Abstract
After protracted low level arsenic exposure, the normal human prostate epithelial cell line RWPE-1 acquires a malignant phenotype with DNA hypomethylation, indicative of disrupted methyl metabolism, and shows arsenic adaptation involving glutathione overproduction and enhanced arsenic efflux. Thus, the interplay between methyl and glutathione metabolism during this progressive arsenic adaptation was studied. Arsenic-treated cells showed a time-dependent increase in LC50 and a marked increase in homocysteine (Hcy) levels. A marked suppression of S-adenosylmethionine (SAM) levels occurred with decreased methionine adenosyltransferase 2A (converts methionine to SAM) expression and increased negative regulator methionine adenosyltransferase B, suggesting reduced conversion of Hcy to SAM. Consistent with Hcy overproduction, activity and expression of S-adenosylhomocysteine hydrolase (converts S-adenosylhomocysteine to Hcy) were both increased. Expression of cystathionine β-synthase, a key gene in the transsulfuration pathway, and various glutathione production genes were increased, resulting in a 5-fold increase in glutathione. Arsenic efflux increased along with expression of ATP-binding cassette protein C1, which effluxes arsenic as a glutathione conjugate. Evidence of genomic DNA hypomethylation was observed during early arsenic exposure, indicating that the disruption in methyl metabolism had a potential impact related to oncogenesis. Thus, cellular arsenic adaptation is a dynamic, progressive process that involves decreased SAM recycling and concurrent accumulation of Hcy, which is channeled via transsulfuration to increase glutathione and enhance arsenic efflux but may also impact the carcinogenic process.
Arsenic is a metalloid widely distributed in the environment mainly found in inorganic forms. Inorganic arsenic is a human carcinogen inducing cancers of the skin, lung, and urinary bladder and possibly liver, prostate, and kidney (1–4). Indeed, several studies indicate a link between inorganic arsenic exposure and human prostate cancer (1–4). Our laboratory has developed an in vitro prostate cancer model in which the normal human prostate epithelial cell line RWPE-1 is malignantly transformed after chronic, low level arsenite exposure (1, 5). The transformed cells, called CAsE-PE, form malignant carcinoma after inoculation into nude mice and overexpress prostate-specific antigen (1, 5), similar to human prostate carcinoma. Chronic, low level arsenite exposure also induces adaptation to arsenic, in which cells become resistant to acute arsenic toxicity (6–8). This arsenic adaptation occurs through various metabolic changes, including overexpression of efflux transport proteins and increased glutathione production (6–8).
Arsenic can be enzymatically modified to form mono- and dimethylarsenic compounds via arsenic methyltransferases using SAM2 as the methyl donating cofactor (9). Because this methylation may generate toxic intermediates, it is no longer thought to be detoxicating of inorganic arsenic (9–11). SAM is involved in most cellular methyltransferase reactions, including DNA methylation (12). DNA methylation is a key controlling factor in expression of various genes (13), and hypomethylation can lead to overexpression of several genes linked to oncogenesis (14). However, RWPE-1 cells have a very poor capacity to methylate arsenic and would not biochemically compete for SAM through arsenic biomethylation (15). Nonetheless, after arsenic-induced malignant transformation and concurrent adaptation, these cells show clear evidence of genomic DNA hypomethylation (15).
The mechanism of arsenic-induced oncogenic transformation is not fully understood, but a comparison of different models reveals potentially common patterns. Altered gene expression associated with genomic or gene specific DNA hypomethylation has been observed in vitro (15–17) and in vivo (18–21) during arsenic exposure at potentially oncogenic levels. Alterations in critical metabolic pathways, particularly the overproduction of glutathione (6) or decreased levels of SAM (16, 17) are seen after chronic arsenic exposure. In fact, it is suspected that the DNA hypomethylation observed after arsenic exposure may be, in part, a result of a competition for SAM, because SAM is required for both DNA methylation and the biomethylation of inorganic arsenic, at least in arsenic methylation-competent cells (16, 17). However, such a competition for SAM in methylation reactions would not occur in cells that poorly methylate arsenic, such as RWPE-1 cells and its malignant transformant, CAsE-PE cells (15). Thus, in these cells arsenic may impact methyl metabolism through some as yet undefined indirect mechanism.
