Abstract
Hsf-1 (heat shock factor-1) is a transcription factor that is known to regulate cellular heat shock response through its binding with the multispecific transporter protein, Ralbp1. Results of present studies demonstrate that Hsf-1 causes specific and saturable inhibition of the transport activity of Ralbp1 and that the combination of Hsf-1 and POB1 causes nearly complete inhibition through specific bindings with Ralbp1. Augmentation of cellular levels of Hsf-1 and POB1 caused dramatic apoptosis in non-small cell lung cancer cell line H358 through Ralbp1 inhibition. These findings indicate a novel model for mutual regulation of Hsf-1 and Ralbp1 through Ralbp1-mediated sequestration of Hsf-1 in the cellular cytoskeleton and Hsf-1-mediated inhibition of the transport activity of membrane-bound Ralbp1.
In response to heat stress, human cells respond by activation of Hsf-1 (heat shock factor-1), a transcription factor that binds to NGAAN repeats of the promoter of heat shock genes, augmenting transcription (1-5). Considered the master regulator of the heat shock response (1-3), Hsf-1 binds DNA constitutively, and its binding affinity is based upon its phosphorylation in response to heat shock (1-5). In the unstressed state, Hsf-1 is sequestered in a complex with tubulin, HSP90, and Ralbp1 (6). Stress or constitutively active Ral-GTP binding to Ralbp1 triggers the release of Hsf-1 and its migration to the nucleus, where its transcription factor activity is important for the expression of heat shock proteins (6, 7). Although these studies focused on Ralbp1 present in the cytoplasm bound to the cytoskeleton and nuclear membrane, several previous and subsequent reports have clearly demonstrated the presence of Ralbp1 in nuclear as well as plasma membranes (8-12). In several recent studies, we have conclusively demonstrated that Ralbp1 is a transmembrane protein with a defined cell surface domain (8-11) and that it catalyzes in ATP hydrolysis-dependent trans-membrane anti-gradient efflux of toxic xenobiotics as well as endogenous metabolites. The preferred physiological substrates for transport by Ralbp1 are glutathione-electrophile conjugates of electrophilic lipid metabolites that arise from stress or heat shock-induced lipid peroxidation (13). The cell surface domain of Ralbp1 can be targeted by highly specific antibodies that inhibit the transport activity of Ralbp1 and result in dramatic regression of tumor in syngeneic and xenograft models of melanoma, lung cancer, and colon cancer (14, 15). The membrane functionality of Ralbp1 is also evident from its crucial role in endocytosis as a rate-regulatory element (12, 16-18).
An endocytosis-linked protein POB1 that binds Ralbp1 in a similar region as Hsf-1 has been shown to be a specific and saturable inhibitor of the glutathione-electrophile conjugates and doxorubicin (DOX)2 transport activity of membrane-reconstituted purified Ralbp1 (19). We proposed that just as POB1 could function as an inhibitor of the transport activity of Ralbp1, Hsf-1 could also function as a transport inhibitor. This model predicts that under heat stress, dissociation of Hsf-1 from Ralbp1 would release inhibition on the efflux of the proapoptotic glutathione conjugates of lipid peroxidation products generated as an unavoidable consequence of heat shock or other stress. We have addressed this prediction in H358 non-small cell lung cancer cell line (NSCLC) by examining the effect of Hsf-1 and POB1 on the transport of and cellular accumulation and efflux of DOX and on apoptosis. POB1 and Hsf-1 were found to inhibit Ralbp1 at independent binding sites and together almost completely abrogated its transport and anti-apoptotic activities. The results suggest a novel framework for viewing the chief regulatory mechanisms of the heat shock and stress response pathways.
EXPERIMENTAL PROCEDURES
Materials—The human full-length Hsf-1 cDNA was a gift of Dr. Nahid F. Mivechi (Institute of Molecular Medicine and Genetics and Department of Radiology, Medical College of Georgia, Augusta, GA). Polyclonal anti-Hsf1 IgG was from Stressgen. Bacterial strains (DH5α and BL21(DE3)) were purchased from Invitrogen. pET30a(+), the T7 promoter-based expression vector, was purchased from Novagen, Inc. (Madison, WI). Restriction enzymes, thermophilic DNA polymerase (Vent polymerase), and DNA ligase were from New England Biolabs (Beverly, MA). dNTPs were from Applied Biosystems (Foster City, CA). High pressure liquid chromatography-grade oligonucleotides were synthesized by Biosynthesis, Inc. (Lewisville, TX). DOX (Adriamycin) was obtained from Adria Laboratories (Columbus, OH). 14C-DOX (specific activity 44.8 Ci/mmol), [3H]LTC4 (specific activity 146 Ci/mmol) and [3H]colchicine (specific activity 74.3 Ci/mmol) were purchased from PerkinElmer Life Sciences. Rabbit polyclonal antibodies to histone H3 and manganese-superoxide dismutase were purchased from Sigma and Stressgen Bioreagent (Ann Arbor, MI), respectively. Secondary antibody for Western blots, goat anti-rabbit IgG horseradish peroxidase conjugate, was purchased from Sigma. Polyclonal rabbit anti-human reconstituted Ralbp1 IgG, anti-human reconstituted POB1 IgG, and preimmune IgG were prepared and purified as described elsewhere (8, 19). cDNA of POB1 (2200 bp) was kindly provided by Prof. Leen J. Blok (Erasmus University Rotterdam, The Netherlands). Fluorescein isothiocyanate-labeled Annexin V conjugate and a TdT-mediated dUTP nick end labeling (TUNEL) fluorescence detection kit were purchased from Invitrogen and Promega (Madison, WI), respectively. Dinitrophenyl-S-glutathione (DNP-SG) was synthesized from CDNB and GSH according to the method described by us previously (20). The sources of other chemicals used in this study were the same as described previously (10, 11).
Cell Lines and Cultures—Human NSCLC line H358 (bronchioalveolar) from ATCC (Manassas, VA) was used in these studies. Cells were cultured at 37 °C in a humidified atmosphere of 5% CO2 in RPMI 1640 medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 1% (v/v) penicillin/streptomycin solution, 2 mm l-glutamine, 10 mm HEPES, 1 mm sodium pyruvate, 4.5 g/liter glucose, and 1.5 g/liter sodium bicarbonate.
Cloning, Prokaryotic Expression, and Purification of His-tagged Hsf-1—The human full-length Hsf-1 cDNA was a gift of Dr. Nahid F. Mivechi. The cDNA of Hsf-1 was used as a template for PCR amplification of the Hsf-1 coding sequence. The upstream (5′-GGCGGTACCATGGAAGATTATACCAAAATAGAG-3′) and downstream (5′-CCGCTCGAGCTACATCTTCTTAATCTGATTGTC) primers were designed to introduce a KpnI restriction site (underlined) immediately upstream of the initiator codon and XhoI site (underlined) immediately downstream of the stop codon of the Hsf-1 open reading frame. The PCR amplification was performed under the following incubation conditions: DNA template, 500 ng; primers, 30 pmol each; dNTPs, 2.5 μm; thermopol buffer, 1×; bovine serum albumin, 1×; Vent polymerase 2.5 units. PCR cycles were at 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, and 1 min at 72 °C and a final extension at 72 °C for 7 min. The PCR product purified by using the Qiagen PCR purification kit was digested with KpnI/XhoI restriction enzymes. The cleaved PCR products were ligated into pET30a(+) previously digested with the same restriction enzymes.
The ligated products were expressed into the DH5α-competent cells, and plasmid DNA was purified from the overnight culture of single colony using the Qiagen DNA purification kit. Techniques for restriction enzyme digestion, ligation, transformation, and other standard molecular biology manipulations were based on methods described by Sambrook et al. (21). The sequence of Hsf-1 was confirmed by DNA sequencing. Following verification of the sequence, the pET30a(+) plasmid containing the full-length Hsf-1 was used to transform Escherichia coli strain BL21(DE3), and protein was expressed in E. coli BL21(DE3) grown at 37 °C after induction with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside.
Bacterially expressed human Hsf-1 was purified by metal affinity chromatography over Ni2+-nitrilotriacetic acid Superflow resin (Qiagen) with slight modifications, as described below. E. coli BL21(DE3) expressing Hsf-1 was lysed in 20 mm Tris-HCl containing 250 mm NaCl, 100 μm PMSF, and 5 mm imidazole, pH 7.9, sonicated, and incubated for 4 h at 4 °C with gentle shaking, followed by centrifugation at 13,000 rpm for 30 min. The supernatant was mixed with Ni2+-nitrilotriacetic acid Superflow resin preequilibrated with the same buffer. The resin was incubated overnight at 4 °C with gentle shaking and washed with wash buffer (20 mm Tris-HCl, 300 mm NaCl, 20 mm imidazole, and 100 μm phenylmethylsulfonyl fluoride, pH 7.9) until A280 was zero. The bound protein from the resin was eluted with elution buffer (20 mm Tris, 500 mm NaCl, 400 mm imidazole, and 100 μm phenylmethylsulfonyl fluoride, pH 7.9) and was dialyzed against 10 mm Tris-HCl, pH 7.4, 100 μm EDTA, and 100 μm phenylmethylsulfonyl fluoride.
Prokaryotic Expression and Purification of POB1 and POB1-(1-512)—The 2200-bp full-length long version cDNA of POB1 was a gift from Prof. Leen J. Blok (Erasmus University, Rotterdam, The Netherlands). The bacterially expressed His-tagged POB1 and truncated mutant POB1-(1-512) (encoding aa 1-512) proteins were cloned, expressed, and purified as described previously (19).
Cloning, Prokaryotic Expression, and Purification of Ralbp1—Purified Ralbp1 protein was obtained from E. coli BL21(DE3) expressing the pET30a(+) plasmid containing full-length cDNA corresponding to the sequence of RLIP76, the 76.4-kDa (655-aa) splice variant encoded by the human gene, RALBP1. The purification was carried out using DNP-SG affinity chromatography as described previously (8), and purity was confirmed by SDS-PAGE and Western blot analyses.
Liposome Preparation—The proteoliposome preparation procedures used here have been described in detail previously (8, 11). Proteoliposomes containing either Ralbp1, Hsf-1, or POB1 were prepared by the addition of the corresponding purified recombinant protein to a sonicated emulsion of 1:4 cholesterol and phospholipids (soybean asolectin, 95% purity; Sigma) in liposome reconstitution buffer (10 mm Tris-HCl, pH 7.4, 2 mm MgCl2, 1 mm EGTA, 100 mm KCl, 40 mm sucrose, 2.8 mm β-mercaptoethanol, 0.05 mm butylated hydroxytolune, and 0.025% polidocanol). The final protein concentration in the liposome reconstitution mixture was 50 μg/ml. Liposome formation was initiated by the addition of SM2 Biobeads (200 mg/ml). We have shown that vesiculation is complete within 4 h and yields primarily unilammelar vesicles with a median diameter of 0.25 μm and intravesicular/extravesicular volume ratio of 18 μl/ml (8, 22). The liposomes reconstituted with purified recombinant Hsf-1, POB1, and Ralbp1 proteoliposomes were used for delivery of these proteins to cells in culture. Efficiency of delivery for Ralbp1 and POB1 proteoliposomes has been established previously (8, 19). Successful delivery of all three proteins to cells was confirmed in the present studies.
