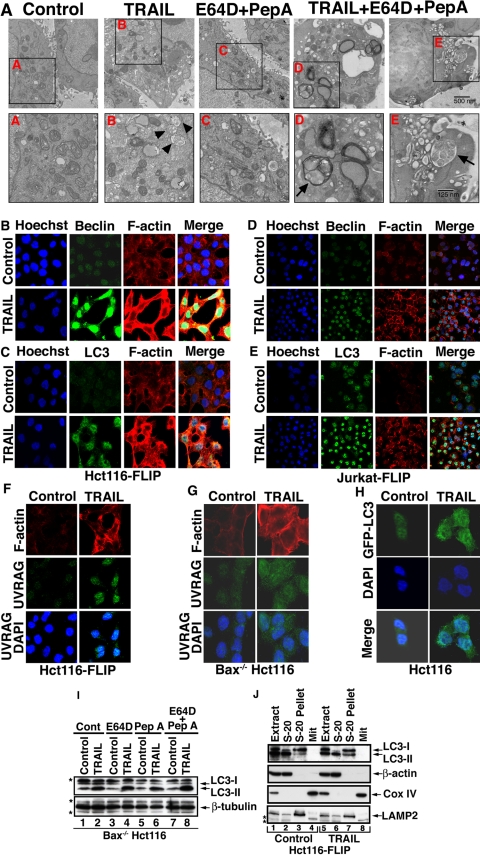
Since Beclin-1 is an autophagic protein, we assessed the possibility that TRAIL induces an autophagic response in such TRAIL apoptosis-resistant cells. Beclin-1 up-regulation in the response of FLIP-overexpressing cells to TRAIL was associated with increased autophagosome formation as detected by electron microscopy (Fig. 2A). The increased accumulation of autophagosomes was further confirmed by treatment of the cells with TRAIL in the presence of two lysosomal protease inhibitors, E64D, a membrane-permeable inhibitor of cathepsins B, H, and L, and pepstatin A, an inhibitor of cathepsins D and E (29). Such a combined treatment by TRAIL and cathepsin inhibitors, but not with the inhibitors alone, gave rise to numerous large autophagosomes having a significant amount of enclosed undigested content, including multiple organelles. Using confocal microscopy, we confirmed the increase in the expression level of Beclin-1 in FLIP-overexpressing Hct116 or Jurkat cells (Fig. 2, B and D). As previously reported (30), in Hct116-FLIP cells, Beclin-1 localized both to nuclei and mitochondria (Fig. 2B). A TRAIL-mediated increase in MAP-LC3/Atg8 expression and a change in its subcellular distribution from cytoplasm to membranous autophagosomes or lysosomes was confirmed for both endogenous (Fig. 2, C and E) and transfected GFP-LC3 (Fig. 2H) (31). The punctate appearance of endogenous LC3 is more clearly evident in the higher magnification of Fig. 2C shown in the supplemental materials (Fig. S1). Likewise, increased expression of UVRAG was observed in TRAIL-treated Hct116-FLIP or Bax-/- Hct116 cells (Fig. 2, F and G). Similar to LC3, the increased expression of UVRAG had a punctate appearance and cytoplasmic localization (Fig. 2, F and G). LC3 is initially synthesized as an unprocessed form, proLC3, which is converted to a proteolytically processed form lacking C-terminal amino acids, LC3-I, and is finally modified into the phycoerythrin-conjugated form, LC3-II. LC3-II is the only protein marker that is reliably associated with completed autophagosomes (32). To further investigate the ability of TRAIL to induce an autophagic flux in apoptosis-resistant cells, we assessed by immunoblotting the level of LC3-II in Bax-/- Hct116 cells treated with TRAIL in the presence of E64D and/or pepstatin A (29). An increased expression level of LC3-II was observed in TRAIL-treated cells, particularly in cells treated in the presence of both inhibitors that together cover a wide range of cathepsins (Fig. 2I). To determine the subcellular localization of TRAIL-induced LC3-II, we performed a subcellular fractionation experiment for control and TRAIL-treated Hct116-FLIP cells. The increased expression in LC3-II was detected in the pellet of the S-20 cytosolic fraction, which includes autophagosomes and lysosomes (Fig. 2J).
FIGURE 2.
TRAIL-mediated autophagy in apoptosis-resistant tumor cells. A, electron microscopy of Hct116-FLIP cells treated with TRAIL in the presence or absence of the cathepsin inhibitors E64D and pepstatin A. The bottom shows magnified images of the indicated areas A-E in the top. The arrowheads depict autophagosomes containing recognizable cellular materials; arrows depict autophagosomes in the presence of E64D and pepstatin A. Scale bar in the top, 500 nm; scale bar in the bottom, 125 nm. B-E, TRAIL-mediated induction of Beclin-1, MAP-LC3, and F-actin in Hct116 (B and C) and Jurkat (D and E) clonal cell lines stably transfected with FLIP. The indicated clonal cell lines were treated with TRAIL (100 ng/ml, 24 h). Adherent Hct116 cells (in a chamber culture) or Jurkat cytospins were then fixed, permeabilized, blocked, and stained with Hoechst (blue), anti-Beclin-1, or anti-MAP-LC3 Ab (green) and rhodamine phalloidin (polymerized F-actin). Cells were visualized by confocal microscopy. F and G, up-regulation of UVRAG in Hct116-FLIP (F) or Bax-/- Hct116 cells (G) treated with TRAIL as described in B. H, punctate appearance of extranuclear LC3 in the TRAIL-resistant Hct116 clonal cell line stably transfected with GFP-LC3. (The mechanism of TRAIL resistance in this clone that was selected from WT Hct116 is not known). I, TRAIL-mediated autophagic flux in Bax-/- Hct116 cells. The autophagic flux is indicated by increased detection of LC3-II in TRAIL-treated cells (lanes 2, 4, and 6) and particularly in the presence of both E64D and pepstatin A (lane 8) as compared with controls (lanes 1, 3, 5, and 7). The immunoblotting was performed with MBL anti-LC3 Ab. J, increased level of LC3-II in lysosomic fraction present in the postnuclear and postmitochondrial S-20 pellet. Control or TRAIL-treated Hct116-FLIP cells were Dounce-homogenized and subcellularly fractionated to obtain the mitochondrial pellet (Mit), S-20 supernatant (cytosol), and S-20 pellet. The proteins in the various fractions were separated by SDS-PAGE and assessed by immunoblotting for the indicated proteins. Increased LC3-II expression (detected by MBL anti-LC3 Ab) is noted in the S-20 pellet in TRAIL-treated cells (lane 7 versus lane 3). LAMP2 (lysosomal-associated membrane protein 2) serves as a marker for the S-20 pellet. β-Actin and Cox IV serve as markers for the cytosol and mitochondria, respectively. The asterisks indicate nonspecific protein bands.