Glutathione is one of the most important anti-oxidant molecules of the cell and has critical functions in various detoxication processes (22). For instance, arsenic will form glutathione conjugates that can be effluxed by transport proteins (7, 8), and glutathione depletion greatly enhances arsenic toxicity in various model systems (6, 7, 23, 24). In fact, arsenic efflux as a glutathione conjugate is thought to be a key component of cellular arsenic adaptation (7, 8). It is noteworthy that SAM pathway and glutathione production are linked through the transsulfuration pathway (see Scheme 1), with Hcy as the nodal compound. In fact, Hcy is irreversibly catabolized via the transsulfuration pathway, making it unavailable for further SAM production. Furthermore, disrupted glutathione homeostasis has clearly been shown to be a result of altered SAM metabolism (25).
SCHEME 1.
Connections between the SAM cycle, transsulfuration pathway, and glutathione production. MTR, 5-methyltetrahydrofolate homocysteine methyltransferase; CBS, cystathionine β-synthase; CTH, cystathionase; GCLC/M, glutamate-cysteine ligase catalytic/modulatory subunit; GSS, glutathione synthetase; CH3THF, methyltetrahydrofolate.
Thus, in the present work, we use an established cell model system to test the hypothesis that arsenic adaptation may have an impact on methyl metabolism by shifting it toward increased glutathione production. Such a response may enhance short term survival by allowing greater arsenic efflux but could be harmful in the long run by disruption of cellular methyl homeostasis and precipitation of oncogenic events. In this regard, RWPE-1 cells were selected because, at the point of malignant transformation with arsenic (∼30 weeks), they show evidence of disrupted methyl metabolism (DNA hypomethylation), as well as a clear adaptation to arsenic (5, 6, 15). In addition, because RWPE-1 cells do not methylate arsenic, results would not be complicated by potential direct competition for SAM. Early time points were studied to assess key events in the acquisition of metabolic adaptation.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents—Sodium arsenite (NaAsO2) was obtained from Sigma. Keratinocyte serum-free medium, epidermal growth factor, bovine pituitary extract, and 100× penicillin G-streptomycin-amphotericin mixture were purchased from Invitrogen. The following antibodies were used: mouse monoclonal antibody to β-actin purchased from Calbiochem; rabbit polyclonal antibody to S-adenosylhomocysteine hydrolase (AHCY) purchased from Proteintech (Chicago, IL); rabbit polyclonal antibody to MAT2A purchased from Santa Cruz (Santa Cruz, CA); and rabbit polyclonal antibody to glutamate-cysteine ligase catalytic (GCLC) purchased from Abcam (Cambridge, MA). Horseradish peroxidase-conjugated secondary antibodies were purchased from Amersham Biosciences. The Bradford protein assay was purchased from Bio-Rad.
Cell Culture and Conditions of Exposure—RWPE-1 cells were derived from normal human prostate epithelial cells and are immortalized but not tumorigenic (26, 27). The cells were grown at 37 °C in a humidified atmosphere containing 5% CO2 in complete medium (keratinocyte serum-free medium containing 50 μg/ml bovine pituitary extract, 5 ng/ml epidermal growth factor, and 1× antibiotic-antimycotic mixture) and passaged weekly. Chronic arsenic treatment consisted of a continuous exposure to 5 μm arsenite for up to 16 weeks. These cells are termed CAsE-PE for chronic arsenic exposed prostate epithelial cells. Cells grown in medium without arsenic provided passage-matched controls. Similar arsenic concentrations result in malignant transformation in these cells at ∼30 weeks of exposure (5).
Acute Arsenite Lethality—Control RWPE-1 or cells cultured for different times in arsenite were seeded at 5 × 104 cells/well in 6-well plates and incubated at 37 °C and 5% CO2 in arsenic-free medium. After 24 h, the medium was replaced with medium containing acutely cytholethal levels (≤75 μm) of sodium arsenite. The cells were incubated for an additional 48 h, at which point they were rinsed in phosphate-buffered saline three times and harvested. Cell viability was measured by trypan blue dye exclusion and cell counting.
Gene Expression Analysis—Total RNA was extracted from cells with TRIzol reagent (Invitrogen), followed by a purification step with an RNeasy column (Qiagen). RNA was reverse-transcribed with murine leukemia virus reverse transcriptase and oligo(dT) primers (Applied Biosystems, Foster City, CA). The ABI Primer Express software (Applied Biosystems) was used to design the forward and reverse gene-specific primers (supplemental Table S1). The primers were synthesized by Sigma-Genosys (Woodlands, TX). The SYBR green master mix (ABgene, Rochester, NY) was used for real time PCR analysis. Sample-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels were used to normalize values, and the results are expressed as percentages of adjusted control expression.