Transport Activity of Ralbp1 and Inhibition by Hsf-1 or POB1—In addition to being used to deliver Ralbp1 to cells, Ralbp1 liposomes were used in isolated cell-free systems to measure the transport activity of Ralbp1 toward model radiolabeled substrates, including 14C-DOX, [3H]DNP-SG, [3H]LTC4, and [3H]COL. The ATP-dependent specific uptake of each of these compounds was quantified as described previously (8, 23, 24) by using a 96-well plate filtration manifold to separate the extravesicular drug from that taken up by the vesicles. Uptake was measured in parallel in Ralbp1 proteoliposomes and control liposomes (prepared in the absence of protein) in the absence or presence of 4 mm ATP at a fixed time point of 5 min at 37 °C. In the transport reaction, the final concentration of Ralbp1 protein (in the form of Ralbp1 liposome) was 8.3 μg/ml. We have demonstrated previously that this method estimates the initial velocity of ATP-dependent transport (8). Background drug binding to the 0.45-μm membrane in the filtration manifold was adjusted, and ATP-dependent uptake was calculated by subtracting out uptake in the proteoliposomes in the absence of ATP.
To study the effect of purified Hsf-1 or POB1 protein on transport activity of Ralbp1, purified Hsf-1 or POB1 protein (without reconstitution in proteoliposomes) was added to the transport reaction mixture containing Ralbp1 proteoliposomes or control liposomes. The concentration of POB1 or Hsf-1 in the transport reaction mixture ranged from 0 to 13.3 μg/ml. In the controls for inhibition studies, Hsf-1 or POB1 protein was replaced with an equal amount of bovine serum albumin.
Inhibition of Transport of 14C-DOX, [3H]COL, [3H]DNP-SG, and [3H]LTC4 by POB1—Crude membrane inside-out vesicles (IOVs) were prepared from Ralbp1+/+, Ralbp1+/-, and Ralbp1-/- MEFs using established procedures as described by us for the human erythrocytes (25) and K562 cells (8). For these experiments, a fixed amount of IOV protein (20 μg/30 μl) was reconstituted in proteoliposomes along with varying amounts (0-300 ng/30 μl) of either purified reconstituted POB1 or its deletion mutant (POB1-(1-512)), and the transport rates of 14C-DOX, [3H]COL, [3H]DN-PSG, and [3H]LTC4 were measured as described above. In one control, POB1 proteins (full-length POB1 or its deletion mutant) were excluded, whereas in another control, an equivalent amount of heat-inactivated respective protein was reconstituted in proteoliposomes. Each determination was performed in triplicate.
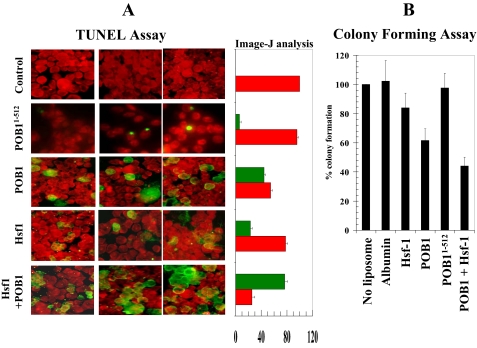
Effect of Full-length Hsf-1 and POB1 on Apoptosis by TUNEL Assay—In these studies, we assessed whether cellular levels of Hsf-1 and POB1 could be augmented by proteoliposome-mediated delivery to cultured lung cancer cells and assessed whether uptake is correlated with apoptosis by the TUNEL assay. The H358 non-small cell lung cancer cells (1 × 106 cells) were grown on the coverslips. The cells were treated with equal amounts of liposomes reconstituted with 40 μg/ml (final concentration) recombinant POB1-(1-512), POB1, Hsf-1, and POB1 plus Hsf-1 protein. After a 24-h incubation, the medium was removed, and cells were washed with PBS four times. A TUNEL assay was performed using the Promega fluorescence detection kit. Briefly, cells were fixed by immersing slides in freshly prepared 4% paraformaldehyde solution for 30 min at 4 °C, followed by washing with PBS. The cells were permeabilized by immersing the slides in 0.2% Triton X-100 solution in PBS for 5 min followed by washing with PBS. Cells were equilibrated with equilibration buffer (provided by Promega) for 10 min. The equilibrated areas were blotted with tissue paper, and TdT incubation buffer was added to the cells and placed in the humidified chamber. The chamber was covered with aluminum foil to protect from direct light. Slides were incubated at 37 °C for 60 min, and the reaction was terminated by immersing the slides in 2× SSC buffer for 15 min followed by washing with PBS. The slides were stained in propidium iodide solution for 10 min in the dark and washed with distilled water several times. Slides were analyzed under a fluorescence microscope using a standard fluorescein filter set to view the green fluorescence at 520 nm and red fluorescence of propidium iodide at >620 nm. Fluorescence micrographs were taken using a Zeiss LSM 510 META (Germany) laser-scanning fluorescence microscope at ×400 magnification.
Effects of Hsf-1 and POB1 Liposomes on 14C-DOX Accumulation in Lung Cancer Cell—H358 cells were harvested and washed with PBS, and aliquots containing 5 × 106 cells (in triplicate for each time point) were inoculated into fresh medium. After overnight incubation, the cells were pelleted and resuspended in 80 μl of medium containing 4 μg of Hsf-1 or POB1 proteoliposomes or control liposomes and incubated at 37 °C for 24 h. After 24 h of incubation, 20 μl of 14C-DOX (final concentration 3.6 μm, specific activity 8.5 × 104 cpm/nmol) was then added to the medium and incubated for 5, 10, 20, and 30 min at 37 °C. Drug uptake was stopped by rapid cooling on ice. Cells were centrifuged at 700 × g for 5 min at 4 °C, and medium was completely decanted. Radioactivity was determined in the cell pellet after washing twice with ice-cold PBS.
14C-DOX Efflux Studies—H358 cells were harvested and washed with PBS, and aliquots containing 5 × 106 cells (in triplicate) were inoculated into fresh medium. After overnight incubation, the cells were pelleted and resuspended in 80 μl of medium containing 4 μg of either control, Hsf-1, or POB1 liposomes and incubated at 37 °C for 24 h. After a 24-h incubation, 20 μl of 14C-DOX (final concentration 3.6 μm, specific activity 8.5 × 104 cpm/nmol) was then added to the medium and incubated for 60 min at 37 °C. Cells were centrifuged at 700 × g for 5 min, after which the supernatant was removed completely, and the cell pellet was washed with PBS twice. The pellet was immediately resuspended in 1 ml of PBS. Aliquots of 50 μl of clear supernatant were removed every minute for radioactivity counting for 15 min, and total radioactivity remaining in the transport reaction was quantified at the end of the experiment. The back-added curves of cellular residual VRL versus time were constructed as described previously (11).
Drug Sensitivity Assay—Cell density during the log phase was determined by counting trypan blue-excluding cells in a hemocytometer, and 20,000 cells were plated into each well of 96-well flat bottomed microtiter plates. After 24 h of incubation, the cells were treated with control liposomes or liposomes containing reconstituted Hsf-1 or POB1 (final concentration 40 μg/ml). DOX was added, and IC50 was measured by performing a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay 96 h later, as described previously (19, 26). Eight replicate wells were used for each point in each of three separate measurements of IC50.
Colony-forming Assay—H358 cells (0.1 × 106 cells/500 μl) were incubated with control, POB1-(1-512), POB1, Hsf-1, and POB1 plus Hsf-1 liposomes (40 μg/ml final concentration for each liposome) for 24 h, and then aliquots of 50 and 100 μlin 60-mm Petri dishes, separately, in a total volume of 4 ml by adding medium. After 10 days, control and proteoliposome-treated cells were stained with methylene blue for 30 min, and colonies were counted using an Innotech Alpha Imager.
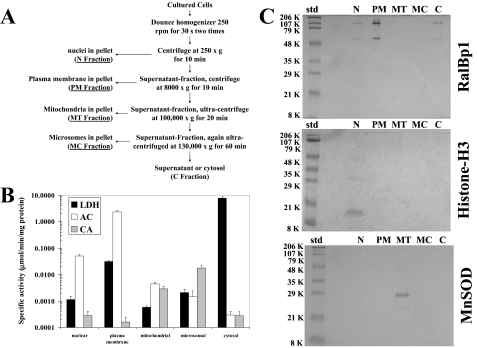
Subcellular Fractionation—Differential fractionation was carried out according to Jia et al. (27) with minor modifications. All steps were performed at 4 °C with 10 mm Tris-HCl, pH 7.4, containing 250 mm sucrose used as fractionation buffer throughout the procedure. About 1 × 109 H358 cells were harvested by centrifugation at 1000 × g for 10 min and washed with PBS. The cell pellet was resuspended in 0.5 ml of buffer and Dounce-homogenized at 250 rpm (30 s × 2). The cell homogenate was then centrifuged at 250 × g for 10 min to isolate nuclei in the pellet (fraction N). The supernatant was centrifuged at 8000 × g for 10 min to isolate plasma membrane in the pellet (fraction PM). The supernatant was centrifuged at 100,000 × g for 20 min to isolate mitochondria in the pellet (fraction M). The supernatant was again centrifuged at 130,000 × g for 60 min to isolate microsomes in the pellet (fraction MC). The final supernatant was called cytosol (fraction C). All pellets were washed with fractionation buffer before lysis in lysis buffer for enzyme activities and Western blot analyses. For heat shock, the cells were exposed to 42 °C for 30 min, brought back to 37 °C, and allowed to recover for 2 h at 37 °C before use in further experiments.
Markers of Subcellular Fractions—Nuclei and mitochondria fractions were characterized by Western blot analyses using histone H3 antibodies and manganese-dependent superoxide dismutase antibodies, respectively (27). Plasma membrane and cytosol fractions were characterized by the activities of acetylcholinesterase and lactate dehydrogenase spectrophotometrically, respectively (28). The microsomal fraction was characterized by the activity of carnitine acyltransferase spectrophotometrically (29).
Hsf-1 siRNA Preparation—Predesigned chemically synthesized siRNA duplex was purchased from Dharmacon Research (Lafayette, CO). A 20-nucleotide-long scrambled siRNA duplex was used as a control. The siRNA duplex was resuspended in 1× universal buffer, provided by Dharmacon Research Laboratory. The targeted cDNA sequence (AAGAACGACAGTGGCTCAGCA) corresponds to nt 809-829. The corresponding sense and antisense siRNA sequences were GAACGACAGUGGCUCAGCAUU and P-UGC-UGAGCCACUGUCGUUCUU, respectively. The sequence of the scrambled siRNA in the sense and antisense directions were GUAACUGCAACGAUUUCGAUGdTdT and CAUCGAAAUCGUUGCAGUUACdTdT, respectively. Transfection of siRNA duplexes was performed using Lipofectamine 2000 transfection reagent kit (Invitrogen) and assay for silencing 48 h after transfection. The heat shock treatment was given as described above.
Immunohistochemical Localization of Ralbp1 and Effects of Heat Shock—Immunohistochemical localization of Ralbp1 was performed on H358 cells by the method described previously with slight modifications (10). Cells were grown on coverslips and were fixed by using methanol/acetic acid in a ratio of 3:1. Nonspecific antibody interactions were minimized by pretreating cells with 10% goat serum in Tris-buffered saline for 60 min at room temperature. The cells were incubated with primary antibody, anti-Ralbp1 IgG, for 2 h at room temperature in a humidified chamber. After washing off the primary antibody with PBS (10 times, 3 min each), rhodamine red-X-conjugated goat anti-rabbit IgG (1:50 dilution in PBS) as secondary antibodies were added and incubated for 1 h at room temperature in a humidified chamber. The unbound secondary antibodies were removed by washing with PBS (10 times, 3 min each), and nuclei were stained using 4′,6-diamidino-2-phenylindole. Coverslips were air-dried and mounted on the slides with Vectashield mounting medium for fluorescence (Vector Laboratories, CA). Slides were photographed at ×400 magnifications using a Leica DMLB fluorescence microscope. Photographs were taken with 1-s integration. For heat shock, the cells were exposed to 42 °C for 30 min, brought back to 37 °C, and allowed to recover for 2 h at 37 °C before use in further experiments.