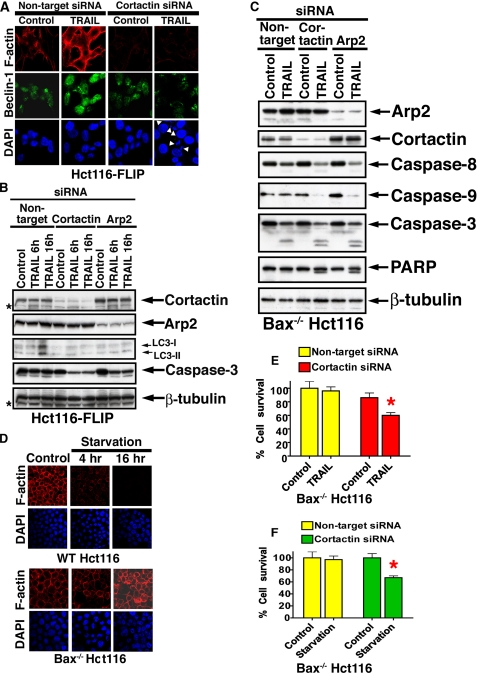
Reversal of TRAIL Resistance by Inhibition of F-actin Polymerization—Surprisingly, we consistently observed increased polymerization of F-actin as detected by rhodaminephalloidine in association with TRAIL-mediated autophagy in either Hct116-FLIP or Bax-/- Hct116 cells (Fig. 2, B-G). To further investigate if the changes we observed in the F-actin dynamics in response to TRAIL were essential for the protective response, we interfered with the F-actin polymerization process by RNAi knockdown of arp2 or cortactin. The actin-related protein 2/3 (Arp2/3) complex is responsible for the dynamics of the actin networks by virtue of its ability to initiate actin filament branches (33). Cortactin regulates the function of the Arp2/3 complex and hence is a key mediator of actin polymerization (34). As expected, cortactin siRNA (Fig. 3A) and also arp2 siRNA (not shown) reduced the basal level of F-actin polymerization. Furthermore, the reduction in expression of either Cortactin or Arp2 blocked the protective autophagic cell response to TRAIL, as indicated by a lack in LC3 up-regulation and LC3-II formation (Fig. 3B). Note that the affinity of different anti-LC3 Abs to various LC3 forms (I and II) determines the strength of the signal detected by immunoblotting (29, 31, 32). AnaSpec anti-LC3 Ab detects an increase in both LC3-I and LC3-II in TRAIL-treated cells, and this increase is reversed by either cortactin or arp2 RNAi. The difference in the level of LC3-II between treatments is considered significant in determination of an autophagic flux. Unexpectedly, we observed induction of caspase activity and the appearance of fragmented nuclei in Hct116-FLIP cells treated with TRAIL and cortactin siRNA but not in cells treated with only the cortactin siRNA alone (Fig. 3A). Increased caspase activity was detected in either Hct116-FLIP or Bax-/- Hct116 cells treated with TRAIL and cortactin or arp2 siRNAs (Fig. 3, B and C). These findings suggest that the up-regulation in F-actin polymerization in the response of apoptosis-defective cells to TRAIL is an essential part of the protective mechanism, since specific interference of F-actin polymerization shifts the cell response to apoptosis. Since increased F-actin polymerization has not been previously connected to autophagy, we further assessed F-actin changes in the Hct116 cell response to amino acid starvation. We observed up-regulated F-actin polymerization in Bax-/- Hct116 but not in WT Hct116 cells incubated under starvation conditions for 4-16 h (Fig. 3D). In Bax-deficient Hct116, increased F-actin in response to starvation was associated with cell survival (5% TUNEL-positive cells), whereas in apoptosis-competent WT Hct116 cells, the starvation conditions reduced the F-actin polymerization level in association with apoptotic cell death (90% TUNEL-positive cells). In sum, cortactin RNAi decreased significantly the survival capability of the Bax-/- Hct116 cells exposed to either TRAIL or starvation (Fig. 3, E and F). The involvement of Cortactin and Arp2 in TRAIL-mediated cytoprotective autophagy further indicates the survival role of F-actin polymerization in this response.
FIGURE 3.
Circumvention of cytoprotective autophagy by cortactin or arp2 RNAi. A, inhibition of TRAIL-mediated up-regulation of Beclin-1 and F-actin by cortactin RNAi. Hct116-FLIP cells were treated with nontargeting or cortactin siRNAs followed by TRAIL (50 ng/ml, 16 h) and assessed by confocal microscopy for the expression of F-actin, Beclin-1, and DAPI. The white arrowheads indicate fragmented nuclei. B, cortactin or arp2 RNAi blocks the TRAIL-mediated induction of LC3 (I and II, as assessed by AnaSpec anti-LC3 Ab) and results in activation of caspase-3 in Hct116-FLIP cells. Hct116-FLIP cells were treated as described in A and assessed by immunoblotting for the expression of the indicated proteins. C, reversal of TRAIL-resistance of Bax-/- Hct116 cells by cortactin or arp2 RNAi. Bax-deficient Hct116 cells were treated with siRNAs as above and then treated with TRAIL (50 ng/ml, 6 h), lysed, and assessed by immunoblotting for the expression of the indicated proteins. β-Tubulin serves as a control for equal loading. D, starvation-induced polymerization of F-actin in apoptosis-defective tumor cells but reduced polymerization in apoptosis-competent cells. WT Hct116 and Bax-/- Hct116 cells were kept in Hanks' balanced salt solution for 4 or 16 h and then assessed for F-actin polymerization as described in A. At 16 h of starvation, WT Hct116 cells, but not the Bax-/- Hct116 cells, were TUNEL-positive (90% versus 5%, respectively; data not shown). E and F, reversal of apoptosis resistance to TRAIL (E) or starvation (F) of Bax-/- Hct116 cells by cortactin RNAi. Bax-/- Hct116 cells were treated with cortactin siRNA and TRAIL or amino acid starvation for additional 6 h. Cell viability was determined by the Titer-Glo luminescent cell viability assay. Data are means ± S.E. of triplicates in one experiment of three performed for TRAIL or starvation. The red asterisks indicate statistical significant difference as compared with control (p < 0.05, Mann-Whitney U test).