Western Blot Analysis—Total protein extract was obtained using M-PER reagent (Pierce) according to the manufacturer's instructions. Protein concentration was determined using the Bradford assay, and 30 μg protein of each sample were electrophoresed and transferred to polyvinylidene difluoride membranes according to the manufacturer's directions (Invitrogen). Immunoblotting was performed using antibody dilutions for AHCY at 1:200, GCLC at 1:500, or β-actin for standardization, at 1:10,000. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody was used at a 1:10,000 dilution and Super-Signal West Pico Chemiluminescent Substrate (Pierce) was used for protein detection. The signals were visualized by exposure to Hyperfilm (Amersham Biosciences). Densitometric analysis was performed using the Image J program (developed by the National Institutes of Health).
Hcy, Glutathione, and SAM Levels—The cells were harvested after trypsinization, and total Hcy levels were assayed at selected time points according to the manufacturer's instruction with a homocysteine microtiter plate assay (Diazyme, San Diego, CA). Total and oxidized glutathione were assayed using a glutathione assay kit from Trevigen (Gaithersburg, MD), and reduced glutathione levels were derived from these values. SAM levels were assayed according to manufacturer instructions using the Bridge-it® SAM fluorescence assay kit (Mediomics, St. Louis, MO).
Enzymatic Activity—Enzymatic activity of glutathione reductase was measured according to the manufacturer's instruction using a glutathione reductase assay kit from Cayman Chemical (Ann Harbor, MI). AHCY enzymatic activity was measured according to the protocol described by Kloor et al. (28).
Arsenite Efflux—Control cells and cells exposed to low level arsenic for 16 weeks were grown in arsenite-free medium to ∼70% confluence. The cells were then pre-exposed to 10 μm arsenite for 24 h, washed three times with phosphate-buffered saline, and placed in fresh arsenic-free medium for 24 h. These cells were then harvested, counted, and digested overnight with 50% perchloric acid:nitric acid (2:1) at 70 °C. As a reflection of effluxed arsenic, the total remaining cellular arsenic levels were determined by graphite furnace atomic absorption spectrophotometry using a PerkinElmer Life Sciences AAnalyst 600 equipped with an auto sampler. The data were normalized to cell number.
Restriction Enzyme Digestions of Genomic DNA—One μg of DNA from control and arsenic-treated cells at 4 weeks of arsenic exposure was separately digested with either 10 units of RsaI, 10 units each of RsaI and the methylation-sensitive restriction enzyme HpaII, or 10 units each of RsaI and MspI (New England BioLabs, Ipswich, MA) at 37 °C for 16 h. This was followed by heat inactivation of the enzymes at 65 °C for 20 min. The digests were then stored at –20 °C. HpaII does not cut DNA if the internal cytosine of its restriction site (CCGG) is methylated. MspI has the same restriction site but can cut when the internal cytosine is methylated.
Methylation-sensitive Arbitrarily Primed PCR—A couple of primers (MGC0, 5′-AACCCTCACCCTAACCGCGC-3′; MGF2, 5′-AACCCTCACCCTAACCCGCG) were used to amplify restriction-digested DNA (200 ng) (29–31). PCRs were performed in a total volume of 50 μl using 2.5 units of platinum Taq polymerase (Invitrogen), the buffer provided by the manufacturer, 1.5 mm MgCl2, 200 μm each of the four deoxynucleotide triphosphates, and 25 pm of the primer. The PCR consisted of 94 °C for 10 min of enzyme activation, followed by five cycles at low stringency conditions of 94 °C for 30 s, 45 °C for 60 s, 72 °C for 90 s, and 35 cycles at higher stringency conditions of 94 °C for 15 s, 55 °C for 15 s, 72 °C for 60 s, and a final elongation of 7 min at 72 °C. PCR products were resolved on a 6% polyacrylamide TBE-urea gel according to the manufacturer's instructions and stained with SilverXpress silver staining kit (Invitrogen). The gels were finally scanned.
Statistical Analysis—The data are expressed as the means ± S.E. and represent at least three separate determinations. The data from arsenic-exposed cells are individually compared with passage-matched concurrent control data. These data were analyzed for statistical significance using Student's t test with a p value of ≤0.05 considered significant. In addition, to help define correlations between progressive changes in LC50 and expression of selected genes (ABCC1 and AHCY) or glutathione, linear regression was performed using mean LC50 data verses individual data for ABCC1, AHCY, or reduced glutathione. The resulting slope was tested against a slope of zero using an unpaired t test, and the p values of these tests are reported.