Chromatin Immunoprecipitation (ChIP)—A ChIP assay was performed to determine whether Ralbp1 binds chromatin after nuclear translocation, using the ChIP-IT kit from Active Motif following the manufacturer's instructions. Briefly, two flasks containing 4.5 × 107 cells each were grown and fixed with 37% formaldehyde. Cells were washed with PBS followed by the addition of 10 ml of glycine stop solution (provided by Active Motif) and washing with PBS. The cells were collected into a 15-ml conical tube by adding 2 ml of ice-cold scraping solution and were pelleted by centrifugation for 10 min at 2000 × g at 4 °C. The pellet was resuspended into 1 ml of ice-cold lysis buffer (supplemented with 5 μl of protease inhibitor mixture and 5 μl of phenylmethylsulfonyl fluoride) and was incubated for 10 min in ice. Cells were transferred to an ice-cold Dounce homogenizer and were Dounce-homogenized with 10 strokes to aid nuclei release, followed by transfer to a 15-ml conical tube and centrifuged for 10 min at 4000 × g at 4 °C. Pellet was resuspended into 1.0 ml of shearing buffer (supplemented with 5 μl of protease inhibitor mixture) and was sheared with 10 pulses using a Sonics Vibracell VC 130 sonicator. The sheared DNA sample was centrifuged at 10,000 × g in a 4 °C microcentrifuge for 12 min. The supernatant was collected into a microcentrifuge tube and was precleaned with protein G beads to reduce nonspecific background. Aliquots of 200 μl were incubated with control IgG as well as negative control IgG (provided by Active Motif) and anti-Ralbp1 IgG overnight on a rotator at 4 °C. Aliquots of 100 μl of protein G beads were added into each of the antibody chromatin incubations and were incubated for 2 h at 4 °C. The beads were pelleted by centrifugation at 4000 × g for 2 min, followed by washing, and eluted with 100 μl of ChIP elution buffer (provided by Active Motif). The eluted DNA were purified using the DNA purification minicolumns (provided by Active Motif) and were amplified by PCR using the control primers as well as negative control primers (provided by Active Motif) and Ralbp1 primers. PCR products were quantified by running on 1% agarose gel. For heat shock, the cells were exposed to 42 °C for 30 min, brought back to 37 °C, and allowed to recover for 2 h at 37 °C before fixing with 37% formaldehyde and use in further experiments.
Protein Cross-linking and Immunoprecipitation—The protein cross-linking and immunoprecipitation assay was performed according to the method of Aumais et al. (30). Briefly, purified recombinant Ralbp1, POB1, and Hsf-1 proteins (10 μg each) were cross-linked by incubation with 0.1 mm N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP; from Sigma) in a total volume of 0.5 ml in 10 mm sodium phosphate buffer, pH 7.4, for 30 min. Excess SPDP was removed by passing the solution through a Sephadex G-50 spin column preequilibrated with sodium phosphate buffer (pH 7.4). The sample was treated with 0.5 mm N-ethylmaleimide for 10 min to block all free SH groups that could prematurely cleave cross-links. Protein complexes were immunoprecipitated by incubating with anti-Ralbp1 IgG for 12 h followed by with protein A-Sepharose in radioimmune precipitation assay buffer (50 mm Tris, pH 7.4, 4 mm EDTA, 150 mm NaCl, 1% Triton X-100, and 0.1% SDS) for 2 h. Samples were sedimented by centrifugation at 10,000 × g, washed three times with radioimmune precipitation assay buffer, and then resuspended in 100 μl of SDS-PAGE sample buffer. Cross-linked proteins were analyzed by SDS-PAGE, followed by Western blots against anti-Ralbp1, anti-POB1, and anti-Hsf-1 IgG.
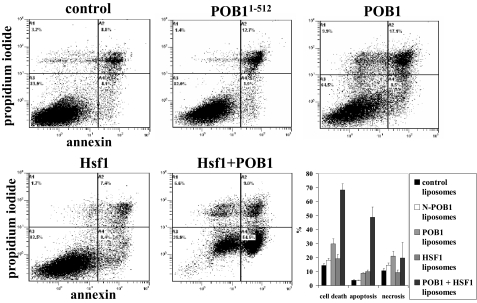
Annexin V Apoptosis Assays; Effect of Hsf1, POB1, POB1-(1-512) and Hsf-1 Plus POB1 on Apoptosis by Fluorescein Isothiocyanate-labeled Annexin V Conjugate Assay—Early apoptosis was analyzed by studying the loss of membrane asymmetry using Annexin V staining analyzed by flow cytometry according to the method of Hammill et al. (31). Briefly, H358 cells (0.5 × 106 cells/ml) were grown in a 6-well plate and were treated with control liposomes or liposomes reconstituted with 40 μg/ml (final concentration) recombinant Hsf-1, POB1, POB1-(1-512), or both (Hsf-1 and POB1) protein. After a 24-h incubation, the cells were harvested and centrifuged at 1500 × g for 5 min. Cells were washed once with PBS and resuspended in 400 μl of cold annexin binding buffer containing 5 μl of Annexin V-fluorescein isothiocyanate and 5 μl of 0.1 mg/ml propidium iodide. Cells were incubated at room temperature for 10 min in the dark and were analyzed by flow cytometry using a Beckman Coulter Cytomics FC500 flow cytometer. Results were processed using CXP2.2 analysis software from Beckman Coulter.
RESULTS
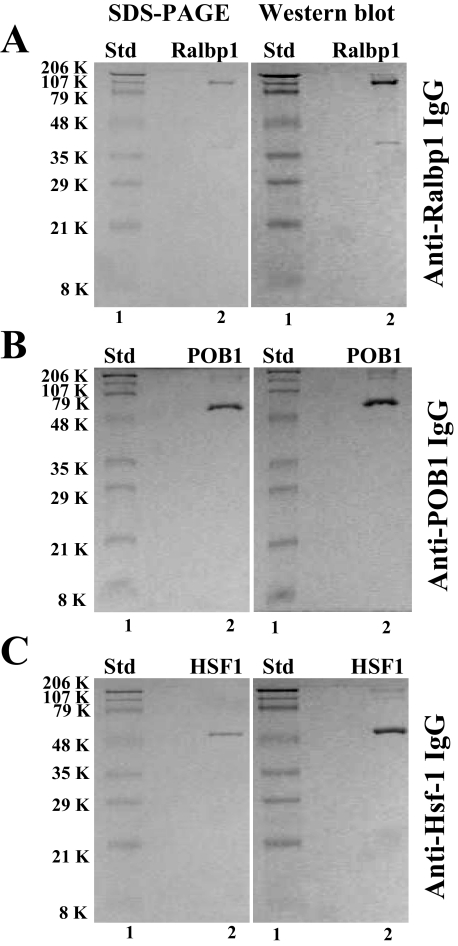
Purification of Recombinant Ralbp1, Hsf-1, and POB1—Recombinant human Ralbp1 protein was purified by DNPSG affinity chromatography, as described previously (8). The SDS-PAGE and anti-Ralbp1 Western blot (Fig. 1A) demonstrated largely intact and pure protein at the 95 kDa position typical for the aberrant behavior in SDS-PAGE of this 76-kDa protein (8). The faint band at 38 kDa is a truncated C-terminal peptide (aa 410-655), which has an actual mass of 27 kDa, apparently due to aberrant behavior in SDS-PAGE. This 38 kDa band arises due to inherent proteolyic susceptibility of Ralbp1, as indicated by the demonstrated identity of the 38-kDa protein found in recombinant preparations with that purified from human tissues and previously designated DNPSG ATPase (25).
FIGURE 1.
Purity of recombinant human Ralbp1, POB1, and Hsf-1. DNPSG affinity-purified reconstituted Ralbp1 (5 μg) and Ni2+-nitrilotriacetic acid Superflow resin (Qiagen)-purified reconstituted POB1 (5 μg) and reconstituted Hsf-1 (5 μg) were applied to SDS-PAGE and subjected to Western blot analyses against rabbit-anti-Ralbp1 IgG (A), rabbit anti-POB1 IgG (B), and rabbit anti-Hsf-1 IgG (C). SDS-polyacrylamide gels were stained with Coomassie Brilliant Blue R250, and Western blots were developed using horseradish peroxidase-conjugated goat anti-rabbit IgG as secondary antibody. Std, standard.
Ni2+-nitrilotriacetic acid affinity-purified Hsf-1 and POB1 were similarly examined and found free of significant impurities by SDS-PAGE, and specific recognition by the respective antibodies demonstrated the identity of the purified protein. The purified His-tagged POB1 and Hsf-1 proteins were seen in both Western blots and SDS-PAGE at the expected Mr of 78 and 58 kDa, respectively (Fig. 1, B and C, respectively).
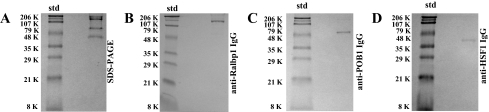
Evidence for Complex Formation between Ralbp1, Hsf-1, and POB1—Oosterhoff et al. (32) cloned POB1 (partner of Ralbp1) as the first known binding partner of Ralbp1 using a yeast two-hybrid method using Ralbp1 as bait and clearly demonstrated specific binding and complex formation between Ralbp1 and POB1. More recent studies by Hu and Mivechi (6) have shown that Ralbp1 is found complexed with Hsf-1 as well. However, complex formation between all three proteins had not been previously demonstrated. In the present studies, we addressed the question of whether a trimeric complex exists between these three proteins by using protein cross-linking and immunoprecipitation using the established method of Aumais et al. (30) (Fig. 2). To solutions containing all three authenticated purified recombinant proteins, the cross-linking agent SPDP was added, followed by immunoprecipitation using anti-Ralbp1 antibody alone. The precipitate was solubilized and subjected to SDS-PAGE and also Western blot analyses against anti-Ralbp1 as well as anti-POB1 and anti-Hsf-1. The SDS-PAGE of solubilized immunoprecipitated proteins (Fig. 2A) revealed the presence of all three proteins, at 95, 78, and 58 kDa, corresponding to the expected migration in SDS-PAGE of Ralbp1, POB1, and Hsf-1, respectively. The identity of these bands was confirmed by Western blotting of the immunoprecipitated protein fraction against anti-Ralbp1 (Fig. 2B), anti-POB1 (Fig. 2C), and anti-Hsf-1 (Fig. 2D). These findings strongly indicated the formation of a ternary complex between Ralbp1, Hsf-1, and POB1.
FIGURE 2.
Ternary complex formation between Ralbp1, Hsf-1, and POB1. Purified recombinant Ralbp1, POB1, and Hsf-1 proteins (10 μg each) were cross-linked by incubation with 0.1 mm SPDP in a total volume of 0.5 ml in 10 mm sodium phosphate buffer, pH 7.4, for 30 min. Excess SPDP was removed by passing the solution through a Sephadex G-50 spin column. Sample was treated with 0.5 mm N-ethylmaleimide for 10 min to block all free SH groups, and protein complexes were immunoprecipitated by incubating with anti-Ralbp1 IgG for 12 h, followed by protein A-Sepharose in radioimmune precipitation assay buffer for 2 h. Samples were sedimented by centrifugation at 10,000 × g and washed with radioimmune precipitation assay buffer and then resuspended in 100 μl of SDS-PAGE sample buffer. Cross-linked proteins were analyzed by SDS-PAGE (A), followed by Western blots against anti-Ralbp1 IgG (B), anti-POB1 IgG (C), and anti-Hsf-1 IgG (D). Std, standard.