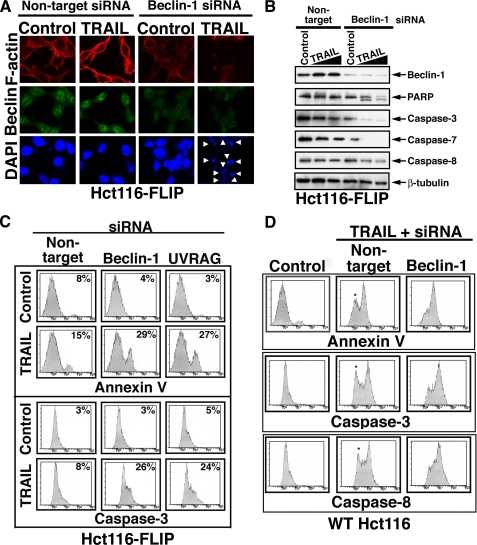
Inhibition of Autophagy Shifts the TRAIL Response of Apoptosis-resistant Cells from Autophagy to Apoptosis—To investigate if the observed apoptotic response was a result of specific inhibition of the autophagic process, we targeted by RNAi two established autophagic proteins, Beclin-1 and UVRAG. In Hct116-FLIP cells treated with TRAIL following treatment with beclin-1 or UVRAG siRNAs, but not following nontargeting siRNA treatment, we observed significant cell loss as assessed by trypan blue exclusion dye (not shown), by the presence of fragmented nuclei (Fig. 4A), and by flow cytometry detection of membranous phosphatidylserine exposure (Fig. 4C). In addition, the induction of caspase activity was indicated by reduced detection by immunoblotting of the caspase-3, -7, and -8 prodomains (Fig. 4B), by PARP cleavage, and by flow cytometry detection of active caspase-3 (Fig. 4C). These results suggest that the TRAIL-mediated autophagic response in Hct116-FLIP cells is a cell-protective mechanism whose inhibition enables new apoptotic avenues despite the block mediated by FLIP overexpression.
FIGURE 4.
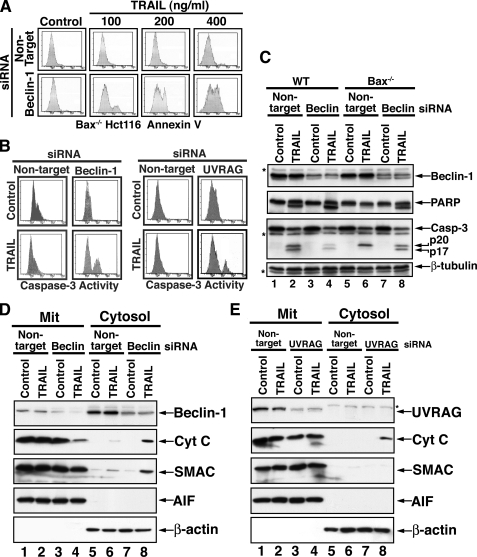
Knockdown of beclin-1 or UVRAG in FLIP-overexpressing cells shifts the autophagic response to apoptosis. A and B, inhibition of TRAIL-mediated autophagy in Hct116-FLIP cells by beclin-1 RNAi induces caspase activity and nuclei fragmentation. Hct116-FLIP cells were treated with nontargeting control or beclin-1 siRNA and TRAIL as described under “Materials and Methods.” The cells were then either assessed by confocal microscopy for the staining of F-actin, Beclin-1, and DAPI (A) or lysed and assessed by immunoblotting for the indicated proteins (B). The arrowheads in A indicate fragmented nuclei. C, inhibition of TRAIL-mediated autophagy in Hct116-FLIP cells by beclin-1 or UVRAG RNAi induced apoptosis, as assessed by the presence of Annexin V-positive cells and caspase activation. A proof for effective UVRAG RNAi is included in Fig. 5. D, beclin-1 RNAi induces caspase activation in TRAIL-nonresponsive cells within the WT Hct116 cell line. Hct116 cells were treated with siRNAs and TRAIL and then assessed by flow cytometry for phosphatidylserine exposure and activation of caspase-3 and -8. Untreated cells and those treated with only nontargeting siRNA gave a similar pattern, and the latter are shown as control. The asterisks in the middle column panels indicate the TRAIL-nonresponding cells that exhibit a shift to the response area following treatment with beclin-1 siRNA (right column panels).
In WT Hct116 cells that were assessed under similar experimental conditions, the apoptotic response to TRAIL was efficiently developed only in ∼60% of the cells, whereas the remaining cells did not undergo phosphatidylserine exposure and did not exhibit caspase activation. Interestingly, beclin-1 RNAi in WT Hct116 sensitized the nonresponsive cells to TRAIL-mediated apoptosis (Fig. 4D). These findings suggest that naturally occurring apoptotic defects capable of affecting the cell apoptotic response to TRAIL contribute to the initiation of a protective autophagic response, whose inhibition ultimately renders the cells susceptible to TRAIL-mediated apoptosis.
Although TRAIL-mediated apoptosis is a death receptor-initiated cascade, in certain cell types, the productive propagation of this cascade is mitochondria-dependent (18, 19). Thus, Bax knock-out in Hct116 cells rendered these cells completely resistant to TRAIL-mediated apoptosis. In these Bax-/- Hct116 cells, the initial phase of the TRAIL signal, which includes caspase-8 activation at the DISC and subsequent Bid cleavage, is executed, but due to the lack of mitochondrial involvement, the apoptotic cascade is not productive. Thus, Bax-deficient Hct116 cells maintain their uninterrupted proliferation despite an upstream activation of caspase-8 and subsequent processing of caspase-3 prodomain to its p20 subunit, which remains inactive in the absence of mitochondria-derived SMAC or cytochrome c (18, 27). To further investigate a potential involvement of protective autophagy in the apoptotic resistance to TRAIL of Bax-/- Hct116 cells, we assessed the effects of beclin-1 or UVRAG knockdown in these cells. Thus, knockdown of beclin-1 allowed the development of an apoptotic cascade that was TRAIL dose-dependent (Fig. 5A). Once Beclin-1 is down-regulated, a TRAIL-dependent apoptotic response takes place, and up to 50% of the otherwise TRAIL-resistant cells succumb to apoptotic cell death. These findings suggest that Beclin-1 (serving as either an essential component of the autophagic process or on its own) inhibits the productive propagation toward apoptosis of TRAIL signal transduction. In addition, RNAi of either beclin-1 or UVRAG enabled the induction of caspase-3 activity by TRAIL in a significant fraction of the treated cells (Fig. 5B). Indeed, we observed that beclin-1 knockdown allowed the further processing of caspase-3 p20 to its p17 form (Fig. 5C). The possibility that beclin-1 RNAi was associated with an off-target effect on the expression levels of IAP proteins, XIAP, c-IAP-1, c-IAP-2, or survivin, was excluded (data not shown). Furthermore, no potential binding between Beclin-1 and either caspase-3 or XIAP was detected by co-immunoprecipitation (data not shown). Since the autoprocessing of caspase-3 p20 into its p17 subunit has been demonstrated to be mitochondria-dependent (18, 19, 27), we investigated whether knockdown of beclin-1 rendered the Bax-/- Hct116 mitochondria sensitive to the upstream signals delivered through the TRAIL-Rs. Surprisingly, we observed that the reduced expression of Beclin-1 enabled Bax-/- mitochondria to release a significant portion of their cytochrome c, as detected by the loss of cytochrome c in the mitochondrial pellets and its presence in the cytosolic fraction (Fig. 5D). SMAC, but not AIF, was co-released with cytochrome c. UVRAG RNAi also enabled some release of mitochondrial cytochrome c in response to TRAIL but at a much lower magnitude when compared with beclin-1 RNAi (Fig. 5, D and E). Interestingly, in Bax-/- Hct116, UVRAG was mainly associated with the mitochondria (Fig. 5E), and thus, its knockdown is demonstrated mainly in the mitochondrial fraction. UVRAG is a positive regulator of Beclin-1; therefore, the individual knockdown of either beclin-1 or UVRAG may impinge on the same cellular event.