RESULTS
Our prior work showed that chronic, low level in vitro arsenic exposure for ∼30 weeks results in malignant transformation of the human prostate epithelial cell line RWPE-1 (5). Additionally, these cells show stable adaptation to arsenic during this transformation involving overproduction of glutathione (6), concurrently with DNA hypomethylation (15). Based on the hypothesis that these changes may be linked, we studied the impact of arsenic exposure during the early stages of adaptation (≤16 weeks) on cellular glutathione production and methyl metabolism (see Scheme 1 for a pathway overview) as well as potential related oncogenic events (DNA methylation).
Progressive Development of Arsenic Adaptation in RWPE-1 Cells—The progressive acquisition of resistance to acute arsenic cytholethality was assessed in RWPE-1 cells chronically exposed to low level (5 μm) arsenic and compared with passage match control (Fig. 1). Arsenic adaptation occurred as an early event, and even at 4 weeks of low level exposure, the LC50 for acute arsenic exposure was increased compared with control cells (∼2.8-fold). The resistance to acute high dose arsenic cytholethality progressively increased with the time of exposure to low level arsenic in these cells. These data clearly indicate that arsenic adaptation is a dynamic, progressive process in human cells.
FIGURE 1.
Acute cytholethality of inorganic arsenic in RWPE-1 cells chronically exposed to low level arsenite (5 μm). After 48 h of acute exposure to various concentrations of sodium arsenite, the cells were harvested, the viable cells were counted, and the LC50 values were derived from survival curves. The data are expressed as LC50 and represent the means ± S.E. of three separate experiments. An asterisk indicates a significant difference (p ≤ 0.05) from control.
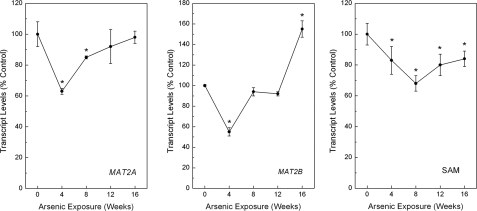
Altered SAM Recycling during Arsenic Adaptation—To determine the impact of arsenic adaptation on cellular methyl metabolism, the expression levels of several key components of the SAM cycle (see Scheme 1) were assessed. Because RWPE-1 cells do not readily methylate arsenic (15), the results would not be complicated by the need for SAM in arsenic methylation. Transcript levels of genes encoding for key enzymes in the SAM recycling system were initially studied (Fig. 2). The expression of methionine adenosyltransferase 2A (MAT2A), the enzyme responsible for the final step in SAM synthesis, was initially decreased during arsenic adaptation at 4 and 8 weeks (maximum decrease, ∼40%) but returned to normal levels by 12 weeks. MAT2B, the negative regulatory subunit that, by binding to MAT2A, inhibits its enzymatic activity, although initially decreased at 4 weeks, returned to control levels at 8 and 12 weeks but then was significantly increased by 50% after 16 weeks of continuous low level arsenic treatment. These data suggest an overall reduction of SAM production occurred first by a reduction of synthesis via reduced MAT2A and then by an increase in MAT2B related inhibition. Indeed, SAM levels fortify this notion as they were persistently decreased during arsenic adaptation, with reductions starting by 4 weeks of treatment, as great as 35% (Fig. 2, right panel).
FIGURE 2.
Levels of MAT2A (left panel) and MAT2B (middle panel) transcript and cellular SAM (right panel) in RWPE-1 cells during progressive arsenic adaptation. The data are expressed as means ± S.E. (n ≥ 3) and given as percentages of control (100%) expression after normalization to GAPDH. An asterisk indicates a significant difference (p ≤ 0.05) from control.
AHCY transcript, which encodes for an enzyme that produces Hcy from SAH, showed a clear, time-dependent increase to maximal levels of 240% of control by 16 weeks of low level arsenic exposure (Fig. 3, top left panel). In fact, there was a very strong positive relationship between increasing AHCY transcript and increasing LC50 (linear regression p = 0.0063). Assessment of protein levels of AHCY showed a strong increase at 12 weeks of exposure that correlated with increased transcript levels (Fig. 3, top right panel). AHCY enzymatic activity was similarly increased (Fig. 3, bottom left panel). Furthermore, when Hcy levels were assessed, they were also markedly increased (Fig. 3, bottom right panel). This increase in Hcy is consistent with Hcy overproduction via up-regulation of AHCY at the transcript, protein, and enzymatic activity levels during arsenic adaptation.
