Abstract
Structure maintenance of chromosome 1 (SMC1) is phosphorylated by ataxia telangiectasia-mutated (ATM) in response to ionizing radiation (IR) to activate intra-S phase checkpoint. A role of CK2 in DNA damage response has been implicated in many previous works, but the molecular mechanism for its activation is not clear. In the present work, we report that SMC3 is phosphorylated at Ser-1067 and Ser-1083 in vivo. Ser-1083 phosphorylation is IR-inducible, depends on ATM and Nijmegen breakage syndrome 1 (NBS1), and is required for intra-S phase checkpoint. Interestingly, Ser-1067 phosphorylation is constitutive and is not induced by IR but also affects intra-S phase checkpoint. Phosphorylation of Ser-1083 is weakened in cells expressing S1067A mutant, suggesting interplay between Ser-1067 and Ser-1083 phosphorylation in DNA damage response. Consistently, small interfering RNA knockdown of CK2 leads to attenuated phosphorylation of Ser-1067 as well as intra-S phase checkpoint defect. Our data provide evidence that phosphorylation of a core cohesin subunit SMC3 by ATM plays an important role in DNA damage response and suggest that a constitutive phosphorylation by CK2 may affect intra-S phase checkpoint by modulating SMC3 phosphorylation by ATM.
DNA damage response (DDR)3 is a signal transduction pathway that coordinates cell cycle arrest, DNA repair, and apoptosis in the presence of damaged DNA (1). Genetic and biochemical research has established a conceptual framework for DNA damage at the molecular level that includes sensors, transducers, and effectors (2). Loss or mutation of DDR genes causes many cancer-prone disorders. Thus, understanding of DDR has a profound impact on mechanisms of cancer development and treatment.
In response to double-stranded DNA breaks (DSB), the central checkpoint kinase ATM activates the G1, intra-S, and G2/M checkpoints by phosphorylation of downstream effectors. The central theme of DDR is to turn dormant proteins into active effectors to execute the cell cycle arrest, DNA repair, and apoptosis; phosphorylation is a major mechanism for such activation. How constitutively active proteins contribute to checkpoint control is enigmatic. The CK2 kinase is such an example. CK2 has long been implicated in DDR, but its role in regulation of cell cycle checkpoint is not well understood. CK2 is defined as a messenger-independent protein kinase found in complex with two catalytic subunits (α and/or α′) and two regulatory β subunits (3). In contrast to ATM, CK2 is an essential and abundant kinase that is constitutively active in the absence of exogenous DNA damage. CK2 regulates many seemingly unrelated cellular processes including cell division (4), transcription (5), DNA repair (6, 7), proliferation (8), and apoptosis (9, 10). CK2 phosphorylates proteins with a consensus sequence of (S/T)XX(D/E) (where X denotes any amino acid) and is active toward almost any protein with this consensus in vitro. As a result, more than 300 in vitro substrates are known (11). Accumulating biochemical and genetic data implicate CK2 in DDR. In response to DNA damage, CK2 regulates polymerase I/III gene expression (12), phosphorylates histone H4 (13), and controls checkpoint adaptation (14). In mammalian cells, CK2 phosphorylates the tumor suppressor p53 in response to UV (15). CK2 is important for DNA single strand break repair (6) as it phosphorylates the key single strand break repair protein XRCC1 to create docking sites for binding of Aprataxin that contains a phospho-peptide binding Forkhead-associated (FHA) domain (7). Phosphorylation of XRCC1 by CK2 is constitutive and is not stimulated by DNA damage.
The failure of the intra-S checkpoint is manifested by the inability of cells to slow down DNA synthesis in response to IR, a phenotype generally referred to as radio-resistant DNA synthesis (RDS). One of the intra-S checkpoint pathways is mediated by Mre11/Rad50/NBS1 (MRN)-SMC1 (16–18). SMC1 phosphorylation at Ser-957 and Ser-966 is induced by IR in an ATM- and NBS1-dependent manner, which is important for IR sensitivity and suppression of RDS. SMC1 associates with SMC3, and together with SCC-1/Rad21 and SCC-3 (including SA-1 and SA-2 in human), form the cohesin complex (19, 20). The cohesin complex is indispensable for many, if not all, aspects of chromosomal metabolism, including sister chromatid cohesion in the S and G2 phase of the cell cycle (21), DNA repair (22, 23), and gene transcription (24). Since SMC1 is an important effector in the intra S-phase checkpoint pathway, we are interested in whether other cohesion subunits are also involved. We herein report that SMC3 is phosphorylated at Ser-1067 and Ser-1083. Although Ser-1083 phosphorylation is IR-inducible and depends on ATM, Ser-1067 phosphorylation is constitutive and depends on CK2 but is not further enhanced by IR. However, both Ser-1067 phosphorylation and Ser-1083 phosphorylation are required for intra-S phase checkpoint. Furthermore, phosphorylation of SMC3 at Ser-1083 is weakened in cells expressing SMC3-S1067A. Thus, the CK2 kinase may affect IR-induced S-phase checkpoint through SMC3 Ser-1083 phosphorylation.