FIGURE 5.
Involvement of Beclin-1 and UVRAG in the TRAIL resistance of Bax-deficient cells. A, TRAIL dose-dependent apoptotic response of Bax-/- Hct116 cells following beclin-1 knockdown. The cells were treated with nontargeting or Beclin-1-specific siRNA for 60 h followed by treatment with the indicated doses of TRAIL for 6 h. Flow cytometry assessment of Annexin-V-positive cells is shown. B, flow cytometry assessment of caspase-3 activity in beclin-1 or UVRAG knockdown Bax-/- Hct116 cells treated with TRAIL. C, TRAIL-mediated caspase-3 activity and PARP cleavage in Bax-/- Hct116 cells following beclin-1 RNAi. The cells were treated as in B, lysed, and assessed by immunoblotting for the indicated proteins. Reduced detection of active caspase-3 subunits (lane 4) is typical for a late stage of full-blown apoptosis. TRAIL induction of Beclin-1 requires a longer treatment time than the 6 h presented. β-Tubulin serves as a control for equal loading. The asterisks indicate unidentified protein bands. D and E, TRAIL-mediated release of mitochondrial apoptogenic proteins in Bax-/- Hct116 cells following knockdown of beclin-1 or UVRAG. The cells were treated with either nontargeting or beclin-1 siRNA (D) or UVRAG siRNA (E) and TRAIL. The cell extracts were fractionated to obtain mitochondrial and cytosolic fractions. The fractions were then assessed by immunoblotting for the indicated proteins.β-Actin and AIF serve as loading controls and as fractionation markers. A significant loss in cytochrome c is noted in the mitochondrial pellet of cells treated with beclin-1 siRNA and TRAIL (D, lane 4). Since UVRAG was mainly detected in the mitochondrial pellet (E, lanes 1 and 2), the mitochondrial pellet (E, lanes 3 and 4) was utilized to demonstrate the effective knockdown of UVRAG by RNAi.
3MA is an inhibitor of Vps34 (class III phosphatidylinositol 3-kinase), which has been demonstrated to block the formation of autophagosomes. Although inhibition by 3MA may not be exclusively autophagy-specific, it has been shown that treatment with 3MA in conjunction with other stress stimuli shifted the autophagic response to apoptosis (9). Likewise, treatment of either Jurkat-FLIP (Fig. 6, A and B) or Hct116-FLIP (Fig. 6C) cells with TRAIL in the presence of 3MA resulted in a shift from an autophagic to an apoptotic cell response. The apoptotic response of these FLIP-overexpressing cells was documented by the presence of fragmented nuclei (Fig. 6, A and C), TUNEL-positive cells (Fig. 6C), and caspase activity (Fig. 6B). There was a concomitant loss in cell survival features, including lack of TRAIL-mediated up-regulation of Beclin-1 (Fig. 6C) and F-actin (Fig. 6A).
FIGURE 6.
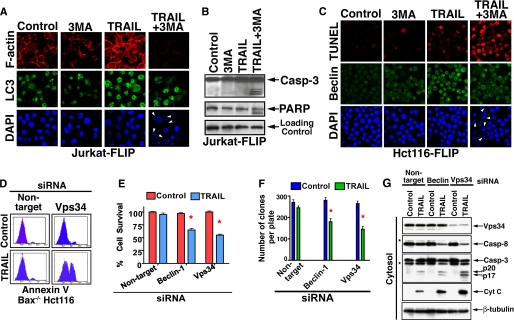
Inhibition of TRAIL-mediated autophagy by 3MA or vps34 RNAi induces apoptosis. A and B, inhibition of TRAIL-mediated autophagy in Jurkat-FLIP by 3MA results in nuclei fragmentation (A) and caspase activation (B). A clonal Jurkat cell line stably transfected with FLIP was treated with TRAIL (100 ng/ml) and/or 3MA (10 mm) for 6 h. Jurkat-FLIP cells were assessed by confocal microscopy for the expression of F-actin, LC3, and DAPI (A). Control and treated Jurkat-FLIP cells were lysed and assessed by immunoblotting for the presence of active caspase-3 and PARP cleavage (B). C, inhibition of TRAIL-mediated autophagy in Hct116-FLIP by 3MA results in a marked increase in the presence of TUNEL-positive cells. Hct116-FLIP cells were assessed by confocal microscopy for TUNEL, Beclin-1, and DAPI. The presence of fragmented nuclei in TRAIL/3MA-treated cells is indicated by white arrowheads (A and C, right columns, bottom). D, RNAi of vps34 sensitizes Bax-/- Hct116 cells to TRAIL-mediated apoptosis. The experimental details are as described above. 46% of the cells were Annexin V-positive when treated with vps34 siRNA and TRAIL at 100 ng/ml for 6 h. D-F, TRAIL-apoptotic susceptibility of Bax-/- Hct116 cells treated with Beclin-1 or vps34 siRNA as assessed by loss in cellular ATP content (E) and by a clonogenic assay (F). E, experimental details are as described above, and ATP content was determined by the fluorescent Cell-Titer Glo assay. The data in E (means ± S.E. of quadruplicate determinations) represent one of three experiments with similar results. F, colonies developed during 14-day culture in methylcellulose-based medium (Stem Cell Technologies) were counted using an inverted microscope and gridded screen. The data (means ± S.E. of triplicate 35-mm plates) represent one of three experiments with similar results. The red asterisks indicate a statistically significant difference as compared with control (p < 0.05, Mann-Whitney U test). G, induction of caspase activity and cytochrome c release by vps34 RNAi. The cells utilized in D-F were Dounce-homogenized, separated into cytosolic and mitochondrial fractions, and assessed by immunoblotting for the indicated proteins. The cytosolic fraction is shown, and the asterisks indicate unidentified protein bands. β-Tubulin served as a control for equal loading.
In mammalian cells, Beclin-1 functions as a part of a complex with Class III phosphatidylinositol 3-kinase and Vps34 (35), and this complex is further regulated by UVRAG. Since the classical pharmacological inhibitor of autophagy, 3MA, blocks Vps34 activity (36), we utilized vps34 RNAi knockdown to substitute for the nonselective inhibition by 3MA with a specific targeting agent. Indeed, we observed a significant apoptotic shift in the response of Bax-/- Hct116 cells to TRAIL (Fig. 6D). The apoptotic shift by vps34 RNAi was further confirmed by cellular ATP loss (Fig. 6E), by reduced cell clonogenicity (Fig. 6F), and by induced caspase activity and cytochrome c release (Fig. 6G).