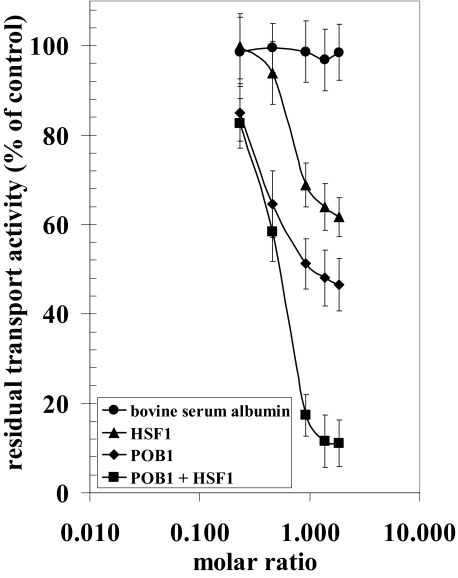
Inhibition of the DOX Transport Activity of Ralbp1 by POB1 and Hsf-1—POB1 was cloned originally by others in a two-hybrid yeast screen with Ralbp1 as bait, thus the name, partner of Ralbp1 (32). These studies also partially elucidated the binding regions; the C-terminal amino acids, 499-655, were shown to be necessary for POB1 binding. In previous studies, we had demonstrated that POB1 inhibited the DOX transport activity of Ralbp1 in a saturable manner and that a maximum of about 50% inhibition was achievable with POB1 (19). Because Hsf-1 has been shown to bind Ralbp1 in a similar region (aa 440-655), we hypothesized that Hsf-1 would inhibit the transport activity of Ralbp1. To test this postulate, we used purified recombinant Ralbp1 reconstituted into artificial asolectin/cholesterol liposomes for measurement of ATP-dependent transport activity using 14C-DOX, without or with the addition of purified Hsf-1 or POB1 recombinant protein to the transport medium. 14C-DOX uptake into the proteoliposomes in the absence of ATP was subtracted from that in the presence of ATP to obtain ATP-dependent uptake. The increasing molar ratio of albumin added to the transport reaction did not affect ATP-dependent 14C-DOX transport catalyzed by Ralbp1. In contrast, POB1 caused a saturable inhibition of 14C-DOX transport with maximal inhibition of about 50% (Fig. 3). The addition of Hsf-1 also caused saturable inhibition of transport, with a maximum of ∼40%. In the presence of both proteins, ∼90% inhibition of 14C-DOX transport was observed (Fig. 3). The additive nature of transport inhibition is consistent with distinct but perhaps overlapping binding sites for POB1 (aa 499-655) and Hsf-1 (aa 440-655), both being present in the C-terminal region (6, 19). These striking findings predict that simultaneous augmentation of POB1 and Hsf-1 in cells would very effectively inhibit Ralbp1 transport activity in cells.
FIGURE 3.
Inhibition of the 14C-DOX transport in Ralbp1 liposomes by POB1 or Hsf-1. 14C-DOX transport activity was measured as ATP-dependent DOX uptake in artificial soybean-asolectin liposomes reconstituted with purified reconstituted Ralbp1 using established methods for quantifying 14C-DOX uptake by vesicles (8, 11). Controls included liposomes prepared without Ralbp1, liposomes prepared with heat-inactivated Ralbp1, and absence of ATP. Purified Hsf-1 and/or POB1 protein were added in varying molar ratios to the transport reaction mixture. Results presented are representative of three separate experiments, each with triplicate determinations.
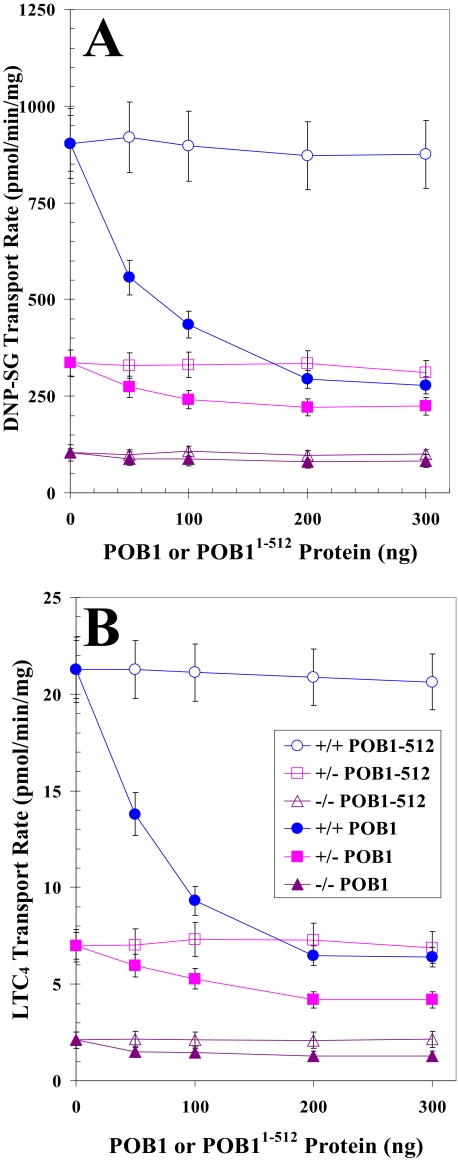
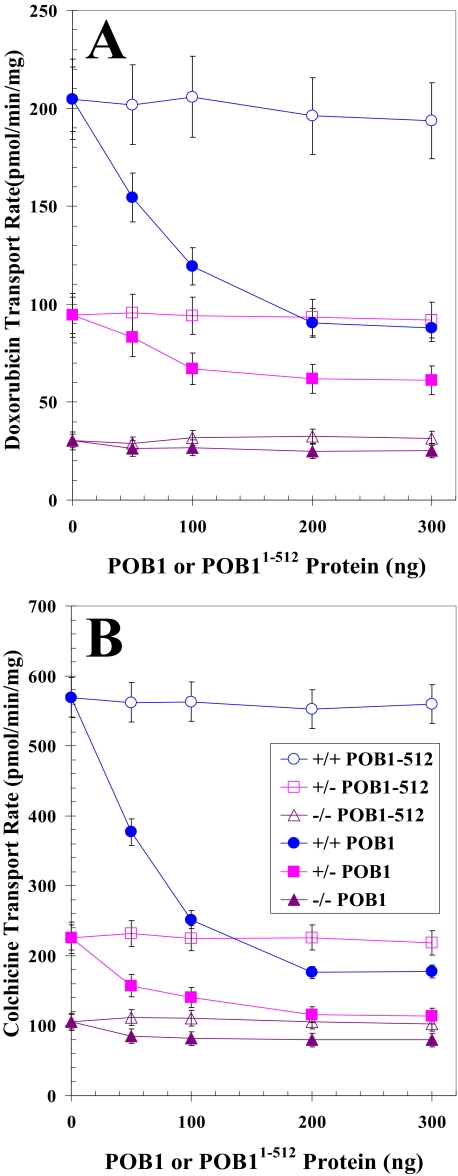
Differential Effect of POB1 on Transport of Glutathione Conjugates and Drugs by Plasma Membrane Vesicles from Ralbp1 Knock-out MEFs—If POB1 functioned to inhibit Ralbp1 in cells, the transport of glutathione conjugate substrates would be inhibited in cells augmented with POB1 only if Ralbp1 were present. To address this postulate, we compared the rate of transport for the glutathione conjugates DNP-SG and LTC4 and model substrate drugs DOX and COL in crude plasma membrane inside-out vesicles from Ralbp1+/+, Ralbp1+/-, and Ralbp1-/- MEFs. Purified full-length POB1 or the Ralbp1 non-binding mutant Ralbp1-(1-512) was added to the transport reaction mixture at varying concentrations. Total DNP-SG or LTC4 transport capacity (Fig. 4, A and B) and total DOX or COL transport capacity (Fig. 5, A and B) were unaffected by increasing the concentration of POB1-(1-512). In contrast, the addition of POB1 protein caused significant concentration-dependent and saturable inhibition of transport. Ralbp1+/- and Ralbp1-/- had significantly lower total transport capacity for both conjugates and both drugs than Ralbp1+/+. POB1 did not significantly inhibit the residual transport capacity in Ralbp1-/-, consistent with our prediction. Results of these studies show that the presence of Ralbp1 in plasma membrane vesicles is necessary for observing the transport inhibitory activity of POB1.
FIGURE 4.
Comparing POB1-mediated inhibition of glutathione conjugate efflux in Ralbp1+/+, Ralbp1+/-, or Ralbp1-/- MEFs. The transport activity of Ralbp1 toward DNP-SG (A) and LTC4 (B) was measured in crude membrane IOVs prepared from Ralbp1+/+ (circles), Ralbp1+/- (squares), and Ralbp1-/- (triangles) MEFs. The effect of full-length POB1 (closed symbols) or POB1-(1-512) (open symbols) at varying molar ratios was examined by including varying concentrations of these proteins in the transport medium. Transport medium contained IOV protein (20 μg/30 μl), 100 nm [3H]LTC4 (specific activity 1.5 × 103 cpm/pmol), or 100 μm [3H]DNP-SG (specific activity 3.8 × 103 cpm/nmol), without or with 4 mm ATP (three experiments, each in triplicate; n = 9). Heat-inactivated POB1 protein was also used for additional control.
FIGURE 5.
Comparing POB1-mediated inhibition of drug efflux in Ralbp1+/+, Ralbp1+/-, or Ralbp1-/- MEFs. The transport activity of Ralbp 1 toward DOX(A) and COL (B) was measured in crude membrane IOVs prepared from Ralbp1+/+ (circles), Ralbp1+/- (squares), and Ralbp1-/- (triangles) MEFs. The effect of full-length POB1 (closed symbols) or POB1-(1-512) (open symbols) at varying molar ratios was examined by including varying concentration of these proteins in the transport medium. Transport medium contained IOV protein (20 μg/30 μl), 3.6 μm 14C-DOX(specificactivity 8.7×104cpm/nmol), or 5μm[3H]COL(specificactivity 1.3× 104 cpm/nmol) without or with 4 mm ATP (three experiments, each in triplicate; n = 9). Heat-inactivated POB1 protein was also used for additional control.
Augmenting Cellular Ralbp1, POB1, or Hsf-1 Using Liposomal Delivery in Cultured Cells—Since augmentation of POB1 in lung cancer cells has previously been shown to increase cytotoxicity of DOX, increase DOX-accumulation in cells, decrease rate of efflux of DOX from cells, and increase apoptosis even in the absence of DOX (19), these effects should be more pronounced with a combination of Hsf-1 and POB1. We tested these predictions using an experimental model in which cellular levels of Hsf-1 and POB1 were augmented acutely by treating cells with proteoliposomes containing purified recombinant Hsf-1 or POB1. We have standardized and used this experimental model to demonstrate and quantify delivery of Ralbp1 and POB1 to cells in culture previously (19).
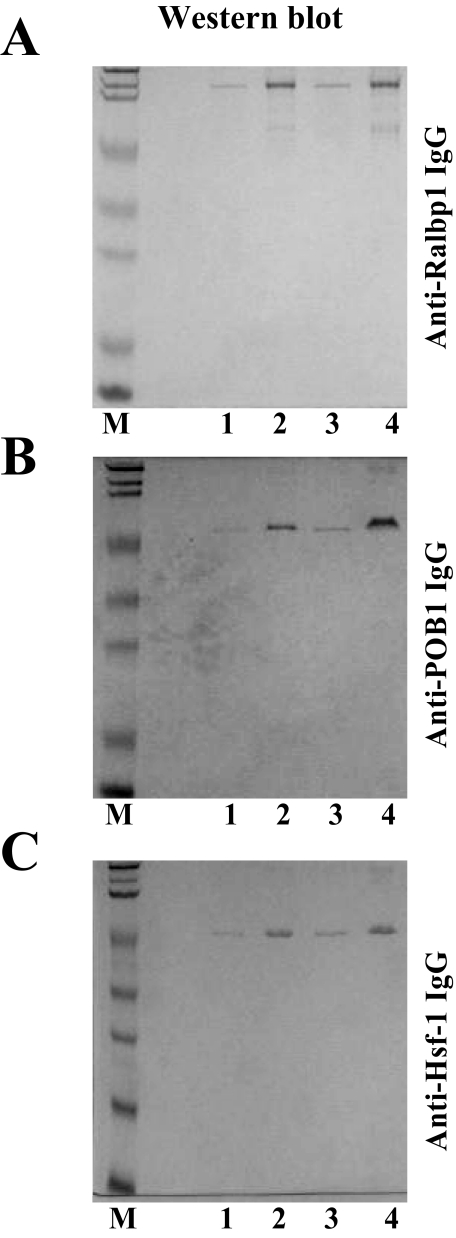
Effective entrapment of the three purified recombinant proteins was demonstrated by comparing Western blot for the three proteins in the supernatant versus pellet fraction of the proteoliposome vesiculation mixture subjected to 104,000 × g for 1 h (Fig. 6, A-C, lanes 1 and 2, respectively). The majority of protein added to the vesiculation reaction mixture was found in the pellet (proteoliposome) fraction.