MATERIALS AND METHODS
Plasmids, Recombinant Proteins, and Generation of Stable Cell Line—Full-length cDNA encoding wild-type human SMC3 was generated by PCR using a cDNA pool (reverse transcription products from RNA insolated from HeLa S3 cells) as a template. PCR product was cloned into pcDNA3.1D/V5-His-TOPO (Invitrogen) to produce C terminus V5 tagged pcDNA-SMC3 plasmid. Subsequently, site-directed mutagenesis was performed to substitute alanine to serine (SMC3-S1067A, SMC3-S1083A) by using the QuikChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA). The resulting mutant plasmid was confirmed by DNA sequencing. Plasmid encoding GST-SMC3 (amino acids 881–1218) was cloned into pGEX-4T-1 (Amersham Biosciences). The fusion protein was expressed and purified according to the standard procedures. Stable cell lines that harbor a “tet-on” promoter to regulate SMC3-WT, SMC3-S1067A, and SMC3-S1083A expression were generated using “Flp-In™ T-Rex™” system (Invitrogen). Two tandem FLAG tags were added to the C terminus of SMC3 to distinguish them from endogenous SMC3 by SDS-PAGE mobility. Forty-eight hours after transfection, hygromycin (200 μg/ml) and blasticidin (10 μg/ml) were added into medium for selection. Single clones were isolated for each SMC3 construct after 3 weeks of selection. FLAG-SMC3 expression was induced by doxycycline at a concentration of 1 μg/ml. Clones that express comparable exogenous FLAG-SMC3 were used for functional analysis. SV40 T-antigen immortalized NBS1 fibroblasts were established from an NBS patient (25). NBS1-LBI cell lines with retroviral expression of NBS1-WT were produced as described (16).
Cell Culture, Antibodies, in Vitro Kinase Assay and Transfections—Human cervical cancer cell line HeLa cells and human embryonic kidney cell line 293T cells were purchased from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Rabbit polyclonal SMC3, V5, CK2α, CK2α′, and pS1083-SMC3 phospho-antibodies were from Bethyl Laboratories. ATM and NBS1 antibodies were from GeneTex (San Antonio, TX). Immunoprecipitation and Western blotting were carried out as described (26). In vitro kinase assays were carried out as described (27).
Small interference RNA (siRNA) for ATM, CK2α, CK2α′, and Vimentin were purchased from Dharmacon Research as SMARTPOOLs. HeLa cells were transfected with siRNAs by Oligofectamine (Invitrogen) according to the manufacturer's protocol. Seventy-two hours after transfection, cells were treated with IR and recovered for 1 h or for the indicated times before harvest. Transient transfection of plasmids encoding SMC3 and ATM was carried out in 293T cells using Lipofectamine (Invitrogen) and calcium phosphate, respectively. Forty to sixty hours after transfection, cells were treated with IR and recovered for 1 h before harvest.
RDS Assay—RDS assay was carried out as described (16). Briefly, 293T stable cell lines expressing WT, S1067A, and S1083A SMC3 were labeled with 10 nCi/ml of [14C]thymidine (PerkinElmer Life Sciences) for 36 h to normalize the total amount of DNA among different samples. Cells were irradiated with 10 Gy of IR and recovered for 1 h. They were then pulse-labeled with 1 μCi/ml [3H]thymidine (PerkinElmer Life Sciences) for 30 min, washed with PBS, fixed with methanol, and lysed with 0.5 m NaOH. The lysates were counted in a liquid scintillation counter. Radio-resistant DNA synthesis was calculated using the ratio of radioactivity of 3H/14C. Overlapping 3H and 14C emissions were corrected with quenched 3H and 14C standards. Five replicas were measured for each sample.
Identification of Phosphorylation Sites Using Mass Spectrometry—Identification of phosphorylation sites with mass spectrometry was carried out as described previously with minor modifications (28). Both a MALDI (matrix-assisted laser desorption) linear ion trap mass spectrometer (vMALDI-LTQ, Thermal Finnigan, San Jose, CA) and a capillary HPLC (75-μm ID column)-electrospray linear ion trap mass spectrometer (Proteomix, Thermal Finnigan) were used. The cohesin complex was immunoprecipitated using antibodies against SMC1 or SMC3 from nuclear extract made from HeLa S3 cells 2 h after 20 Gy of γ-irradiation and resolved in a 4–20% SDS-PAGE. Trypsin digests of SMC1, SMC3, and SCC3 (they co-migrate on the SDS-PAGE gel), and SCC1 bands were subject to mass spectrometric analysis to identify phosphorylation sites as described previously (28).