FIGURE 3.
Transcript (top left panel), protein (top right panel), and activity (bottom left panel) levels of AHCY as well as Hcy levels (bottom right panel) in RWPE-1 cells during progressive arsenic adaptation. The levels of AHCY protein and cellular Hcy were determined at 12 weeks of exposure. The data are expressed as means ± S.E. (n ≥ 3) and given as percentages of control (100%) after normalization to GAPDH (transcript), β-actin (protein), or by μg of protein (activity). An asterisk indicates a significant difference (p ≤ 0.05) from control.
Betaine-homocysteine methyltransferase (BHMT) and 5-methyltetrahydrofolate-homocysteine methyltransferase are key genes encoding for enzymes involved in the conversion of Hcy back into methionine and, eventually, in the production of SAM. At the transcript level BHMT transcript was significantly reduced over 60% during the initial phase of arsenic adaptation (data not shown), indicating that the use of Hcy in the SAM cycle may be reduced during the early adaptation period. 5-Methyltetrahydrofolate-homocysteine methyltransferase transcript levels were unchanged, indicating at this point in SAM recycling that major changes did not occur with arsenic adaptation.
The Transsulfuration Pathway in Arsenic Adaptation—Transcript levels for genes encoding for major enzymes of the transsulfuration pathway, which irreversibly utilizes Hcy to produce cysteine as a precursor of glutathione, were assessed. Transcript levels of cystathionine β-synthase (CBS), which encodes for the key enzyme involved in the first, irreversible step in the transsulfuration pathway and plays a key role in the regulation between transsulfuration and remethylation, progressively increased with duration of arsenic exposure. This increase in CBS (154 ± 13%) was statistically significant compared with control (100 ± 13%) at 16 weeks of exposure. No changes were observed for cystathionine γ-lyase, the second step in the pathway (data not shown). Given an overproduction of Hcy combined to a suppression of SAM, it appears that the transsulfuration has been activated during arsenic adaptation.
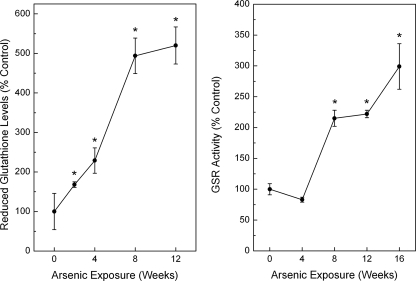
Levels of Glutathione and Glutathione Synthesis Enzymes during Arsenic Adaptation—The transcript levels of genes encoding for enzymes in the glutathione synthesis pathway were markedly increased during arsenic adaptation (Fig. 4). The catalytic subunit of the enzyme involved in the rate-limiting step of glutathione synthesis, namely the GCLC and the positive modulatory subunit glutamate-cysteine ligase modulatory, were markedly increased to a maximum of 750 and 300%, respectively, with chronic, low level arsenic exposure. Because the production of both GCL subunits was enhanced, this likely indicates an increase in catalytic activity. That protein levels of GCLC were increased 6-fold compared with passage matched control at 16 weeks of arsenic exposure fortifies this notion. Glutathione synthetase, the gene encoding for the enzyme involved in the final step of glutathione synthesis, was increased by up to 280% with arsenic exposure. Indeed, total glutathione levels were rapidly and progressively increased during chronic, low level arsenic exposure to a maximum of 325% after 12 weeks of arsenic exposure (data not shown). Likewise, reduced glutathione levels were consistently elevated up to 500% of control (Fig. 5, left panel), indicating that the increase observed in total glutathione was mainly caused by an augmentation of the reduced glutathione pool. Reduced glutathione showed very strong, positive relationship with increasing LC50 to arsenic (linear regression p = 0.0003). Glutathione reductase activity was also increased, starting at 8 weeks of exposure, by up to 2.5-fold of control (Fig. 5, right panel).
FIGURE 4.