FIGURE 6.
Liposomal reconstitution and delivery of Ralbp1, POB1, and Hsf-1 in cells. Purified Hsf-1, Ralbp1, and POB1 proteins (5 μg/ml) were added to liposome vesiculation mixtures containing cholesterol/asolectin (1:4, w/w), and polidocanol. Vesiculation was started by the addition of SM-2 beads, which sequester polidocanol and cause formation of primarily unilamellar vesicles with a median diameter of 0.5 μm (22). The SM-2 beads were separated by centrifugation, and the supernatant fraction was applied to an ultracentrifuge at 104,000 × g for 1 h at 4 °C. The supernatant and pellet fractions were applied to SDS-PAGE, trans-blotted, and subjected to Western blot analysis against the respective antibody to determine the amount of each protein entrapped by liposomes (A-C, lanes 1 and 2, supernatant and pellet fractions, respectively). The proteoliposomes recovered from the pellet were diluted in buffer and added to cultured H358 cells (5 × 106 cells) to a final protein concentration of 40 μg/ml. Shown are Western blot analyses comparing cells treated with control liposomes or proteoliposomes containing Hsf-1, POB1, or Ralbp1 (A-C, lanes 3 and 4, respectively) to determine whether cellular content of these proteins was increased after incubation with the respective proteoliposomes.
Human NSCLC cell line NCI-H358 was used to test whether the liposomally entrapped proteins could be delivered successfully to cells. H358 is a very well characterized NSCLC cell line that, typical for other non-small cell lung cancer cell lines, is relatively resistant to DOX (33). Liposomal delivery of protein to intact H358 NSCLC cells was tested by incubating these cells for 2 h with control liposomes (without protein) versus proteoliposomes containing one of the three recombinant proteins. An increase in total cellular content of each protein by ∼3-fold (as quantified by scanning densitometry) was observed under the present conditions in cells incubated with respective proteoliposomes (Fig. 6, A-C, lanes 3 and 4, respectively). These studies confirmed previous results demonstrating that proteoliposomes can deliver these proteins into cells (8).
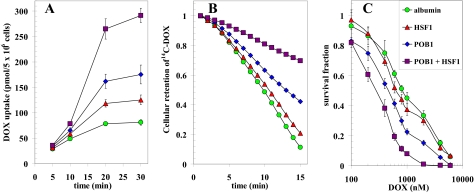
Effect of POB1 and Hsf-1 on Accumulation, Efflux, and Cytotoxicity of DOX in H358 NSCLC—Because inhibition or depletion of Ralbp1 has been shown previously to selectively target certain cancer cell types for apoptosis and sensitize them to chemotherapy drug toxicity (14, 26), we reasoned that augmenting cellular POB1 or Hsf-1 should inhibit efflux of DOX from cells, resulting in increased accumulation in cells and thus increased cytotoxicity of DOX. H358 cells were incubated with 14C-DOX for varying time intervals, and drug uptake in cells was quantified. Cells were pretreated for 24 h with proteoliposomes prepared in the presence of either albumin or purified recombinant POB1, Hsf-1, or both. The result of these drug-accumulation studies (Fig. 7A) showed that the uptake was increased substantially by Hsf-1, was increased more by POB1, and was almost 5 times as high when both proteins were added. To confirm that the increased accumulation was due to inhibited transport, drug efflux studies were carried out in cells pretreated in the same manner. The drug efflux curve was steepest with albumin, affected slightly by Hsf-1, affected more by POB1, and markedly slowed when both proteins were present (Fig. 7B). The decreased efflux and increased accumulation caused by Hsf-1, POB1, or the combination translated into increased sensitivity to DOX in a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cytotoxicity assay done at 96 h after drug exposure (Fig. 7C). Taken together, these findings demonstrated that POB1 and Hsf-1 are effective inhibitors of DOX efflux in these NSCLC cells known to express Ralbp1 and that the greater drug accumulation translates to greater cytotoxicity.
FIGURE 7.
Effect of POB1 and Hsf-1 supplementation on DOX-cytotoxicity, uptake, and efflux. H358 NSCLC cells were treated with proteoliposomes containing 40 μg/ml albumin (green circles), Hsf-1 (red triangles), POB1 (blue diamonds), or both Hsf-1 and POB1 (purple squares). Uptake of 14C-DOX (specific activity 8.5 × 104 cpm/nmol) by cells was determined by incubating the cells for varying periods of time with radiolabeled 14C-DOX and measuring radioactivity in the cell pellet (A). For efflux studies, cells were loaded with radiolabeled 14C-DOX for 60 min, followed by rapid dilution in 1 ml of buffer. Aliquots of the buffer were taken at 1-min intervals, and cellular drug content was calculated by back-addition from the residual DOX in cells at the end of the experiment (B) (11). The cytotoxic effects of DOX were measured by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay 96 h after drug exposure (C) (26).
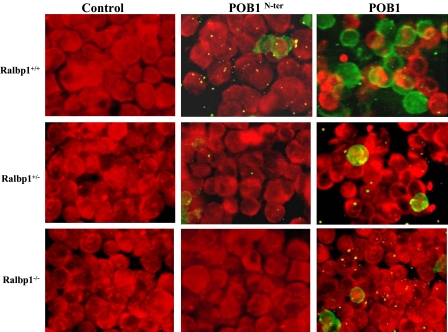
Loss of Ralbp1 Abrogates the Proapoptotic Effects of POB1—The present studies show that augmentation of POB1 in lung cancer cells inhibits the efflux of drugs and glutathione conjugates and triggers cytotoxicity through apoptosis, with consequent decreased colony forming potential. Because the transport studies indicate that the inhibitory effects of POB1 are absent in the absence of POB1, if the transport activity is also a major determinant of apoptosis, the apoptotic effect of POB1 augmentation should be absent in Ralbp1-/- cells. To test this prediction, we compared the appearance of apoptosis by TUNEL assay between Ralbp1+/+, Ralbp1+/-, and Ralbp1-/- MEFs treated with proteoliposomes containing either full-length POB1 or POB1-(1-512). The studies shown above have demonstrated the ability of the proteoliposomes to successfully deliver proteins to cells. The results of TUNEL studies (Fig. 8) demonstrated that although the control liposomes caused no apoptosis, POB1 did cause apoptosis. Although POB1-(1-512) induced apoptosis in a small population of Ralbp1+/+ cells (not statistically significantly different from controls), its apoptotic effect was absent in Ralbp1-/-. Furthermore, the apoptotic effect of full-length POB1 was attenuated in Ralbp1+/- cells and nearly absent in Ralbp1-/- cells. These studies demonstrate that Ralbp1 is required for at least the majority of total apoptotic effect of POB1, and in context of the effect of POB1 on transport activity, these findings are most consistent with idea that the apoptotic activity of POB1 is mediated through its effects on inhibition of transport.
FIGURE 8.
Loss of Ralbp1 results in loss of POB1-mediated apoptosis. The effect of POB1 overloading on apoptosis in MEFs was examined by a TUNEL assay. Ralbp1+/+, Ralbp1+/-, and Ralbp1-/- MEFs were grown on coverslips incubated with liposomes containing either no POB1, POB1-(1-512), or full-length POB1 (40 μg/ml final concentration) for 24 h prior to the TUNEL assay using a Promega fluorescence detection kit and examined using a Zeiss LSM 510 META (Germany) laser-scanning fluorescence microscope with filters of 520 and >620 nm. Photographs taken at identical exposure at ×400 magnification are presented. Apoptotic cells showed green fluorescence and characteristic of cell shrinkage.
Effect of Proteoliposome-mediated Hsf-1 and POB1 Augmentation on Apoptosis and Colony Forming Activity—In previous studies, we have demonstrated that Ralbp1 inhibition, in the absence of cytotoxic drugs, causes apoptosis in lung and several other types of cancer cells while sparing nonmalignant cell types (14). The mechanism of apoptosis has been postulated to be through the direct toxicity of lipid hydroperoxide metabolites that accumulate upon the inhibition of Ralbp1 (34) as well as through effects on stress defense pathway proteins, including c-Jun N-terminal kinase and AP1, and that apoptosis proceeds though a caspase-dependent mechanism (13, 35). In the present studies, we tested the effects of the combination of Hsf-1 and POB1 in causing apoptosis by flow cytometry and immunohistochemistry.
Flow cytometry was performed to analyze the presence of apoptosis or necrosis induced by augmenting cells with purified recombinant Hsf-1 or POB1 by liposomal delivery. Dual staining with Annexin V and propidium iodide staining was used to quantify apoptosis and necrosis, respectively (Fig. 9). The Annexin V-positive fraction of cells was unaffected by either control liposomes (no protein) or by proteoliposomes containing N-terminal (aa 1-512) POB1 recombinant protein, which lacks the Ralbp1 binding region. Intact POB1, in contrast, caused a significant increase in both apoptosis and necrosis. In contrast, Hsf-1 alone caused an increase in the apoptotic fraction but did not affect necrosis. Cells treated with liposomes containing both Hsf-1 and POB1 were affected in a supra-additive manner with a marked increase in cell death, contributed largely by an increase in apoptosis. These findings were mirrored in the results of apoptosis assay by TUNEL, which reports the appearance of DNA fragmentation, a late event in apoptosis (Fig. 10A). Quantitation of red and green fluorescence by image analysis confirmed the qualitative findings that the combination of Hsf-1 and POB1 was most effective at inducing apoptosis and that the N-terminal POB1, which cannot bind Ralbp1 (19), also is not effective in inducing apoptosis or necrosis.
FIGURE 9.
Effect of Hsf-1 and POB1 on apoptosis and necrosis by flow cytometry. H358 cells (0.5 × 106 cells/ml) were grown in a 6-well plate and were treated with control liposomes or liposomes reconstituted with 40 μg/ml (final concentration) recombinant Hsf-1, POB1, POB1-(1-512), or both (Hsf-1 and POB1) protein. After a 24-h incubation, the cells were harvested and centrifuged at 1500 × g for 5 min. Cells were washed once with PBS and resuspended in 400 μl of cold annexin binding buffer containing 5 μl of Annexin V-FITC and 5 μl of 0.1 mg/ml propidium iodide. Cells were incubated at room temperature for 10 min in the dark and were analyzed by flow cytometry using a Beckman Coulter Cytomics FC500 flow cytometer. Results were processed using CXP2.2 analysis software from Beckman Coulter.
FIGURE 10.
Effect of POB1 and/or Hsf-1 on apoptosis and colony forming efficiency in H358 NSCLC cells. A, the effect of POB1 and Hsf-1 overloading on the TUNEL apoptosis assay. The H358 cells were grown on coverslips incubated for 24 h with control liposomes and proteoliposomes (40 μg/ml final concentration) containing either POB1-(1-512), POB1, Hsf-1, or both (POB1 + Hsf-1) for 24 h prior to the TUNEL assay using the Promega fluorescence detection kit and examined using a Zeiss LSM 510 META laser-scanning fluorescence microscope with filters of 520 and >620 nm. Photographs taken at identical exposure at ×400 magnification are presented. Apoptotic cells showed green fluorescence and characteristic of cell shrinkage. Data were also analyzed by Image-J, and results are presented in the adjacent bar diagram (live cells are red, and apoptotic cells are green). The colony-forming assay was performed by staining the cells with methylene blue, and the colonies were counted using an Innotech Alpha Imager (B). Values are means ± S.D. of three separate experiments.