RESULTS
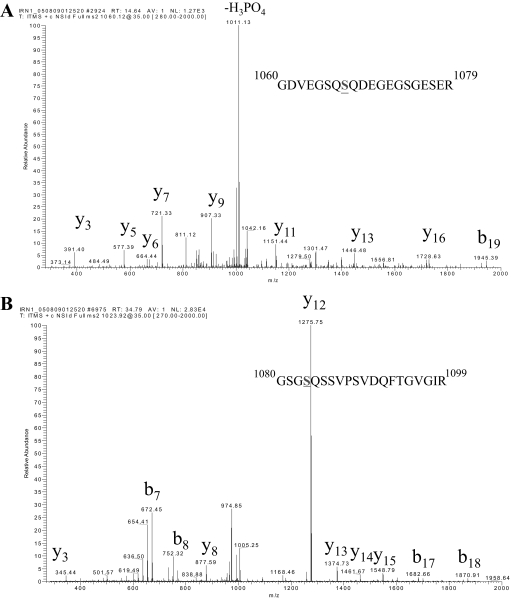
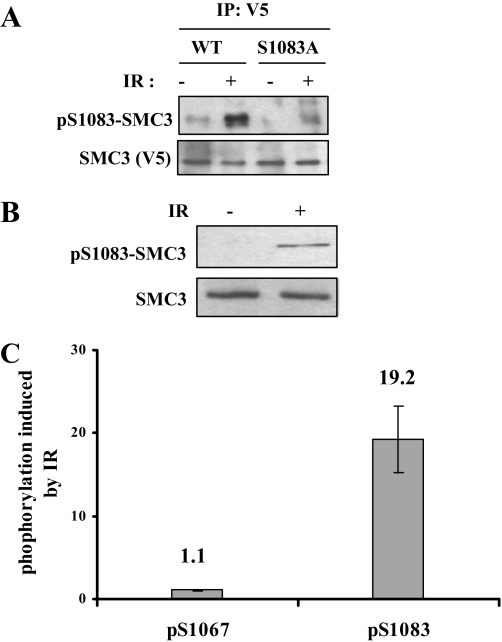
Mapping SMC3 Phosphorylation Sites—We used mass spectrometry to analyze phosphorylation of the cohesin complex, which includes SMC1, SMC3, SCC-1, and SCC-3 (including SA-1 and SA-2), in cycling and IR-treated cells. The cohesin complex was immunoprecipitated with SMC1 antibody from HeLa cells treated with 20 Gy of IR and recovered for 2 h. The immunoprecipitates were resolved on SDS-PAGE. Tryptic peptides resulting from in-gel digestion of the individual cohesin subunit were exhaustively analyzed with mass spectrometry. To achieve a more thorough analysis, we used both MALDI-linear ion trap mass spectrometry (vMALDI-LTQ) and capillary HPLC-electrospray-linear ion trap mass spectrometry. In addition to the previously reported Ser-957 and Ser-966 of SMC1, Ser-1067 and Ser-1083 of SMC3 were also phosphorylated (Fig. 1, A and B). We raised a phospho-specific antibody against pS1083 of SMC3. As shown in Fig. 2A, this antibody specifically recognized phosphorylated SMC3 at Ser-1083 as the S1083A mutation abolished the Western blot signal. Using this antibody, we confirmed that endogenous SMC3 Ser-1083 phosphorylation is induced by IR (Fig. 2B).
FIGURE 1.
SMC3 is phosphorylated at Ser-1067 and Ser-1083. MS/MS spectra that identify Ser-1067 (A) and Ser-1083 (B) as phosphorylated. SMC3 was immunoprecipitated from nuclear extracts made from HeLa cells and further isolated on SDS-PAGE. The SMC3 band was in-gel digested with trypsin and analyzed with capillary liquid chromatography-MS/MS.
FIGURE 2.
SMC3 is phosphorylated at Ser-1067 and Ser-1083 in cycling and IR-treated cells. A, characterization of pS1083-SMC3 phospho-specific antibody. V5-SMC3-WT and V5-SMC3-S1083A were transfected in 293T cells, and cells were irradiated with 10 Gy IR. SMC3 was immunoprecipitated (IP) with a V5 antibody and Western blotted with pS1083-SMC3 antibody. B, phosphorylation of the endogenous SMC3 at Ser-1083. HeLa cells were irradiated with 10 Gy of IR and allowed to recover for 1 h. Whole cell lysates were Western blotted with antibodies against pS1083-SMC3 and SMC3. C, quantitative mass spectrometric measurement of Ser-1067 and Ser-1083 phosphorylation induction in response to IR. HeLa cells were labeled with light or heavy isotope and treated with or without IR, respectively. Equal amounts of two isotope-labeled cells were mixed for the subsequent processing. Phosphorylation of Ser-1067 and Ser-1083 in light labeled SMC3 (IR-treated) are normalized to those in heavy labeled SMC3 (untreated) to calculate phosphorylation induced by IR.
We noticed in the mass spectrometry measurement that in contrast to that of Ser-1083, the intensity of the phospho-peptide containing Ser-1067 did not change in response to IR. Since we failed to obtain the phospho-specific antibody against pS1067, we carried out quantitative mass spectrometry using stable isotope-labeled amino acids in cultured cells (29) to determine whether phosphorylation of Ser-1067 is IR-inducible (supplemental Fig. 1). Cells were grown in culture medium that contain “light” or “heavy” isotope-labeled Arg and Lys (resulting in mass increases of 10 and 6 Da for Arg- and Lys-containing peptides, respectively). Light isotope-labeled cells were treated with IR, whereas heavy isotope-labeled cells were not treated. They were mixed and then underwent identical manipulation during cell lysis, immunoprecipitation, and MS analysis. Peptides labeled with isotopes are chemically identical but distinguished by mass difference so that their mass spectrometric intensities can be directly compared, and when normalized to an unmodified peptide of different sequence (serving as a loading control), the ratio of induction can be measured. Such a quantitative measurement showed that phosphorylation of Ser-1067 did not increase significantly (∼10%) after IR, whereas phosphorylation of Ser-1083 increases by nearly 20-fold (Fig. 2C). We concluded that Ser-1067 is constitutively phosphorylated and is not further induced by IR.