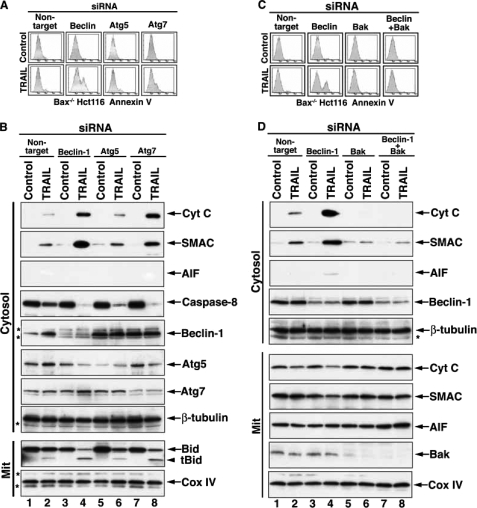
To further assess if mitochondrial apoptotic involvement in response to the knockdown of autophagic genes is unique to components of the Beclin-1·UVRAG·Vps34 complex that is involved in the autophagy initiation phase, we performed similar experiments with the knockdown of atg5 and atg7. Interestingly, atg7 knockdown and, to a lesser extent, the knockdown of atg5 also rendered TRAIL sensitivity to certain Bax-/- Hct116 cells, as demonstrated by the presence of Annexin V-positive cells (Fig. 7A). Furthermore, Bax-/- Hct116 cells responded to TRAIL by the release of cytochrome c and SMAC, but not AIF, when beclin-1 or atg7 was knocked down (Fig. 7B). In accordance with its induction of only a low level of phosphatidylserine exposure, RNAi of atg5 caused a reduced level of release of cytochrome c and SMAC, as compared with that mediated by beclin-1 or atg7 RNAi. These results suggest that inhibition of autophagy by targeting certain essential components of this process overcomes Bax deficiency as a TRAIL resistance mechanism.
FIGURE 7.
Apoptotic response of Bax-/- Hct116 cells to TRAIL is enabled by inhibition of autophagy and is Bak-dependent. A, RNAi of beclin-1 or atg7 (and to a lesser extent atg5) sensitizes Bax-/- Hct116 cells to TRAIL-mediated apoptosis. The cells were treated with the indicated siRNAs and TRAIL as described under “Experimental Procedures.” The cells were then assessed by flow cytometry for the presence of Annexin V-positive cells. B, TRAIL-mediated release of mitochondrial cytochrome c and SMAC in Bax-/- Hct116 cells that underwent RNAi for beclin-1, atg5, or atg7. The cell extracts obtained from cells treated with the indicated siRNAs and TRAIL were fractionated to obtain mitochondrial and cytosolic fractions. Evidence for the knockdown of beclin-1 is shown in lanes 3 and 4, for atg5 in lanes 5 and 6, and for atg7 in lanes 7 and 8. Atg5 has been reported to be cleaved during the apoptotic response, which helps explain its down-regulation in cells treated with TRAIL and either beclin-1 (lane 4) or atg7 (lane 8) siRNAs that have been demonstrated by us to shift the autophagic response to apoptosis. C, bak RNAi blocks the TRAIL-mediated phosphatidylserine exposure in cells treated with beclin-1 siRNA. Bax-/- Hct116 cells were treated with nontargeting, beclin-1, bak, or combined beclin-1 and bak siRNAs and then with TRAIL. The cells were then assessed by flow cytometry for staining by Annexin-V-fluorescein isothiocyanate. D, cytochrome c release in response to TRAIL and beclin-1 RNAi in Bax-/- Hct116 cells is inhibited by Bak RNAi. Bax-/- Hct116 cells were treated with the various siRNAs and TRAIL as described in C. Cell extracts were then prepared and separated into mitochondrial and cytosolic fractions. The fractions were assessed by immunoblotting for the indicated proteins. β-Tubulin serves as a loading control for the cytosolic fraction, and Cox IV serves as loading control for the mitochondrial fractions. Proof for effective knockdown of Beclin-1 is shown in the cytosol fraction; proof for effective bak RNAi is demonstrated in the mitochondrial fraction. The asterisks indicate unidentified protein bands.
Molecular Elucidation of the Cellular Players Involved in the Autophagy-to-Apoptosis Shift in Response to TRAIL—Since cytochrome c release is mainly Bax/Bak-dependent, we investigated the possibility that (in the absence of Bax) mitochondrial Bak is activated by the specific inhibition of autophagy employed in these experiments. To this end, we assessed whether the knockdown of bak in Bax-/- Hct116 cells attenuates the release of cytochrome c in response to a combined treatment with TRAIL and beclin-1 siRNA. Indeed, bak knockdown completely blocked TRAIL-mediated apoptosis that was enabled by beclin-1 siRNA treatment in Bax-/- cells (Fig. 7C). Furthermore, it also completely inhibited the beclin-1 RNAi-enabled mitochondrial release of cytochrome c and SMAC in response to TRAIL in the Bax-/- Hct116 cells (Fig. 7D). These results suggest that beclin-1 RNAi enables mitochondrial Bak activation in response to TRAIL in Hct116 Bax-/- cells, implying that there is a cross-talk (direct or indirect) between Beclin-1 and Bak. Beclin-1 is mainly a cytosolic protein, but it is also associated with mitochondria and the nuclei (30, 37). Thus, the cross-talk may take place either upstream or at the mitochondrial level. We employed co-immunoprecipitation experiments to investigate whether the cross-talk between Beclin-1 and Bak is mediated by a direct binding. We did not observe co-immunoprecipitation of Bak and Beclin-1 from either quiescent Hct116 Bax-/- cells or following TRAIL treatment (the immunoprecipitation was performed from either whole cell lysates or purified mitochondria). Therefore, we conclude that the cross-talk between Beclin-1 and Bak is indirect (data not shown).
To investigate whether the cross-talk between beclin-1 knockdown and mitochondrial bak activation involves caspase activity, we assessed whether the silencing of atg genes has an enhancing effect on caspase-8 activity. Indeed, we detected increased caspase-8 activity upon RNAi silencing of beclin-1, atg7, and also atg5 (Fig. 7B). Concomitantly, we also detected an increased presence of tBid in the corresponding mitochondrial pellets.