The cytotoxic effects of POB1 and Hsf-1 were further quantified by colony-forming assays on cells treated with either no proteoliposome or proteoliposomes containing either albumin, POB1, Hsf-1, or both POB1 and Hsf-1 (Fig. 10B). The albumin or POB1-(1-512) proteoliposomes did not significantly affect colony forming capacity. In contrast, Hsf-1 proteoliposomes had a small but significant (p < 0.05) effect as compared with controls. The effect of POB1 was greater, with almost 40% inhibition of colony formation. The combination of the two proteins had additive effects with ∼60% reduction in colony forming capacity of NSCLC cells (Fig. 10B).
Taken together, the results of the present studies confirmed that both Hsf-1 and POB1 bound to Ralbp1 and for the first time demonstrated the formation of a ternary complex. The formation of this complex inhibited the transport activity of Ralbp1 in isolated purified protein systems; this inhibition was also apparent in intact cells, where POB1 and Hsf-1 caused increased cellular accumulation of DOX due to decreased efflux rate. The increased DOX accumulation caused increased cytotoxicity as expected.
The combination of POB1 and Hsf-1 increased apoptosis, even in the absence of any DOX. The combination of Hsf-1 and POB1, which together completely inhibit the transport activity of Ralbp1 (Fig. 3), caused apoptosis to a similar extent as seen previously with anti-Ralbp1 antibodies at a sufficient concentration to completely inhibit Ralbp1 transport activity (36) or with complete depletion of Ralbp1 by siRNA or antisense in both cell and xenograft models of H358 and other cancers (14, 15, 26). Our previous studies in cancer cells as well as Ralbp1-/- MEFs have shown that Ralbp1 inhibition, depletion, or absence causes increased intracellular accumulation of toxic and proapoptotic lipid aldehydes (33, 34, 36, 37), which are normally metabolized and excreted from cells as glutathione conjugates. Inhibition of glutathione conjugate and DOX transport activity of Ralbp1 by POB1 in the present studies also strongly support the model that the proapoptotic effect of Ralbp1 inhibition is mediated primarily by its inhibition of the transport activity of membrane-bound Ralbp1.
Effects of Hsf-1 Overexpression in Heat Shock Response—Our previous studies have shown that transient exposure to oxidative and radiant stress, both of which cause an increase in cellular lipid-derived proapoptotic alkenals, results in increased cellular content of Ralbp1 protein and accompanying resistance to stress after a short recovery period. This poststress resistance is completely abrogated by inhibition of the transport activity of Ralbp1 by the addition of anti-Ralbp1 antibodies (33, 36) that have been shown to inhibit the transport activity of Ralbp1 through specific binding to cell-surface epitopes composed of aa 171-185 in the N-terminal region of Ralbp1 (10). These results indicate that Ralbp1 is an important effector necessary for protection of cells from stressors that increase the cellular concentration of lipid peroxidation-derived proapoptotic moieties (9, 13, 35).
Like Ralbp1, Hsf-1 is also responsive to stress signaling. Hsf-1 is found in an inactive state in the unstressed state in complex with Ralbp1, and because stress signals cause the activation of Hsf-1 as a result of dissociation of this complex, allowing its translocation to the nucleus, where it functions as a master transcription factor for the heat shock response, Ralbp1 could function as an inhibitor of the function of Hsf-1. In the context of present findings demonstrating inhibition of transport activity of Ralbp1 by Hsf-1, an alternative model emerges in which Ralbp1 and Hsf-1 are mutually inhibitory, Ralbp1 inhibiting the transcription activity of Hsf-1 and Hsf-1 inhibiting the transport activity of Ralbp1 as well as sequestering it in a cytoplasmic complex. Implications of this model were explored in context of the effects of heat shock and Hsf-1 overexpression or depletion in Ralbp1+/+ versus Ralbp1-/- MEFs.
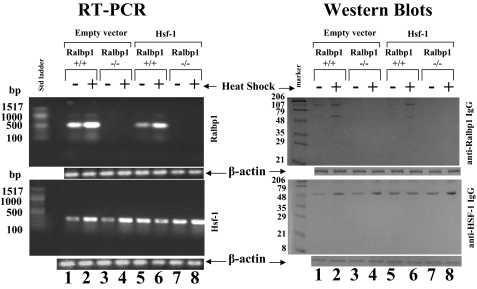
We reasoned that if Ralbp1 were necessary for the function of Hsf-1, the response to Hsf-1 overexpression in Ralbp1+/+ should be distinctly different from that in Ralbp1-/- MEFs. MEFs were prepared from fetal mice (37), and the fourth passage of cells was used for the present studies. We examined the effect of 20-min 42 °C heat shock on the expression of Ralbp1 and Hsf-1 in both genotypes of MEFs transiently transfected with either empty vector or Hsf-1. Ralbp1 and Hsf-1 mRNA and protein were examined by RT-PCR and Western blot, respectively, after a 2-h postshock recovery period at 37 °C.
The MEF model was validated by results showing an absence of Ralbp1 in Ralbp1-/- cells as judged by RT-PCR or Western blots (Fig. 11, lanes 3, 4, 7, and 8). Hsf-1 mRNA and protein levels were similar in Ralbp1+/+ and Ralbp1-/- MEFs (Fig. 11, lane 1 versus lane 3), indicating that Ralbp1 does not participate in mechanisms that regulate Hsf-1 expression. Successful transient transfection of Hsf-1, with slightly increased mRNA and protein level, was confirmed in both Ralbp1+/+ and Ralbp1-/- cells (Fig. 11, lane 1 versus lane 5 and lane 3 versus lane 7, respectively).
FIGURE 11.
Effect of Hsf-1 overexpression in Ralbp1+/+ and Ralbp1-/- MEFs. Ralbp1+/+ and Ralbp1-/- MEFs were cultured from fetal mice, as described previously (37), and all present studies were performed within the first six passages. Heat shock was applied by incubation of cells at 42 °C for 30 min, followed by a 2-h recovery period at 37 °C. Transient transfection of Hsf-1 was carried out using a pcDNA3.1 eukaryotic expression vector containing full-length Hsf-1 cDNA, and empty vector was used as control. Expression of mRNA in Ralbp1+/+ and Ralbp1-/- MEFs was evaluated by RT-PCR analysis. RNA was prepared by the RNeasy kit (Qiagen). For quantitative RT-PCR of Hsf-1, the forward primer was nt 337-348, and reverse primer was nt 635-655 (318-bp product). For Ralbp1, the forward primer was nt 1496-1515, and reverse primer was nt 1948-1968 (472-bp product). RT-PCR was performed using the Ready-to-use RT-PCR beads according to the manufacturer's instructions (Amersham Biosciences). For Western blot analyses, 200 μg of crude membrane homogenate protein was applied in each lane for SDS-PAGE, and after trans-blotting, Western blots were done using rabbit anti-human Ralbp1 polyclonal antibodies and rabbit anti-human Hsf-1 polyclonal antibodies. Secondary antibodies were goat anti-rabbit IgG peroxidase-conjugated. The blots were developed using 4-chloro-1-naphol as the chromogenic substrate. β-Actin was used as an internal control.
Heat shock caused an increase in both mRNA and protein of both Ralbp1 and Hsf-1. The degree of increase in Hsf-1 after heat shock was similar between Ralbp1+/+ and Ralbp1-/- MEFs, suggesting that the presence of Ralbp1 was not necessary for the increase in Hsf-1 seen in response to heat shock. Overexpression of Hsf-1 caused a decrease in Ralbp1 mRNA and protein in Ralbp1+/+ MEFs (Fig. 11, lane 1 versus lane 5). In Ralbp1+/+ cells, overexpression of Hsf-1 blunted the heat shock response in terms of Hsf-1 levels; in contrast, in Ralbp1-/- cells, an heat shock-induced increase in Hsf-1 was preserved. These results suggest that Ralbp1 and Hsf-1 may function as mutual inhibitors, each functioning to suppress the heat shock-induced increase in the other.
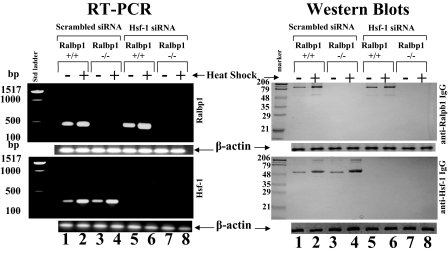
Effects of siRNA-mediated Depletion of Hsf-1 on Heat Shock Response—If the above postulate were true, we would expect that, in an experiment with a similar design, if Hsf-1 were depleted, the level of Ralbp1 would increase, and the heat shock-mediated response in Ralbp1 levels would be augmented. The results of experiments to test this speculation are presented in Fig. 12. As in Fig. 11, Ralbp1 was absent in the Ralbp1-/- MEFs. Also confirming the previous result, heat shock caused an increase in both mRNA and protein of both Ralbp1 and Hsf-1. The heat shock response in Hsf-1 level was similar in both Ralbp1+/+ and Ralbp1-/- MEFs, confirming that the Ralbp1 itself does not play any crucial role in the basal or heat shock-induced expression of Hsf-1. Successful complete depletion of HSF-1 mRNA and protein was accomplished in both Ralbp1+/+ and Ralbp1-/- cells (Fig. 12, lanes 5-8, lower panels). Hsf-1 depletion by siRNA slightly increased basal Ralbp1 mRNA and significantly increased its postheat shock. In contrast to the mRNA result, the Ralbp1 protein level was not suppressed by Hsf-1. These results suggest that Hsf-1 negatively regulates Ralbp1 mRNA level, probably at the transcriptional level. No suppressive effect of Ralbp1 on Hsf-1 basal or heat shock levels was evident.
FIGURE 12.
Effect of Hsf-1 depletion by siRNA in Ralbp1+/+ and Ralbp1-/- MEFs. Ralbp1+/+ and Ralbp1-/- MEFs were transfected with scrambled and Hsf-1 siRNA using the Lipofectamine 2000 reagent kit (Invitrogen). Expression of mRNA in Ralbp1+/+ and Ralbp1-/- MEFs was evaluated by RT-PCR analysis, and band intensities were checked by densitometry. RT-PCR was performed using Ralbp1 gene-specific primers (1496-1515 bp (upstream) and 1948-1968 (downstream)) (472-bp product) and Hsf-1 gene-specific primers (337-348 bp (upstream) and 635-655 (downstream)) (318-bp product). RT-PCR was performed using the Ready-to-use RT-PCR beads according to the manufacturer's instructions (Amersham Biosciences). Comparisons of Ralbp1 and Hsf-1 protein levels were performed by Western blot analyses. 200-μg aliquots of crude membrane extracts were applied to SDS-PAGE and Western blotting against Ralbp1 IgG and Hsf-1 IgG. Scanning densitometry was used to measure the intensities of the bands. β-Actin was used as an internal control.
Subcellular Fractionation of Ralbp1—Results of recent studies by Hu and Mivechi (6) have shown that Ralbp1 and Hsf-1 are sequestered in the cytoplasm in the unstressed state bound to α-tubulin and HSP90 and released upon activated Ral binding. In those studies, Ralbp1 was shown to migrate to the nuclear membrane under stress. Now we concluded that Ralbp1 was predominantly a cytosolic protein; no direct analyses of membrane fractions was performed.