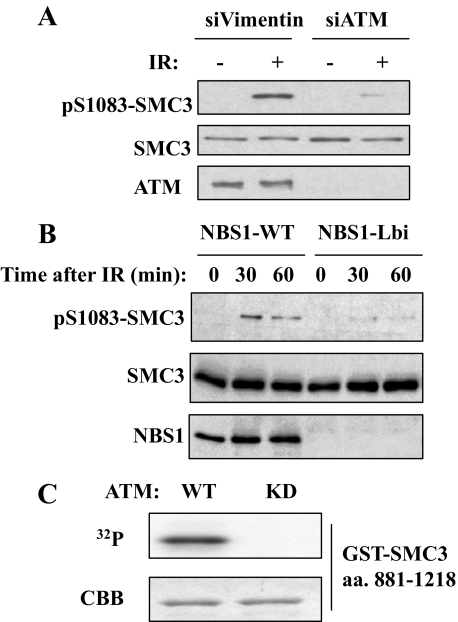
Ser-1083 Phosphorylation Depends on ATM and NBS1—Since Ser-1083 conforms to ATM phosphorylation consensus (30) and its phosphorylation is IR-inducible, we tested whether phosphorylation of Ser-1083 depends on ATM. Knockdown ATM by siRNA significantly reduced Ser-1083 phosphorylation, indicating that Ser-1083 phosphorylation depends on ATM in vivo (Fig. 3A). A similar dependence on ATM was obtained in an immortalized A-T fibroblast cell line that was stably transfected with either a vector (AT22IJE-T) or a fulllength ATM cDNA (pEBS7-YZ5), which has partially restored ATM function (data not shown).
FIGURE 3.
The ATM-NBS1 pathway regulates Ser-1083 phosphorylation of SMC3 in response to IR. A, Western blotting of SMC3 phosphorylation of Ser-1083 in siVimentin- and siATM-transfected in HeLa cells that were irradiated with 10 Gy IR and recovered for 1 h. B, requirement of NBS1 and its phosphorylation for IR-induced phosphorylation of SMC3 at Ser-1083. NBS cells complemented with NBC1 (NBS1-WT) or an empty vector (NBS1-Lbi) were treated with 10 Gy of IR and harvested at the indicated times and Western blotted for SMC3 Ser-1083 phosphorylation. C, in vitro phosphorylation of SMC3 by ATM. FLAG-ATM overexpressed in 293T cells was immunoprecipitated by anti-FLAG antibody and incubated with a GST-SMC3 fragment (amino acids 881–1218) and γ-32P-labeled ATP. In vitro phosphorylation was detected by autoradiography. The amount of SMC3 fragment on SDS-PAGE gel was shown by Coomassie Brilliant Blue (CBB) staining. KD, kinase-dead.
Next, we examined the dependence of SMC3 phosphorylation on NBS1. When compared with the vector-transfected NBS cell line (NBS-Lbi), stable expression of WT-NBS1 rescues Ser-1083 phosphorylation (Fig. 3B). Thus, Ser-1083 phosphorylation appears to be downstream in the ATM-MRN pathway.
To determine whether ATM can phosphorylate SMC3 in vitro, we carried out in vitro kinase assays using immunoprecipitated ATM expressed in 293T cells and a GST-SMC3 fragment encompassing amino acids 881–1218 expressed in Escherichia coli. As shown in Fig. 3C, WT ATM, but not kinase-dead ATM, phosphorylates the SMC3, demonstrating that ATM is capable of phosphorylating SMC3 in vitro. Taken together, these results suggest that ATM phosphorylates Ser-1083 of SMC3 both in vivo and in vitro.
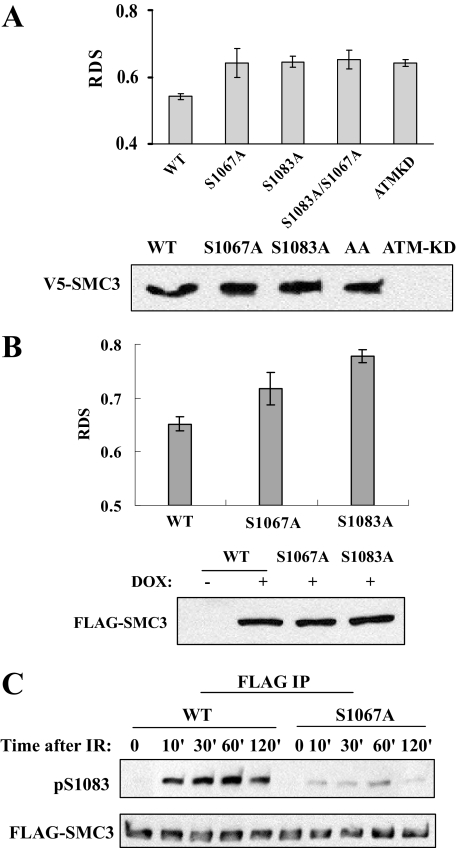
Phosphorylation of Ser-1083, as Well as Ser-1067 of SMC3, Is Required for Intra S-phase Checkpoint—We tested whether SMC3 phosphorylation is required for intra-S phase checkpoint. We transiently expressed V5-SMC3 proteins, including WT, S1067A, S1083A, or S1067A/S1083A mutants in 293T cells, and measured RDS. Because phosphorylation of Ser-1067 is not IR-induced, we expected that it would not regulate the intra-S phase checkpoint. Thus, the S1067A mutant was used as a negative control to demonstrate that not all phosphorylation sites regulate intra-S phase checkpoint. As expected, DNA synthesis in cells expressing SMC3-WT was decreased to 55% of that in cycling cells after IR; cells expressing SMC3-S1083A exhibited RDS phenotype, in which the DNA synthesis rate is 64% of that in cycling cells. Unexpectedly, cells expressing the SMC3-S1067A mutant also displayed RDS despite the fact that phosphorylation of Ser-1067 is not IR-induced. Cells expressing SMC3-S1067A/S1083A double mutant also showed RDS (Fig. 4A). The RDS level of cells expressing SMC3 mutants is similar to that of positive control cells that expressed kinase-dead ATM. These data suggest that phosphorylation of both Ser-1067 and Ser-1083 is required for intra-S phase checkpoint activation. To confirm this conclusion, we established stable cell lines that harbor a tet-on promoter to induce FLAG-tagged SMC3-WT, SMC3-S1067A, and SMC3-S1083A expression in 293T cells. Comparable levels of SMC3-WT and mutants are expressed as early as 24 h after the addition of doxycycline (Fig. 4B), and the induced SMC3 protein is incorporated into the cohesin complex as FLAG-SMC3 can be co-precipitated with endogenous SMC1 and RAD21 (data not shown). Consistent with data from the transient transfection experiment, small but reproducible RDS was observed in the S1067A-expressing cell line. Although DNA synthesis in cells expressing SMC3-WT was decreased to 65% of that in cycling cells after IR (Fig. 4B), it is decreased to 72 and 78% in cells expressing SMC3-S1067A and SMC3-S1083A, respectively (Fig. 4B). Taken together, these data suggest that phosphorylation of Ser-1083 is required for intra-S phase checkpoint and that Ser-1067 phosphorylation also affects intra-S phase checkpoint, although its phosphorylation is not IR-induced.