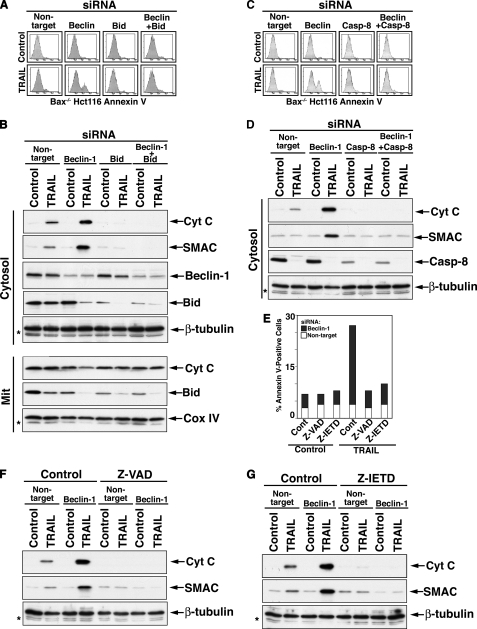
To investigate the potential involvement of Bid upstream of mitochondrial Bak activation, we assessed the effects of its knockdown on the autophagy-to-apoptosis shift in response to TRAIL following beclin-1 knockdown. bid RNAi significantly inhibited the apoptotic response to TRAIL of Bax-/- Hct116 cells that was enabled by beclin-1 RNAi (Fig. 8A). Since Bid cleavage could be mediated by caspase activation downstream of mitochondria, we also assessed the effect of bid knockdown on the mitochondrial release of apoptogenic proteins in response to TRAIL following beclin-1 RNAi. The inhibition of cytochrome c and SMAC release by bid knockdown suggests that the involvement of Bid is upstream of mitochondria (Fig. 8B). To further investigate whether the beclin-1 knockdown-mediated autophagy-to-apoptosis shift in response to TRAIL was induced by upstream caspase activity, we assessed the effects of caspase-8 RNAi as well as that of pharmacological caspase inhibitors on the execution of such a shift. Thus, inhibition of caspase-8 by either caspase-8 RNAi, Z-IETD-fmk, a selective caspase-8 inhibitor, or Z-VAD-fmk, a broad range caspase inhibitor, inhibited the execution of the beclin-1 RNAi/TRAIL shift from autophagy to apoptosis (Fig. 8, C-G). These findings suggest that Beclin-1 down-regulation enhances the level of TRAIL-mediated caspase activity upstream of mitochondria to a level that is sufficient for the tBid-dependent engagement of Bak in the absence of Bax. The exact molecular mechanism responsible for the enhanced caspase-8 activity in this experimental setting remains to be determined and is currently under investigation.
FIGURE 8.
Involvement of Bid and caspase activity upstream of mitochondria in the execution of the shift from TRAIL-mediated autophagy to apoptosis through beclin-1 RNAi. A, a role for Bid in the execution of the autophagy-to-apoptosis shift in the response of Bax-/- Hct116 cells to beclin-1 RNAi and TRAIL. Bax-/- Hct116 cells were treated as described in the legend to Fig. 7C but with bid siRNA instead of bak siRNA. B, inhibition of beclin-1 RNAi/TRAIL-mediated cytochrome c release by bid knockdown. Experimental details are similar to those described in the legend to Fig. 7D. C and D, inhibition of beclin-1 RNAi/TRAIL-mediated apoptosis and cytochrome c release by caspase-8 knockdown. Experimental details are similar to those described for A and B. E, inhibition of beclin-1 RNAi/TRAIL mediated-apoptosis by pharmacological inhibitors of caspase activity. Bax-/- Hct116 cells were treated with beclin-1 siRNA for 40 h and then with TRAIL in the presence of Z-VAD-FMK or Z-IETD-FMK (50 μm each, added 30 min prior to TRAIL addition). The cells were assessed by flow cytometry for the presence of Annexin V-positive cells. Results from one of three performed experiments with similar results are shown. F and G, inhibition of beclin-1 RNAi/TRAIL-mediated-cytochrome c release by pharmacological inhibitors of caspase activity. The extracts of cells described in E were separated into mitochondrial and cytosolic fractions. The immunoblot analysis of the cytosolic fractions is shown.
DISCUSSION
The current studies demonstrate the involvement of protective autophagy in response to TRAIL of tumor cells with various apoptotic defects. In particular, they demonstrate the existence of a double switch between autophagy and apoptosis that can be exploited for TRAIL therapy. On one hand, inhibition of apoptosis by an array of apoptosis defects can invoke protective autophagy in response to TRAIL, representing a switch from default apoptosis to protective autophagy; on the other hand, prevention of such protective autophagy induces apoptotic cell death, representing a second switch from autophagy back to apoptosis. The possibility of potentially reversing multiple mechanisms of TRAIL resistance by the inhibition of autophagy represents a novel concept for the development of a new approach for TRAIL therapy.
Induction of autophagy via TRAIL-Rs has been previously observed (38, 39), but its cytoprotective effect has not yet been reported. Tumor TRAIL resistance is mediated by a variety of defects that block various phases of the apoptotic cascade, including the initial signal transduction, activation of initiator caspases, mitochondrial events, and activation of effector caspases. Utilizing two distinctive types of cancer cells (leukemic and colon carcinoma) with two divergent blocks at distinct phases of the TRAIL-mediated apoptotic pathway, overexpression of FLIP (DISC level) or Bax deficiency (mitochondrial level), we observed the induction of protective autophagy in response to TRAIL. The importance of an autophagic response to TRAIL in apoptosis-defective tumor cells derives from the generation of a new platform for the reversal of TRAIL resistance. Our studies demonstrate that targeting autophagy can serve as a mechanism for the circumvention of TRAIL resistance in tumor cells with different apoptosis defects. It has been suggested that an ultimate block in the apoptosis machinery, such as that mediated by a Bax/Bak double deficiency, shifts the mechanism of cell death to necrosis or another nonapoptotic death mechanism. However, the combined knock-out of Bax and bak, which was proven to serve as a highly important experimental model to decipher the molecular mechanisms of cell death, is not physiologic. Bax frameshift mutations due to defects in DNA mismatch repair have been described as a mechanism for apoptosis resistance in colorectal carcinoma and certain hematological malignancies (40, 41). In contrast, only a few bak mutations have been noted in tumor cells, whereas a combined deficiency of these proapoptotic proteins has not been reported to occur naturally. Thus, apoptosis defects acquired during tumorigenesis are usually cascade-specific and do not block all apoptotic avenues available in the cell. Therefore, as demonstrated in our studies, the inhibition of TRAIL-induced autophagy in cells with a certain defect(s) in their apoptotic machinery, would still allow for a shift to an unblocked cellular apoptotic cascade(s).