To directly address the question of the cellular distribution of Ralbp1, we performed subcellular distribution analyses of Ralbp1 by differential centrifugation as well as confocal immunohistochemistry in cells fixed to ensure intact plasma membranes. Standard methods for subcellular fractionation and fractional purity determination were followed, as described previously (27-29). Marker enzymes used were lactate dehydrogenase (for cytosol), acetylcholinesterase (for plasma membrane), and carnitine acyltransferase (for microsomes). The mitochondrial fraction was assessed for purity using Western blot analysis against anti-human manganese-dependent superoxide dismutase, and nuclear fraction purity was demonstrated using Western blot against anti-human histone-H3. Results of these studies (Fig. 13, A-C; note log scale) demonstrated excellent separation of fractions. Lactate dehydrogenase activity in the cytosol fraction was much higher than in any other fractions; acetylcholinesterase activity in the plasma membrane fraction was higher than in any other fraction; and carnitine acyltransferase activity was significantly greater in the microsomal fraction. Histone H3 was found only in the nuclear fraction and manganese-dependent superoxide dismutase in the mitochondrial fraction, without significantly detectable cross-contamination (Fig. 13C). These results established that the five fractions were adequately separated. Ralbp1 was found in the plasma membrane fraction (about half of the total) in the unstressed state, with significant amounts also present in cytosol and nuclear fraction and none found in the mitochondrial or microsomal fractions (Fig. 13C).
FIGURE 13.
Subcellular distribution of Ralbp1. The scheme used for fractionating cell homogenate from 1 × 109 H358 cells into the nuclear (N), plasma membrane (PM), mitochondrial (MT), microsomal (MC), and cytosolic (C) fractions (27) is presented (A). Fractions were assayed for activity of lactate dehydrogenase (27), acetylcholinesterase (28), and carnitine acyltransferase (29) (B). The fractions (100 μg each) were subjected to SDS-PAGE, transblotted, and analyzed by Western blot for Ralbp1 as well as for histone-H3 and manganese-dependent superoxide dismutase (MnSOD) to demonstrate separation of nuclear and mitochondrial fractions, respectively (C). Std, standard.
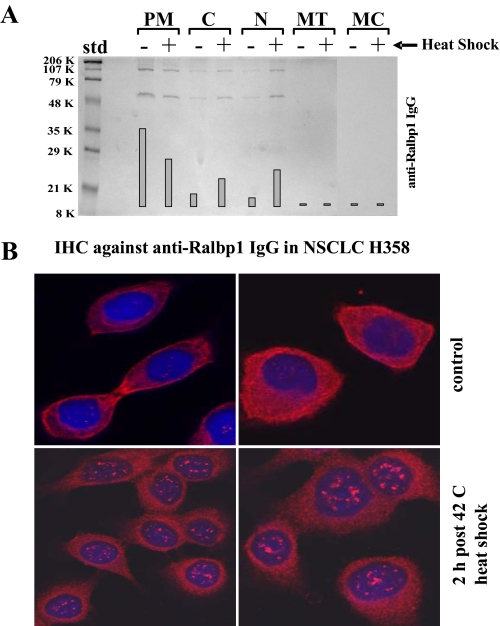
Effects of Heat Shock on the Subcellular Distribution of Ralbp1 and Hsf-1—Upon application of 42 °C heat shock, followed by a 2-h recovery, the subcellular fractionation studies revealed a decrease in Ralbp1 in the membrane fraction, with a shift to the cytosol and more prominently the nucleus (Fig. 14A). The findings of subcellular fractionation were correlated with immunohistochemistry of fixed cells (Fig. 14B). Although cytosolic staining is evident, sharply demarcated membrane features also indicated the presence of Ralbp1 in the membrane.
FIGURE 14.
Effect of heat shock on subcellular distribution of Ralbp1. The effect of heat shock (30 min at 42 °C, followed by a 2-h recovery at 37 °C) on Ralbp1 in each of the fractions is shown by Western blot analysis. Results were quantified by scanning densitometry (A). The cellular distributions of Ralbp1 were also analyzed by immunohistochemistry (B) using polyclonal anti-Ralbp1 antibodies without (top two panels) or with heat shock (bottom two panels). Std, standard.
Plasma membrane binding of Ralbp1 was more apparent when contrasted with cells after heat shock, in which the sharply demarcated plasma membrane staining disappears (Fig. 14B). These findings are consistent with subcellular fractionation results in the present studies, our previous studies (8-11), and results of studies by others showing an important role of Ralbp1 in membrane functions, including endocytosis and receptor-ligand signaling (12, 16-18). Ralbp1 has been shown by others to be an integral component of the clathrin-dependent endocytic apparatus (12). Clathrin-dependent endocytosis normally functions to terminate receptor-ligand signaling by endocytosis and subsequent degradation of the receptor-ligand complex, such as insulin-insulin receptor, epidermal growth factor-epidermal growth factor receptor, or transforming growth factor-transforming growth factor receptor. Other investigators have shown that stressors cause an increase in endocytosis of ligands, such as insulin, possibly contributing to phenomena such as insulin resistance. Our recently studies showing the nearly complete lack of clathrin-dependent endocytosis in Ralbp1-/- mice strongly support an integral and perhaps rate-regulatory role of Ralbp1 in clathrin-dependent endocytosis (38, 39). Because mutants of Ralbp1 that reduce glutathione conjugate binding and transport result in parallel reduction of endocytosis,3 our studies suggest that ATP hydrolysis-dependent efflux of endogenous glutathione conjugates may serve as an energy transduction mechanism required for endocytosis. Results of the present studies showing the stress-mediated translocation of Ralbp1 from the plasma membrane fraction to intracellular compartments are quite consistent with the observation of others that stressors result in a transient increase in clathrin-dependent endocytosis (16-18, 38, 39).
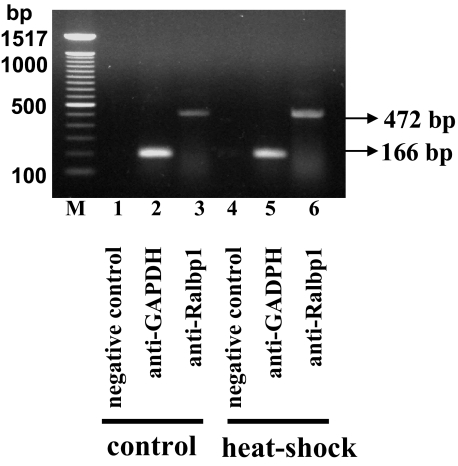
In addition to the shift of Ralbp1 away from the membrane after heat shock, these studies also clearly demonstrated translocation of Ralbp1 to the nucleus in response to heat shock. To determine whether the patchy nuclear staining was due to chromatin binding through its leucine zipper motif in the C-terminal (9), we performed a ChIP assay in nuclear extracts of H358 cells using Ralbp1 antibodies for immunoprecipitation (Fig. 15). No signal was seen upon immunoprecipitation with negative control IgG; as positive control, the antibodies were used toward RNA polymerase II, which is known to bind to the promoter region of the glyceraldehyde-3-phosphate dehydrogenase gene. Neither the positive nor the negative controls were affected by heat shock. In both unstressed and heat-shocked cells, Ralbp1 was found bound to chromatin. Scanning densitometry indicated a 2.7-fold increase in binding to chromatin from heat-shocked cells. These studies indicate that Ralbp1 can bind chromatin, that this binding is enhanced after heat shock, and that perhaps Ralbp1 could function in transcriptional machinery. Detailed studies are being pursued to determine any functional significance of a potentially novel activity of Ralbp1.
FIGURE 15.
Chromatin binding of Ralbp1 by ChIP assay. H358 cells (4.5 × 107) were grown and fixed with 37% formaldehyde. Cells were scraped, and the chromatin was sheared by the protocol given under “Experimental Procedures.” The chromatin was immunoprecipitated using the negative control IgG, positive control IgG (provided by Active Motif), and anti-Ralbp1 IgG followed by binding with protein G beads. The chromatin was eluted from the protein G beads and was amplified by PCR using the control primers as well as negative control primers (provided by Active Motif) and Ralbp1 primers (forward primer, nt 1496-1515; reverse primer, nt 1948-1968), respectively. PCR products were quantified by running on 1% agarose gel. For heat shock experiments, the cells were exposed to 42 °C for 30 min, brought back to 37 °C, and allowed to recover for 2 h at 37 °C before fixing with 37% formaldehyde.
DISCUSSION
The present studies demonstrate that Ralbp1, Hsf-1, and POB1 can form a ternary complex and for the first time show that Hsf-1 inhibits the transport activity of Ralbp1. The presence of Hsf-1 and POB1 is quite effective at abrogating the transport activity nearly completely. Taken in context of known functions and well defined binding interactions of Ralbp1 with Hsf-1 (6) and POB1 (19, 32), these findings have particular significance with respect to the view of the signaling mechanisms involved. Recent studies have shown that Hsf-1 is stored in the cellular cytoskeleton in a complex with α-tubulin, HSP90, and Ralbp1, released upon stress (6). The heat shock-mediated release can be mimicked by Ral-GTP, which binds Ralbp1 and dissociates Ralbp1 and Hsf-1 from the complex. Taken together with previous studies (6), our findings support a refined model in which, under unstressed conditions, Ralbp1 is found both in the cytosol (complexed with Hsf-1) and in the plasma membrane, and the specific Hsf binding to Ralbp1 in the membrane fraction inhibits the transport activity of Ralbp1. This inhibition is incomplete; POB1, which binds a distinct site in the C-terminal domain, inhibits the remaining transport activity.
The consequences of this inhibition were demonstrated clearly in terms of drug efflux, drug accumulation, cytotoxicity, apoptosis, and colony forming activity. The present studies also provide convincing evidence for the distribution of Ralbp1 in the plasma membrane, cytosol, and nucleus and demonstrate for the first time the translocation of Ralbp1 to nuclear chromatin.
Our studies show that heat shock followed by a 2-h recovery results in an increase in cellular Hsf-1 and Ralbp1 mRNA and protein. Although Hsf-1 appears to be a negative regulator of the level of basal and heat-induced Ralbp1 expression, loss of Ralbp1 does not itself affect Hsf-1 levels. These results imply that Hsf-1 may exert some transcriptional control of Ralbp1; the opposite is unlikely.
Mouse gene knock-out studies as well as a number of studies in human cells have shown that Ralbp1 loss causes a dramatic loss of total DOX transport and total glutathione conjugate transport capacity to ∼20% of wild-type level and that P-glycoprotein and MRP1 (multidrug resistance protein) account for the majority of the remaining transport activity (34). Inhibition of Ralbp1 that would occur should cause an accumulation of the endogenous lipid hydroperoxides, alkenals and their glutathione conjugates, as is known to occur upon Ralbp1 gene disruption (34). Among the most potent proapoptotic lipid-aldehydes obligatorily generated during heat stress is 4-hydroxy-t-2-nonenal, and its role in apoptosis triggered by heat shock is established (13, 35). Our studies showed that in the absence of any cytotoxic drug, the combined treatment with Hsf-1 and POB1 triggered massive apoptosis in lung cancer cells under study; our previous studies have shown that in situ inhibition of Ralbp1 using POB1 or specific antibodies or by Ralbp1 depletion causes apoptosis through activation of both extrinsic and intrinsic apoptosis pathways (19, 38-40). Recent studies showing direct interaction of 4-hydroxy-t-2-nonenal with death receptors and subsequent triggering of apoptosis signaling that can be regulated by Daxx (41-43) reinforce the assertion that mechanisms that regulate cellular 4-hydroxy-t-2-nonenal levels are crucial for defending against apoptosis. Studies in Ralbp1-/- animals showing increased tissue lipid hydroperoxides, alkenals, and their glutathione conjugates as well as marked increase in heat shock proteins (9, 34, 38, 44) provide good evidence for the assertion that Ralbp1 functions in removal of endogenous glutathione-lipid conjugates and that because Ralbp1 is unavailable to sequester Hsf-1, its transcriptional activity is increased. With regard to this proposed mechanism, it should be pointed out that Hsf-1 overexpression also inhibits Ralbp1 expression. Thus, the inhibitory effect of Hsf-1 on Ralbp1-mediated efflux activity is probably aided by transcriptional inhibition of Ralbp1 expression. The marked apoptotic effect of Hsf-1 and POB1 augmentation in lung cancer cells suggests a novel targeted therapy in which liposomally encapsulated Hsf-1 and POB1 could be used clinically as a therapeutic agent.