FIGURE 4.
Phosphorylation of both Ser-1067 and Ser-1083 is required for intra-S phase checkpoint activation. A, RDS measurements of cells transiently transfected with WT, mutant SMC3, or kinase-dead ATM (ATMKD) genes. Plasmids encoding genes of interest were transfected into 293T cells with Lipofectamine™, and cells were labeled with 14C 48 h after transfection. Cells were treated with 10 Gy IR and allowed to recover for 1 h. RDS was measured as described under “Materials and Methods.” Western blotting by the V5 antibody indicates the expression levels of exogenous WT and mutant V5-SMC3. B, RDS measurements of FLAG-SMC3 stable cell lines. WT or mutant FLAG-SMC3 cell lines were cultured in Dulbecco's modified Eagle's medium with doxycycline (DOX) for 24 h before 14C labeling. Cells were treated with 10 Gy IR and allowed to recover for 1 h. RDS was measured as described under “Materials and Methods.” Western blotting by the FLAG antibody was used to determine the expression levels of exogenous WT and mutant FLAG-SMC3 after doxycycline addition for 24 h. AA, S1083A/S1067A. C, SMC3 Ser-1083 phosphorylation in a S1067A stable cell line. FLAG-SMC3-WT and -S1067A-expressing 293T cell lines were treated with 5 Gy of IR and recovered for the indicated times. WT and mutant FLAG-SMC3 were immunoprecipitated (IP) from cells lysates with anti-FLAG M2 antibody and Western blotted with phospho-Ser-1083 or FLAG antibody.
Constitutive Phosphorylation of Ser-1067 Facilitates IR-induced Phosphorylation of Ser-1083—Next, we investigated how a constitutive phosphorylation affects IR-induced intra-S phase checkpoint. Because of the proximity of Ser-1067 and Ser-1083, we hypothesize that Ser-1067 phosphorylation may influence Ser-1083 phosphorylation. We compared phosphorylation of Ser-1083 in the stable cell lines that express FLAG-SMC3-WT or -S1067A mutant. FLAG-SMC3 proteins were immunoprecipitated by FLAG antibody, and Ser-1083 phosphorylation was detected by the phospho-specific antibody. As shown in Fig. 4C, phosphorylation of Ser-1083 in FLAG-SMC3-S1067A cells is significantly lower than that in FLAG-SMC3-WT during the time course between 10 min to 2 h in response to 5 Gy of IR. These results suggest that phosphorylation at Ser-1067 may affect intra-S phase checkpoint through modulating Ser-1083 phosphorylation by ATM.
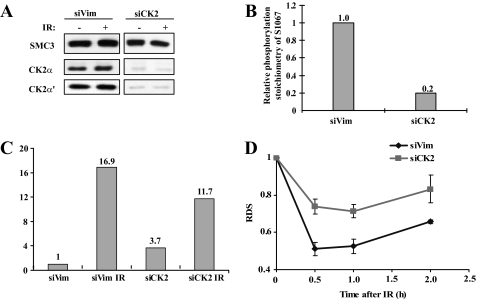
CK2 Phosphorylates Ser-1067 and Promotes Ser-1083 Phosphorylation and Intra-S phase Checkpoint Activation—Inspection of the SMC3 protein sequence revealed that Ser-1067 (1067SQDE1070) conforms to a CK2 phosphorylation consensus site. CK2 has been implicated in DNA damage response, but its role in intra-S phase checkpoint activation is not clear. We tested whether CK2 is required for Ser-1067 phosphorylation and intra-S phase checkpoint. We inactivated CK2 by siRNA knockdown of both catalytic subunits CK2α and CK2α′ (Fig. 5A), a condition known to attenuate its activity (7). Quantitative mass spectrometric analysis showed that the Ser-1067 phosphorylation dropped to 20% in CK2 knockdown cells when compared with that in control cells (Fig. 5B), demonstrating that CK2 is required for constitutive phosphorylation of SMC3 at Ser-1067.
FIGURE 5.