Levels of GCLC transcript (top left panel) and protein (top right panel) as well as glutamate-cysteine ligase modulatory (GCLM; bottom left panel) and glutathione synthetase (bottom right panel) transcripts in RWPE-1 cells during progressive arsenic adaptation. The data are expressed as the means ± S.E. (n ≥ 3) and given as percentages of control (100%) expression after normalization to GAPDH (transcript) or β-actin (protein). An asterisk indicates a significant difference (p ≤ 0.05) from control. The protein data are shown for 16 weeks of exposure only.
FIGURE 5.
Reduced (left panel) glutathione levels during chronic low level arsenic exposure, as well as activity of glutathione reductase (right panel). The data are expressed as the means ± S.E. (n ≥ 3) and given as percentages of control (100%) after normalization. An asterisk indicates a significant difference (p ≤ 0.05) from control.
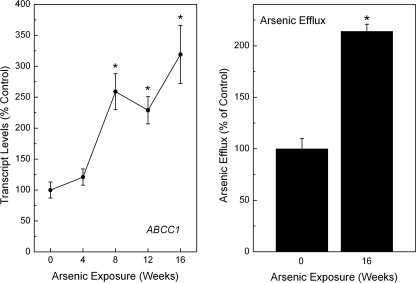
Production of ABCC1 and Efflux during Arsenic Adaptation—Because the transporter ABCC1 is known to be involved in arsenic adaptation through efflux of the metalloid as a glutathione trimer (7), the transcript levels were assessed during progressive arsenic adaptation. As expected, ABCC1 levels showed a strong, progressive increase during arsenic adaptation (Fig. 6, left panel). In fact, ABCC1 transcript levels showed a very strong positive relationship with increasing LC50 values (linear regression p = 0.0002). The efflux capacity during arsenic adaptation was also analyzed (Fig. 6, right panel). In arsenic-adapted cells, arsenic efflux was over 2-fold higher than control, consistent with the role of ABCC1 in arsenic transport and supporting the functional impact of ABCC1 over expression.
FIGURE 6.
Levels of ABCC1 transcript (left panel) in RWPE-1 cells during progressive arsenic adaptation and cellular arsenic efflux (rightpanel). The transcript data are expressed as the means ± S.E. (n ≥ 3) and given as percentages of control (100%) expression after normalization to GAPDH. The efflux data are given as percentages of arsenic effluxed over a 24-h period as compared with control level of efflux over the same period (100%) after normalization to cell number. An asterisk indicates a significant difference (p ≤ 0.05) from control.
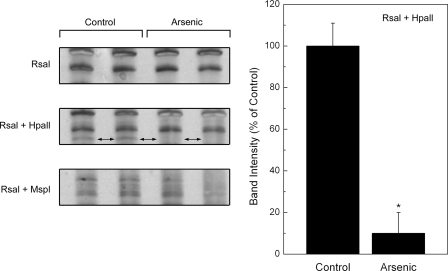
Early Arsenic-induced DNA Methylation Changes—Early arsenic-induced changes in DNA methylation status after 4 weeks of treatment were investigated using methylation-sensitive arbitrarily primed PCR (31). DNA digestions by different sets of restriction enzymes (RsaI, RsaI + HpaII, and RsaI + MspI) were used, and arsenic-related changes in the methylation pattern of PCR products were observed after amplification of DNA digestions by methyl sensitive and insensitive restriction enzymes (Fig. 7). An example is shown in which a band is visible in control after RsaI + HpaII digestion is completely lost in DNA from arsenic treated cells, indicating that the DNA becomes unmethylated after arsenic exposure. This observation reinforces the concept that disruption of methyl metabolism, possibly via the metabolic demands of arsenic adaptation, could have an early impact on genomic DNA methylation. The impact of this early change in DNA methylation on oncogenic transformation requires additional study but is consistent with work showing DNA hypomethylation at the point of malignant transformation in RWPE-1 cells chronically treated with arsenic (15).
FIGURE 7.
Selected areas of DNA methylation in control or arsenic-exposed RWPE-1 cells at 4 weeks of exposure using arbitrarily primed PCR after restriction enzyme digestions. Shown are details of a representative gel with an area of distinct DNA hypomethylation in arsenic-treated DNA (left panels, two-headed arrows) and densitometric analysis (n = 3) of this band (right panel). See methods for details. An asterisk indicates a significant difference (p ≤ 0.05) from control. Unmethylated DNA is not protected from digestion by methylation-sensitive HpaII restriction enzyme resulting in a loss of DNA for PCR amplification. Digestion with RsaI + MspI would also result in no PCR amplification regardless of methylation status. Digestion with RsaI alone and RsaI + MspI were used as controls to determine whether there was differential methylation.