TRAIL-mediated activation of Bak in Bax-deficient Hct116 cells has not been previously reported. Bak in Bax-/- mitochondria has been suggested to be down-regulated in response to TRAIL alone (42) and up-regulated in response to co-treatment with other apoptotic agents (19). In the current study, beclin-1 knockdown alone did not activate Bak; nor did it induce any caspase activity. In contrast, TRAIL treatment of these cells induced apoptotic events upstream of mitochondria, including caspase-8 activation, Bid cleavage, and processing of caspase-3 to its p20 subunit. However, the level of these apoptotic events upstream of mitochondria appears to be below the threshold required for Bak activation, since the Bax-deficient cells maintained their TRAIL resistance. Our analysis of the autophagy-to-apoptosis shift suggests that a combined treatment consisting of beclin-1 RNAi and TRAIL that activates Bak proceeds via a mechanism that is dependent on Bid and caspase-8 upstream of mitochondria. Therefore, it appears that inhibition of the autophagic process by RNAi of beclin-1, vps34, atg7, or atg5 enhances the level of TRAIL-activated caspase-8. These findings suggest that autophagy or certain components of this process keep in check the level of caspase activity upstream of mitochondria. Elimination of such a cap on caspase activity upstream of mitochondria is enabled by the knockdown of beclin-1 or vps34 or by the general inhibition of the autophagic process. Autophagy itself is negatively regulated by a yet unresolved mechanism during full-blown apoptosis; therefore, the negative regulation of caspase activity by autophagy is not apparent in apoptosis-competent tumor cells. The exact molecular mechanism responsible for the augmentation in caspase activity by the knockdown of autophagic proteins remains to be elucidated.
Since tumor cells accumulate a multitude of apoptotic mutations during tumorigenesis, the TRAIL-resistant tumor cell population is expected to be composed of cells with heterogeneous apoptotic defects. If indeed, as our data suggest, various mechanisms of apoptosis resistance shift the default TRAIL response from apoptosis to autophagy, then a common approach for inhibition of autophagy may be suitable to circumvent multiple mechanisms of TRAIL resistance.
In TRAIL-mediated autophagic protection, we observed cytoskeleton changes that were not previously reported in starvation-induced autophagy. In particular, we observed up-regulated F-actin polymerization that was required for the protection effect. The network of F-actin is generated through the activity of the Arp2/3 complex (43). The Arp2/3 complex is regulated by both the Wiscott-Aldrich syndrome protein (WASP) family of proteins and Cortactin (43, 44). We determined the requirement for F-actin polymerization in the TRAILmediated protective response in apoptosis-defective cells by the knockdown of cortactin or arp2/3 that shifted the protective response to caspase-mediated apoptosis. During the revision of this manuscript, Klionsky and co-workers (45) reported that in yeast, Arp2 links the autophagic machinery to the actin cytoskeleton. In particular, they demonstrate that Arp2 regulates the dynamics of Atg9, an integral membrane protein that cycles between intracellular compartments and the vesicle nucleation site (45). Our current findings on the involvement of F-actin and Arp2 in the response of apoptosis-defective cells to TRAIL or starvation are the first to demonstrate cytoskeleton changes in mammalian autophagy. The recent report of Klionsky and co-workers (45) strengthens our conclusion that at least in certain tumor cells, the up-regulation in F-actin polymerization is associated with an autophagic flux.
In summary, these studies suggest that apoptosis-defective tumor cells can survive TRAIL-mediated stress by invoking a protective autophagic process associated with enhanced F-actin polymerization. These findings may serve as the basis for a novel therapeutic approach to potentiate TRAIL efficacy by the inhibition of autophagy. Future studies to elucidate the molecular mechanisms responsible for the shift from protection to apoptosis or to another mechanism of cell death should provide important insights into specific approaches that can toggle between survival and death mechanisms.
Supplementary Material
Acknowledgments
We thank Bert Vogelstein for the clonal cell line Bax-/- Hct116, Jorg Tschopp for the Jurkat-Mock and Jurkat-FLIP clonal cell lines and plasmid pCR3.V64-MET-FLAG-FLIP-L, and Tamotsu Yoshimori for the GFP-LC3 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 CA109285 and RO1 CA111786. This work was also supported by Department of Defense Grant BC063415 (to H. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: Z, benzyloxycarbonyl; IL, interleukin; 3MA, 3-methyladenine; DISC, death-inducing signaling complex; Ab, antibody; DAPI, 4′,6-diamidino-2-phenylindole; fmk, fluoromethyl ketone; TUNEL, terminal dUTP nick-end labeling; RNAi, RNA interference; WT, wild type; MOPS, 4-morpholinepropanesulfonic acid; TRAIL-R, TRAIL receptor.
References
- 1.Levine, B. (2007) Nature 446 745-747 [DOI] [PubMed] [Google Scholar]
- 2.Kondo, Y., Kanzawa, T., Sawaya, R., and Kondo, S. (2005) Nat. Rev. Cancer 5 726-734 [DOI] [PubMed] [Google Scholar]
- 3.Baehrecke, E. H. (2005) Nat. Rev. Mol. Cell Biol. 6 505-510 [DOI] [PubMed] [Google Scholar]
- 4.Levine, B., and Yuan, J. (2005) J. Clin. Invest. 115 2679-2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maiuri, M. C., Zalckvar, E., Kimchi, A., and Kroemer, G. (2007) Nat. Rev. Mol. Cell Biol. 8 741-752 [DOI] [PubMed] [Google Scholar]
- 6.Yu, L., Alva, A., Su, H., Dutt, P., Freundt, E., Welsh, S., Baehrecke, E. H., and Lenardo, M. J. (2004) Science 304 1500-1502 [DOI] [PubMed] [Google Scholar]
- 7.Shimizu, S., Kanaseki, T., Mizushima, N., Mizuta, T., Arakawa-Kobayashi, S., Thompson, C. B., and Tsujimoto, Y. (2004) Nat. Cell Biol. 6 1221-1228 [DOI] [PubMed] [Google Scholar]
- 8.Lum, J. J., Bauer, D. E., Kong, M., Harris, M. H., Li, C., Lindsten, T., and Thompson, C. B. (2005) Cell 120 237-248 [DOI] [PubMed] [Google Scholar]