CK2 is required for phosphorylation of Ser-1067 and regulates Ser-1083 phosphorylation in response to IR and intra-S phase checkpoint. A, CK2 is knocked down by co-transfection of siRNA against both α and α′ subunits. B, quantitative mass spectrometric measurements of Ser-1067 phosphorylation. SMC3 was immunoprecipitated from siCK2 (both CK2α and CK2α′ was knocked down) and siVim (as a control)-transfected cells. The percentage of Ser-1067 phosphorylation was normalized to that in the siVim cell. C, quantitative mass spectrometric measurements of Ser-1083 phosphorylation. SMC3 was purified from siCK2 and siVim cells with or without IR treatment. Phosphorylation of Ser-1083 in indicated conditions was measured and normalized to the level in siVim-transfected cells without IR. D, RDS measurements in siCK2 cells and siVim cells. Sixty hours after siRNA transfection, HeLa cells were labeled with 14C for 36 h and then treated with 10 Gy IR. RDS was measured at the indicated times.
We used stable isotope-labeled amino acids in cultured cells coupled with quantitative mass spectrometry to measure Ser-1083 phosphorylation in CK2 knockdown cells. Consistent with the observation that S1067A impairs Ser-1083 phosphorylation in response to IR, CK2 knockdown also impairs the ability of Ser-1083 phosphorylation to be induced by IR (Fig. 5C). Although 10 Gy of IR induces an ∼17-fold increase in control knockdown cells, it induces only an ∼4-fold increase of Ser-1083 phosphorylation when CK2 is knocked down (Fig. 5C). These results suggest that CK2 is responsible for constitutive phosphorylation of Ser-1067 and promotes IR-induced Ser-1083 phosphorylation.
To determine whether CK2 also regulates intra-S phase checkpoint, we measured RDS in CK2 knockdown cells. As expected, the extent of DNA synthesis inhibition is consistently less in siCK2-transfected cells than in control siRNA-transfected cells during the 2-h period after IR (Fig. 5D), suggesting that CK2 is required for intra-S phase checkpoint activation.
DISCUSSION
We report the identification of two in vivo phosphorylation sites at Ser-1067 and Ser-1083 of SMC3, a core component of the cohesin complex. Using quantitative mass spectrometry and phospho-specific antibody, we show that Ser-1083 is an ATM site both in vitro and in vivo and is required for intra-S phase checkpoint activation. Ser-1067 is constitutively phosphorylated and is not IR-inducible but also modulates intra-S phase checkpoint. Thus, there appears to be interplay between the two phosphorylation sites in that constitutive Ser-1067 phosphorylation promotes IR-induced Ser-1083 phosphorylation, providing an explanation for how a constitutive phosphorylation event regulates DNA damage checkpoint. To our knowledge, this is the first example of a phosphorylation site that is not IR-induced yet plays a role in DNA damage checkpoint. This example provides a cautionary note for the notion that phosphorylation sites, if they are not IR-induced, are not important for DNA damage checkpoints. We also identified CK2 as the kinase that phosphorylates Ser-1067 and found that CK2 is required for suppression of RDS, thus linking CK2 to intra-S phase checkpoint through regulation of Ser-1083 phosphorylation. However, the molecular mechanism by which phosphorylated Ser-1067 regulates Ser-1083 phosphorylation remains unclear.
The Cohesin Complex as a Target of the Intra-S Phase Checkpoint—We and others previously discovered that SMC1 phosphorylation is an important event in the ATM-NBS1-mediated pathway (16, 17). Now our data show that SMC3 is also phosphorylated in response to IR and that its phosphorylation is important for suppression of RDS. SMC1 and SMC3 constitute the core of the cohesin complex. Each SMC subunit is self-folded by antiparallel coiled-coil interaction, creating a rod-shaped molecule with a hinge domain at one end and an ATPase head and tail domain at the other end (31). SMC1 and SMC3 bind to each other tightly through their hinge domains and bind another cohesin component, SCC1, through their head domains to form a ring-like structure that has been proposed to encircle the sister chromatids for establishing and maintaining sister chromatid cohesion (32). Why does the intra-S phase checkpoint target the cohesion complex? We do not know the answer until we identify the downstream targets of SMC phosphorylation. Could the cohesin complex be the elusive intra-S phase checkpoint target that regulates the DNA replication machinery? Cohesion is established in S-phase and is coupled to DNA synthesis (33). Thus, cohesion is proximal to the DNA replication machinery. The formation of the ring-like cohesion structure on chromatin is mediated by ATP hydrolysis (34). SMC ATPase head and tail domain and its flanking coiled-coil region play an important role in cohesion formation. Since Ser-957 and Ser-966 of SMC1 and Ser-1067 and Ser-1083 of SMC3 are located in this region, it is conceivable that phosphorylation of SMC1 and SMC3 may modulate SMC ATPase function to facilitate the slowdown of DNA synthesis when DSB is encountered. Such a model awaits the development of an in vitro ATPase activity assay for the cohesin complex to be tested. Currently, the cohesin complex exhibits very weak ATPase activity with the available in vitro assay (35).
Cohesion is also essential for homologous recombination repair of DSB because the damaged sister chromatid and repair template must be in close proximity (22, 36). Two recent publications reported that in response to a DSB in budding yeast, cohesion is formed de novo near the DSB by loading newly formed cohesin complex that is different from the S-phase cohesin (22, 37). Such an event also depends on the MRN complex (37), an important regulator of the intra-S phase checkpoint pathway mediated by ATM/SMC. An interesting question is whether SMC phosphorylation regulates de novo cohesion formation in response to DNA damage and whether de novo cohesion formation is required for intra S phase checkpoint activation. The cohesion formed de novo must be involved in the homologous recombination events dealing with DSB, including DSB repair. It has been established that SMC1 phosphorylation is essential for survival in response to IR in a mouse knock-in model (18), which makes SMC1 phosphorylation, and possibly SMC3 phosphorylation, the only intra-S phase checkpoint-regulated phosphorylation that is required for both IR survival and checkpoint activation. Note that even NBS1 phosphorylation, an upstream element of the intra-S phase checkpoint pathway, is not required for IR survival, although it is essential for checkpoint. We propose that SMC1 and SMC3 phosphorylation is at the crossroad of checkpoint activation and homologous recombination repair of DSB.