DISCUSSION
In this work, continuous low level inorganic arsenic exposure of normal human prostate cells in vitro resulted in progressive adaptation to the metalloid involving metabolic changes in the SAM cycle, transsulfuration pathway, and glutathione production. A marked overproduction of Hcy, a compound considered at the cross-roads between SAM and glutathione metabolism, also occurred together with persistent suppression of SAM levels during arsenic adaptation. These data indicate that arsenic-adapted cells may overproduce and preferentially channel Hcy into the transsulfuration pathway instead of for remethylation to SAM. Ultimately, these changes appeared to have favored an increase in glutathione synthesis, which likely helped increase arsenic elimination via the efflux pump ABCC1 (7), the levels of which were also increased. Clearly the progressive phenomenon of arsenic adaptation observed in the present work was very strongly correlated with increasing glutathione levels and AHCY and ABCC1 expression. ABCC1 is very likely responsible for the increased efflux capacity of these cells (7). A preferential use of Hcy via transsulfuration toward glutathione synthesis appears responsible for decreased SAM production, reducing the methyl donating cofactor available for various cellular methyltransferases. This loss of SAM to glutathione production could explain why these cells, which only poorly methylate arsenic (15), nonetheless show some evidence of an early loss of DNA methylation in the present work and in prior work show global genomic DNA hypomethylation at a point that coincides with overt arsenic-induced malignant transformation (15). In support of the present work with arsenic, it is clear that glutathione depletion by organic xenobiotics can also reduce DNA methylation, likely via reduced methyl donating SAM cycle components (i.e. methionine and SAM) because of transsulfuration of Hcy on to glutathione (32, 33). Other agents (e.g. cancer chemotherapeutics) or cellular events (e.g. oxidative stress) that also chronically deplete glutathione may have the potential to produce a similar indirect impact on DNA methylation (32, 33). Thus, although RWPE-1 cells do not extensively methylate arsenic, they all the same appear to be subjected to a hypomethylating environment to adapt for survival from a continuous arsenic insult. Hence, arsenic adaptation may actually contribute to oncogenic conversion. In addition, the present results clearly show that arsenic adaptation in human cells is a dynamic, progressive process that impacts a variety of cellular metabolic pathways and products and may trade immediate survival for later issues in cancer development.
The increase in Hcy production is potentially very important because this small molecule is metabolically located at the cross-roads between the SAM cycle and the transsulfuration pathway, which can eventually provide cysteine for glutathione synthesis. Hcy is a breakdown product of SAH, which is the product of the enzymatic transfer of the methyl group from SAM to a methyl acceptor molecule, such as DNA. Elevated Hcy generally indicates increased levels of SAH (34) particularly because the reaction is reversible. SAH is a potent feedback inhibitor of various methyltransferases (35–37), and its removal is key to cellular methyl homeostasis. The marked, progressive overexpression of AHCY, the increases in AHCY enzymatic activity, and the increase in Hcy seen in the present study indicates that these factors are important in arsenic adaptation. Inhibition of AHCY enzymatic activity causes accumulation of SAH and a decrease in cellular glutathione (38), indicating that AHCY, via Hcy, helps modulate glutathione production. The remethylation of Hcy produces methionine for eventual SAM production, whereas the synthesis of cystathionine via CBS is the first, irreversible step of Hcy conversion via the transsulfuration pathway, ultimately leading to cysteine and, potentially, glutathione. An early transcriptional decrease of BHMT, a gene in the Hcy/SAM remethylation pathway, was observed in the present study. BHMT can be up-regulated in response to methionine deficiency (39). Similarly, CBS, a gene encoding for a key enzyme in the transsulfuration of Hcy eventually to glutathione, was markedly increased in the latter stages of arsenic adaptation. Low methionine levels can down-regulate the transsulfuration pathway to enhance remethylation of Hcy into methionine for SAM production (40). However, in the present case during arsenic adaptation, reduced SAM levels occurred despite increased levels of Hcy, indicating that Hcy was being utilized elsewhere. Methylation demand via SAM utilization can be a principal factor in increasing Hcy (41), although this seems unlikely in the present study because RWPE-1 cells have a very low capacity to methylate arsenic (15). In light of the observed reduction in SAM production (associated with increased MAT2B, decreased MAT2A), overproduction of Hcy (likely via increased AHCY expression and activity), increased transsulfuration, and increased production of glutathione seen during arsenic adaptation, it appears that the cells have altered cellular methyl metabolism in favor of enhanced glutathione production and enhanced arsenic efflux for more immediate survival at the potential expense of the long term consequences of methyl (SAM) insufficiency.