Acknowledgments
We thank Xiangle Sun (Core Facility at the University of North Texas Health Science Center, Fort Worth, TX) for helping with flow cytometry and laser capture microdissection (supported by National Institutes of Health Grant ISIORR018999-01A1).
This work was supported, in whole or in part, by National Institutes of Health Grants CA 77495 and CA 104661 (to S. A.). This work was also supported by the Cancer Research Foundation of North Texas (to S. S. S. and S. Y.), the Institute for Cancer Research, and the Joe and Jessie Crump Fund for Medical Education (to S. S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DOX, doxorubicin (Adriamycin); NSCLC, non-small cell lung cancer; COL, colchicine; LTC4, leukotriene C4; SPDP, N-succinimidyl 3-(2-pyridyldithio) propionate; TUNEL, TdT-mediated dUTP nick end labeling; aa, amino acid(s); DNP-SG, dinitrophenyl-S-glutathione; IOV, inside-out vesicle; PBS, phosphate-buffered saline; siRNA, small interfering RNA; nt, nucleotide(s); ChIP, chromatin immunoprecipitation; RT, reverse transcription.
S. Awasthi, unpublished observations.
References
- 1.Mivechi, N. F., Ouyang, H., and Hahn, G. M. (1992) Cancer Res. 52 6815-6822 [PubMed] [Google Scholar]
- 2.Inouye, S., Katsuki, K., Izu, H., Fujimoto, M., Sugahara, K., Yamada, S., Shinkai, Y., Oka, Y., Katoh, Y., and Nakai, A. (2003) Mol. Cell Biol. 23 5882-5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shamovsky, I., Ivannikov, M., Kandal, E. S., Gershon, D., and Nudler, E. (2006) Nature 440 556-560 [DOI] [PubMed] [Google Scholar]
- 4.Kabakov, A. E., Malyutina, Y. V., and Latchman, D. S. (2006) Radiation Res. 165 410-423 [DOI] [PubMed] [Google Scholar]
- 5.Tanaka, K., Namba, T., Arai, Y., Fujimoto, M., Adachi, H., Sobue, G., Takeuchi, K., Nakai, A., and Mizushima, T. (2007) J. Biol. Chem. 282 23240-23252 [DOI] [PubMed] [Google Scholar]
- 6.Hu, Y., and Mivechi, N. F. (2003) J. Biol. Chem. 278 17299-17306 [DOI] [PubMed] [Google Scholar]
- 7.Mivechi, N. F., Shi, X. Y., and Hahn, G. M. (1995) J. Cell Biochem. 59 266-280 [DOI] [PubMed] [Google Scholar]
- 8.Awasthi, S., Cheng, J., Singhal, S. S., Saini, M. K., Pandya, U., Pikula, S., Pikula, J., Singh, S. V., Zimniak, P., and Awasthi, Y. C. (2000) Biochemistry 39 9327-9334 [DOI] [PubMed] [Google Scholar]
- 9.Awasthi, S., Singhal, S. S., Sharma, R., Zimniak, P., and Awasthi, Y. C. (2003) Int. J. Cancer 106 635-646 [DOI] [PubMed] [Google Scholar]
- 10.Yadav, S., Singhal, S. S., Singhal, J., Wickramarachchi, D., Knutson, E., Albrecht, T. B., Awasthi, Y. C., and Awasthi, S. (2004) Biochemistry 43 16243-16253 [DOI] [PubMed] [Google Scholar]
- 11.Stuckler, D., Singhal, J., Singhal, S. S., Yadav, S., Awasthi, Y. C., and Awasthi, S. (2005) Cancer Res. 65 991-998 [PubMed] [Google Scholar]
- 12.Jullien-Flores, V., Mahe, Y., Mirey, G., Leprince, C., Meunier-Bisceuil, B., Sorkin, A., and Camonis, J. H. (2000) J. Cell Sci. 113 2837-2844 [DOI] [PubMed] [Google Scholar]
- 13.Yang, Y., Sharma, A., Sharma, R., Patrick, B., Singhal, S. S., Zimniak, P., Awasthi, S., and Awasthi, Y. C. (2003) J. Biol. Chem. 278 41380-41388 [DOI] [PubMed] [Google Scholar]
- 14.Singhal, S. S., Awasthi, Y. C., and Awasthi, S. (2006) Cancer Res. 66 2354-2360 [DOI] [PubMed] [Google Scholar]
- 15.Singhal, S. S., Singhal, J., Yadav, S., Dwivedi, S., Boor, P. J. Awasthi, Y. C., and Awasthi, S. (2007) Cancer Res. 67 4382-4389 [DOI] [PubMed] [Google Scholar]
- 16.Nakashima, S., Morinaka, K., Koyama, S., Ikeda, M., Kishida, M., Okawa, K., Iwamatsu, A., Kishida, S., and Kikuchi, A. (1999) EMBO J. 18 3629-3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kariya, K., Koyama, S., Nakashima, S., Oshiro, T., Morinaka, K., and Kikuchi, A. (2000) J. Biol. Chem. 275 18399-18406 [DOI] [PubMed] [Google Scholar]
- 18.Morinaka, K., Koyama, S., Nakashima, S., Hinoi, T., Okawa, K., Iwamatsu, A., and Kikuchi, A. (1999) Oncogene 18 5915-5922 [DOI] [PubMed] [Google Scholar]
- 19.Yadav, S., Zajac, E., Singhal, S. S., Singhal, J., Drake, K., Awasthi, Y. C., and Awasthi, S. (2005) Biochem. Biophys. Res. Commun. 328 1003-1009 [DOI] [PubMed] [Google Scholar]
- 20.Awasthi, Y. C., Garg, H. S., Dao, D. D., Partridge, C. A., and Srivastava, S. K. (1981) Blood 58 733-738 [PubMed] [Google Scholar]
- 21.Sambrook, J., Fritsch, E. F., and Miniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., pp. 17.3-17.9, Cold Spring Harbor University, Cold Spring Harbor, NY
- 22.Awasthi, S., Singhal, S. S., Pikula, S., Piper, J. T., Srivastava, S. K., Torman, R. T., Bandorowicz-Pikula, J., Lin, J. T., Singh, S. V., Zimniak, P., and Awasthi, Y. C. (1998) Biochemistry 37 5239-5248 [DOI] [PubMed] [Google Scholar]
- 23.Awasthi, S., Singhal, S. S., Pandya, U., Gopal, S., Zimniak, P., Singh, S. V., and Awasthi, Y. C. (1999) Toxicol. Appl. Pharmacol. 155 215-226 [DOI] [PubMed] [Google Scholar]
- 24.Sharma, R., Singhal, S. S., Wichramarachchi, D., Awasthi, Y. C., and Awasthi, S. (2004) Int. J. Cancer 112 934-942 [DOI] [PubMed] [Google Scholar]
- 25.Awasthi, S., Singhal, S. S., Srivastava, S. K., Zimniak, P., Bajpai, K. K., Saxena, M., Sharma, R., Ziller, S. A., Frenkel, E. P., Singh, S. V., and Awasthi, Y. C. (1994) J. Clin. Invest. 93 958-965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singhal, S. S., Yadav, S., Singhal, J., Zajac, E., Awasthi, Y. C., and Awasthi, S. (2005) Biochem. Pharmacol. 70 481-488 [DOI] [PubMed] [Google Scholar]
- 27.Jia, Z., Moulson, C. L., Pei, Z., Miner, J. H., and Watkins, P. A. (2007) J. Biol. Chem. 282 20573-20583 [DOI] [PubMed] [Google Scholar]
- 28.Leich, F., Stöhr, N., Rietz, A., Ulbrich-Hofmann, R., and Arnold, U. (2007) J. Biol. Chem. 282 27640-27646 [DOI] [PubMed] [Google Scholar]
- 29.Broadway, N. M., and Saggerson, E. D. (1995) Biochem. J. 310 989-995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aumais, J. P., Lee, H. S., Lin, R., and White, J. H. (1997) J. Biol. Chem. 272 12229-12235 [DOI] [PubMed] [Google Scholar]
- 31.Hammill, A. K., Uhr, J. W., and Scheuermann, R. H. (1999) Exp. Cell Res. 251 16-21 [DOI] [PubMed] [Google Scholar]
- 32.Oosterhoff, J. K., Penninkhof, F., Brinkmann, A. O., Grootegoed, J. A., and Blok, L. G. (2003) Oncogene 22 2920-2925 [DOI] [PubMed] [Google Scholar]
- 33.Awasthi, S., Singhal, S. S., Singhal, J., Yang, Y., Zimniak, P., and Awasthi, Y. C. (2003) Int. J. Oncol. 22 721-732 [PubMed] [Google Scholar]
- 34.Awasthi, S., Singhal, S. S., Yadav, S., Singhal, J., Drake, K., Nadkar, A., Zajac, E., Wickramarachchi, D., Rowe, N., Yacoub, A., Boor, P., Dwivedi, S., Dent, P., Jarman, W. E., John, B., and Awasthi, Y. C. (2005) Cancer Res. 65 6022-6028 [DOI] [PubMed] [Google Scholar]
- 35.Cheng, J., Sharma, R., Yang, Y., Singhal, S. S., Sharma, A., Saini, M. K., Singh, S. V., Zimniak, P., Awasthi, S., and Awasthi, Y. C. (2001) J. Biol. Chem. 276 41213-41223 [DOI] [PubMed] [Google Scholar]
- 36.Awasthi, S., Singhal, S. S., Singhal, J., Cheng, J., Zimniak, P., and Awasthi, Y. C. (2003) Int. J. Oncol. 22 713-720 [PubMed] [Google Scholar]
- 37.Singhal, S. S., Yadav, S., Singhal, J., Awasthi, Y. C., and Awasthi, S. (2006) Biochem. Biophys. Res. Commun. 348 722-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singhal, S. S., and Awasthi, S. (2006) in Toxicology of Glutathione Transferases (Awasthi, Y. C., ed) pp. 231-256, CRC Press, Inc., Boca Raton, FL
- 39.Yadav, S., Zajac, E., Singhal, S. S., and Awasthi, S. (2007) Cancer Metastasis Rev. 26 59-69 [DOI] [PubMed] [Google Scholar]
- 40.Awasthi, Y. C., Sharma, R., Yadav, S., Dwivedi, S., Sharma, A., and Awasthi, S. (2007) Curr. Drug Metab. 8 1218-1290 [DOI] [PubMed] [Google Scholar]
- 41.Li, J., Sharma, R., Patrick, B., Sharma, A., Jeyabal, P. V., Reddy, P. M., Saini, M. K., Dwivedi, S., Dhanani, S., Ansari, N. H., Zimniak, P., Awasthi, S., and Awasthi, Y. C. (2006) Biochemistry 45 12253-12264 [DOI] [PubMed] [Google Scholar]
- 42.Sharma, R., Sharma, A., Dwivedi, S., Zimniak, P., Awasthi, S., and Awasthi, Y. C. (2008) Biochemistry 47 143-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, D. M., Tradewell, M. L., Minotti, S., and Durham, H. D. (2007) Cell Stress Chaperones 12 151-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singhal, J., Singhal, S. S., Yadav, S., Suzuki, S., Warnke, M. M., Yacoub, A., Dent, P., Sharma, R., Awasthi, Y. C., Armstrong, D. W., and Awasthi, S. (2008) Int. J. Rad. Oncol. Biol. Phys., in press [DOI] [PMC free article] [PubMed]