Regulation of Intra-S Phase Checkpoint by the CK2 Kinase—A surprising finding of the present work is that the CK2 kinase regulates intra-S phase checkpoint by constitutive phosphorylation of SMC3 at Ser-1067. A role of CK2 in DNA damage response was implicated in many previous works. We show here that CK2 knockdown cells as well as cells overexpressing SMC3-1067A mutant exhibit the RDS phenotype and weakened Ser-1083 phosphorylation. Together these results support the notion that the CK2 kinase is an intra-S phase checkpoint protein and suggest that one possible mechanism for its action is through modulating Ser-1083 phosphorylation.
We do not have sufficient data to explain how phosphorylation of Ser-1067 “primes” the phosphorylation of Ser-1083, but we provide two highly speculative models. First, phosphorylation of Ser-1067 may create a binding site to recruit a DDR protein to facilitate SMC3 phosphorylation. Coincidentally, CK2 phosphorylation creates a binding consensus for the FHA domain (7), a phosphor-peptide binding domain that is important in DNA damage response. Notably, the NBS1 protein contains an FHA domain, which may be recruited to SMC3 by binding to phosphorylated Ser-1067 and facilitates the phosphorylation of SMC3 at Ser-1083 by ATM. The weakened Ser-1083 phosphorylation in NBS cells is consistent with this hypothesis. However, we could not detect such an interaction with biochemical means, probably because such interaction is too transient to be detected. An in vivo phosphorylation-dependent protein-protein interaction assay may provide a better approach to address this issue (38). Second, Ser-1067 phosphorylation of SMC3 may mark a special region of the chromatin that must be dealt with in response to DNA damage so that the nearby Ser-1083 is phosphorylated to activate the checkpoint. An intriguing possibility is that Ser-1067 phosphorylation marks the chromatin that is surrounded by DNA replication late origins. We speculate that creation of the Ser-1067 phosphorylation is coupled with replication origin licensing and DNA replication. Thus, chromatins surrounding all specified DNA replication origins may initially be marked with Ser-1067 phosphorylated SMC3 during replication origin licensing; after firing of the early origins, the protein phosphatase associated with DNA replication machinery (39) removes the phosphorylation mark, leaving the late origins untouched. Such a model can be tested with genome-wide tiling array once a chromatin immunoprecipitation grade Ser-1067 phosphorylation-specific antibody becomes available and will be a direction to pursue in the future.
Supplementary Material
Acknowledgments
We are grateful to Dr. Eva Lee for the reconstituted NBS cell lines. We thank members of the Qin laboratory for discussion and technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA98500 (to J. Q.) and DAMD W81XWH 04-1-0437 (to Y. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and a supplemental figure.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: DDR, DNA damage response; ATM, ataxia telangiectasia-mutated; NBS, Nijmegen breakage syndrome; IR, ionizing radiation; RDS, radio-resistant DNA synthesis; MRN, Mre11/Rad50/NBS1; SMC, structure maintenance of chromosome; GST, glutathione S-transferase; siRNA, small interfering RNA; DSB, double-stranded DNA breaks; WT, wild type; HPLC, high performance liquid chromatography; Gy, grays; MALDI, matrix-assisted laser desorption; MS, mass spectrometry; MS/MS, tandem MS.
References
- 1.Zhou, B. B., and Elledge, S. J. (2000) Nature 408 433–439 [DOI] [PubMed] [Google Scholar]
- 2.Kastan, M. B., and Bartek, J. (2004) Nature 432 316–323 [DOI] [PubMed] [Google Scholar]
- 3.Litchfield, D. W. (2003) Biochem. J. 369 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homma, M. K., and Homma, Y. (2005) Mol. Cell Biochem. 274 47–52 [DOI] [PubMed] [Google Scholar]
- 5.Harvey, E. J., Li, N., and Ramji, D. P. (2007) Arterioscler. Thromb. Vasc. Biol. 27 806–812 [DOI] [PubMed] [Google Scholar]
- 6.Loizou, J. I., El-Khamisy, S. F., Zlatanou, A., Moore, D. J., Chan, D. W., Qin, J., Sarno, S., Meggio, F., Pinna, L. A., and Caldecott, K. W. (2004) Cell 117 17–28 [DOI] [PubMed] [Google Scholar]
- 7.Luo, H., Chan, D. W., Yang, T., Rodriguez, M., Chen, B. P., Leng, M., Mu, J. J., Chen, D., Songyang, Z., Wang, Y., and Qin, J. (2004) Mol. Cell Biol. 24 8356–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebrin, F., Chambaz, E. M., and Bianchini, L. (2001) Oncogene 20 2010–2022 [DOI] [PubMed] [Google Scholar]