- 9.Boya, P., Gonzalez-Polo, R. A., Casares, N., Perfettini, J. L., Dessen, P., Larochette, N., Metivier, D., Meley, D., Souquere, S., Yoshimori, T., Pierron, G., Codogno, P., and Kroemer, G. (2005) Mol. Cell Biol. 25 1025-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Polo, R. A., Boya, P., Pauleau, A. L., Jalil, A., Larochette, N., Souquere, S., Eskelinen, E. L., Pierron, G., Saftig, P., and Kroemer, G. (2005) J. Cell Sci. 118 3091-3102 [DOI] [PubMed] [Google Scholar]
- 11.Amaravadi, R. K., Yu, D., Lum, J. J., Bui, T., Christophorou, M. A., Evan, G. I., Thomas-Tikhonenko, A., and Thompson, C. B. (2007) J. Clin. Invest. 117 326-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carew, J. S., Nawrocki, S. T., Kahue, C. N., Zhang, H., Yang, C., Chung, L., Houghton, J. A., Huang, P., Giles, F. J., and Cleveland, J. L. (2007) Blood 110 313-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almasan, A., and Ashkenazi, A. (2003) Cytokine Growth Factor Rev. 14 337-348 [DOI] [PubMed] [Google Scholar]
- 14.French, L. E., and Tschopp, J. (1999) Nat. Med. 5 146-147 [DOI] [PubMed] [Google Scholar]
- 15.Huang, Y., and Sheikh, M. S. (2007) Toxicol. Appl. Pharmacol. 224 284-289 [DOI] [PubMed] [Google Scholar]
- 16.Kischkel, F. C., Lawrence, D. A., Chuntharapai, A., Schow, P., Kim, K. J., and Ashkenazi, A. (2000) Immunity 12 611-620 [DOI] [PubMed] [Google Scholar]
- 17.Sprick, M. R., Weigand, M. A., Rieser, E., Rauch, C. T., Juo, P., Blenis, J., Krammer, P. H., and Walczak, H. (2000) Immunity 12 599-609 [DOI] [PubMed] [Google Scholar]
- 18.Deng, Y., Lin, Y., and Wu, X. (2002) Genes Dev. 16 33-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeBlanc, H., Lawrence, D., Varfolomeev, E., Totpal, K., Morlan, J., Schow, P., Fong, S., Schwall, R., Sinicropi, D., and Ashkenazi, A. (2002) Nat. Med. 8 274-281 [DOI] [PubMed] [Google Scholar]
- 20.Ravi, R., and Bedi, A. (2002) Cancer Res. 62 1583-1587 [PubMed] [Google Scholar]
- 21.Cheng, J., Hylander, B. L., Baer, M. R., Chen, X., and Repasky, E. A. (2006) Mol. Cancer Ther. 5 1844-1853 [DOI] [PubMed] [Google Scholar]
- 22.Zhang, L., and Fang, B. (2005) Cancer Gene Ther. 12 228-237 [DOI] [PubMed] [Google Scholar]
- 23.Kataoka, T., Schroter, M., Hahne, M., Schneider, P., Irmler, M., Thome, M., Froelich, C. J., and Tschopp, J. (1998) J. Immunol. 161 3936-3942 [PubMed] [Google Scholar]
- 24.Han, J., Goldstein, L. A., Hou, W., and Rabinowich, H. (2007) J. Biol. Chem. 282 16223-16231 [DOI] [PubMed] [Google Scholar]
- 25.Han, J., Goldstein, L. A., Gastman, B. R., Froelich, C. J., Yin, X. M., and Rabinowich, H. (2004) J. Biol. Chem. 279 22020-22029 [DOI] [PubMed] [Google Scholar]
- 26.Han, J., Goldstein, L. A., Gastman, B. R., Rabinovitz, A., and Rabinowich, H. (2005) J. Biol. Chem. 280 16383-16392 [DOI] [PubMed] [Google Scholar]
- 27.Han, J., Goldstein, L. A., Gastman, B. R., and Rabinowich, H. (2006) J. Biol. Chem. 281 10153-10163 [DOI] [PubMed] [Google Scholar]
- 28.Degenhardt, K., Mathew, R., Beaudoin, B., Bray, K., Anderson, D., Chen, G., Mukherjee, C., Shi, Y., Gelinas, C., Fan, Y., Nelson, D. A., Jin, S., and White, E. (2006) Cancer Cell 10 51-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanida, I., Minematsu-Ikeguchi, N., Ueno, T., and Kominami, E. (2005) Autophagy 1 84-91 [DOI] [PubMed] [Google Scholar]
- 30.Liang, X. H., Yu, J., Brown, K., and Levine, B. (2001) Cancer Res. 61 3443-3449 [PubMed] [Google Scholar]
- 31.Mizushima, N., and Yoshimori, T. (2007) Autophagy 3 542-545 [DOI] [PubMed] [Google Scholar]
- 32.Klionsky, D. J., Abeliovich, H., Agostinis, P., Agrawal, D. K., Aliev, G., Askew, D. S., Baba, M., Baehrecke, E. H., Bahr, B. A., Ballabio, A., Bamber, B. A., Bassham, D. C., Bergamini, E., Bi, X., Biard-Piechaczyk, M., et al. (2008) Autophagy 4 151-17518188003 [Google Scholar]
- 33.Goley, E. D., and Welch, M. D. (2006) Nat. Rev. Mol. Cell Biol. 7 713-726 [DOI] [PubMed] [Google Scholar]
- 34.Weed, S. A., and Parsons, J. T. (2001) Oncogene 20 6418-6434 [DOI] [PubMed] [Google Scholar]
- 35.Furuya, N., Yu, J., Byfield, M., Pattingre, S., and Levine, B. (2005) Autophagy 1 46-52 [DOI] [PubMed] [Google Scholar]
- 36.Petiot, A., Ogier-Denis, E., Blommaart, E. F., Meijer, A. J., and Codogno, P. (2000) J. Biol. Chem. 275 992-998 [DOI] [PubMed] [Google Scholar]
- 37.Maiuri, M. C., Le Toumelin, G., Criollo, A., Rain, J. C., Gautier, F., Juin, P., Tasdemir, E., Pierron, G., Troulinaki, K., Tavernarakis, N., Hickman, J. A., Geneste, O., and Kroemer, G. (2007) EMBO J. 26 2527-2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills, K. R., Reginato, M., Debnath, J., Queenan, B., and Brugge, J. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3438-3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, K. J., Lee, S. H., Kim, T. I., Lee, H. W., Lee, C. H., Kim, E. H., Jang, J. Y., Choi, K. S., Kwon, M. H., and Kim, Y. S. (2007) Cancer Res. 67 7327-7334 [DOI] [PubMed] [Google Scholar]
- 40.Abdel-Rahman, W. M., Georgiades, I. B., Curtis, L. J., Arends, M. J., and Wyllie, A. H. (1999) Oncogene 18 2139-2142 [DOI] [PubMed] [Google Scholar]
- 41.Brimmell, M., Mendiola, R., Mangion, J., and Packham, G. (1998) Oncogene 16 1803-1812 [DOI] [PubMed] [Google Scholar]
- 42.Ricci, M. S., Kim, S. H., Ogi, K., Plastaras, J. P., Ling, J., Wang, W., Jin, Z., Liu, Y. Y., Dicker, D. T., Chiao, P. J., Flaherty, K. T., Smith, C. D., and El-Deiry, W. S. (2007) Cancer Cell 12 66-80 [DOI] [PubMed] [Google Scholar]
- 43.Weed, S. A., Karginov, A. V., Schafer, D. A., Weaver, A. M., Kinley, A. W., Cooper, J. A., and Parsons, J. T. (2000) J. Cell Biol. 151 29-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothschild, B. L., Shim, A. H., Ammer, A. G., Kelley, L. C., Irby, K. B., Head, J. A., Chen, L., Varella-Garcia, M., Sacks, P. G., Frederick, B., Raben, D., and Weed, S. A. (2006) Cancer Res. 66 8017-8025 [DOI] [PubMed] [Google Scholar]
- 45.Monastyrska, I., He, C., Geng, J., Hoppe, A. D., Li, Z., and Klionsky, D. J. (2008) Mol. Biol. Cell. 19 1962-1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.