- 9.Ahmad, K. A., Harris, N. H., Johnson, A. D., Lindvall, H. C., Wang, G., and Ahmed, K. (2007) Mol. Cancer Ther. 6 1006–1012 [DOI] [PubMed] [Google Scholar]
- 10.Yamane, K., and Kinsella, T. J. (2005) Clin. Cancer Res. 11 2355–2363 [DOI] [PubMed] [Google Scholar]
- 11.Meggio, F., and Pinna, L. A. (2003) FASEB J. 17 349–368 [DOI] [PubMed] [Google Scholar]
- 12.Johnston, I. M., Allison, S. J., Morton, J. P., Schramm, L., Scott, P. H., and White, R. J. (2002) Mol. Cell Biol. 22 3757–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung, W. L., Turner, F. B., Krishnamoorthy, T., Wolner, B., Ahn, S. H., Foley, M., Dorsey, J. A., Peterson, C. L., Berger, S. L., and Allis, C. D. (2005) Curr. Biol. 15 656–660 [DOI] [PubMed] [Google Scholar]
- 14.Guillemain, G., Ma, E., Mauger, S., Miron, S., Thai, R., Guerois, R., Ochsenbein, F., and Marsolier-Kergoat, M. C. (2007) Mol. Cell Biol. 27 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller, D. M., Zeng, X., Wang, Y., Zhang, Q. H., Kapoor, M., Shu, H., Goodman, R., Lozano, G., Zhao, Y., and Lu, H. (2001) Mol. Cell 7 283–292 [DOI] [PubMed] [Google Scholar]
- 16.Yazdi, P. T., Wang, Y., Zhao, S., Patel, N., Lee, E. Y., and Qin, J. (2002) Genes Dev. 16 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, S. T., Xu, B., and Kastan, M. B. (2002) Genes Dev. 16 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitagawa, R., Bakkenist, C. J., McKinnon, P. J., and Kastan, M. B. (2004) Genes Dev. 18 1423–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haering, C. H., Lowe, J., Hochwagen, A., and Nasmyth, K. (2002) Mol. Cell 9 773–788 [DOI] [PubMed] [Google Scholar]
- 20.Michaelis, C., Ciosk, R., and Nasmyth, K. (1997) Cell 91 35–45 [DOI] [PubMed] [Google Scholar]
- 21.Uhlmann, F., and Nasmyth, K. (1998) Curr. Biol. 8 1095–1101 [DOI] [PubMed] [Google Scholar]
- 22.Strom, L., Lindroos, H. B., Shirahige, K., and Sjogren, C. (2004) Mol. Cell 16 1003–1015 [DOI] [PubMed] [Google Scholar]
- 23.Birkenbihl, R. P., and Subramani, S. (1992) Nucleic Acids Res. 20 6605–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lengronne, A., Katou, Y., Mori, S., Yokobayashi, S., Kelly, G. P., Itoh, T., Watanabe, Y., Shirahige, K., and Uhlmann, F. (2004) Nature 430 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraakman-van der Zwet, M., Overkamp, W. J., Friedl, A. A., Klein, B., Verhaegh, G. W., Jaspers, N. G., Midro, A. T., Eckardt-Schupp, F., Lohman, P. H., and Zdzienicka, M. Z. (1999) Mutat Res. 434 17–27 [DOI] [PubMed] [Google Scholar]
- 26.Wang, Y., Cortez, D., Yazdi, P., Neff, N., Elledge, S. J., and Qin, J. (2000) Genes Dev. 14 927–939 [PMC free article] [PubMed] [Google Scholar]
- 27.Cortez, D., Wang, Y., Qin, J., and Elledge, S. J. (1999) Science 286 1162–1166 [DOI] [PubMed] [Google Scholar]
- 28.Zhang, X., Herring, C. J., Romano, P. R., Szczepanowska, J., Brzeska, H., Hinnebusch, A. G., and Qin, J. (1998) Anal. Chem. 70 2050–2059 [DOI] [PubMed] [Google Scholar]
- 29.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., and Mann, M. (2002) Mol. Cell Proteomics 1 376–386 [DOI] [PubMed] [Google Scholar]
- 30.Maya, R., Balass, M., Kim, S. T., Shkedy, D., Leal, J. F., Shifman, O., Moas, M., Buschmann, T., Ronai, Z., Shiloh, Y., Kastan, M. B., Katzir, E., and Oren, M. (2001) Genes Dev. 15 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Losada, A., and Hirano, T. (2005) Genes Dev. 19 1269–1287 [DOI] [PubMed] [Google Scholar]
- 32.Gruber, S., Haering, C. H., and Nasmyth, K. (2003) Cell 112 765–777 [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, K., Yonekawa, T., Kawasaki, Y., Kai, M., Furuya, K., Iwasaki, M., Murakami, H., Yanagida, M., and Okayama, H. (2000) Mol. Cell Biol. 20 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weitzer, S., Lehane, C., and Uhlmann, F. (2003) Curr. Biol. 13 1930–1940 [DOI] [PubMed] [Google Scholar]
- 35.Gruber, S., Arumugam, P., Katou, Y., Kuglitsch, D., Helmhart, W., Shirahige, K., and Nasmyth, K. (2006) Cell 127 523–537 [DOI] [PubMed] [Google Scholar]
- 36.Schar, P., Fasi, M., and Jessberger, R. (2004) Nucleic Acids Res. 32 3921–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unal, E., Arbel-Eden, A., Sattler, U., Shroff, R., Lichten, M., Haber, J. E., and Koshland, D. (2004) Mol. Cell 16 991–1002 [DOI] [PubMed] [Google Scholar]
- 38.Hu, C. D., and Kerppola, T. K. (2003) Nat. Biotechnol. 21 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, X. H., Walter, J., Scheidtmann, K., Ohst, K., Newport, J., and Walter, G. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.