Arsenic adaptation was correlated with marked, progressive increases in reduced glutathione, activity of glutathione reductase, and expression of various glutathione synthesis genes. This was particularly true for the gene encoding for the catalytic subunit (GCLC) of the limiting first step of glutathione synthesis using cysteine. Similarly, an increase in glutathione reductase transcription and activity has been shown to be frequently involved in the maintenance of reduced glutathione (42). It appears that increases in glutathione can result from both the enhancement of glutathione synthesis and increases in glutathione reduction (43). Clearly the status of the glutathione system was particularly important for adaptation to arsenic in the present study (6). In this regard, enhanced efflux is a fundamental aspect to arsenic adaptation in various cells (7, 8, 44). In the present work, the transcript levels of ABCC1, a gene encoding for an efflux pump involved in transport of xenobiotics conjugated with glutathione, were markedly and progressively increased with arsenic exposure, and this coincided with a much stronger capacity to efflux arsenic in adapted cells. The increase expression of ABCC1 is of particular importance because it is involved in the efflux of a triglutathione-arsenical complex (7), which would require significant and sustained amounts of cellular glutathione for activity. In fact, arsenic-adapted RWPE-1 cells show a loss of arsenic resistance after exposure to inhibitors of ABCC1 transport or after glutathione depletion (6), clearly indicating that both play key roles in arsenic efflux and adaptation. The efflux of three glutathione molecules with each atom of arsenic (7) would very likely precipitate the need for enhanced glutathione production, a concept fortified by results of the present work. It is of interest that this glutathione overproduction appears linked to overproduction of Hcy, and in turn to reduced SAM, in arsenic adaptation.
Reports indicate SAM levels can be depleted after arsenic exposure, which could be the basis for arsenic-induced DNA hypomethylation (16, 17). In the present study, expression of MAT2A, which synthesizes SAM from methionine, was reduced during the initial phase of arsenic adaptation. MAT2B, the negative regulatory subunit of MAT2, increased in the later stages, suggesting an enhanced inhibition of the catalytic subunit. Marked increases were also seen in CBS expression as a late event in arsenic adaptation, which could reduce Hcy availability for the SAM cycle. Thus, at the gene expression level, there is a clear basis for the observed SAM depletion. Indeed, in this study, SAM levels showed rapid and sustained reductions, suggesting that, in this model as in others (16, 17), chronic inorganic arsenic exposure impacts methyl metabolism. This appears true even in cells that do not methylate arsenic, such as the RWPE-1 cells (15). In this regard, SAM supplementation reverses arsenic-induced micronucleus formation, an effect thought to be, at least in part, because of reversal of arsenic-induced SAM depletion (45). SAM depletion is associated with human and rodent cancer and can cause DNA hypomethylation (12, 46, 47) and occurs when cells undergo arsenic-induced malignant transformation (15, 16). In the present work with arsenic exposure, there was early evidence of a loss in DNA methylation, indicating that the imbalance created in methyl metabolism for the sake of arsenic adaptation have the potential for genome wide effects. Indeed, in transformed RWPE-1 cells (CAsE-PE) global hypomethylation occurs concurrently with the acquisition of a malignant phenotype (15). Alterations of DNA methylation, an essential mechanism in gene regulation (48), are frequently observed in oncogenesis (49). Several studies have shown that global (15–18, 21) or gene-specific (19–21) DNA hypomethylation can occur after arsenic exposure in vitro or in vivo. DNA hypomethylation has been associated with acquisition of arsenic-induced malignant phenotype in several cell lines in vitro (15, 16) and with arsenic-induced liver tumors in mice (20). Thus, even when cells do not directly use SAM for arsenic methylation, SAM depletion can still occur, apparently as part of the metabolic events involved in arsenic adaptation. Our data suggest that MAT2A and 2B are altered in a way that would reduce SAM production, and SAM levels are, in fact, reduced. Under normal homeostatic conditions this might suggest an eventual decrease in downstream SAM cycle components, including Hcy, which is inconsistent with the marked increase in Hcy levels that we observed. However, the events occurring with arsenic, although adaptive, also occur as part of the progressive acquisition of a disease state, specifically a malignant phenotype, and normal homeostatic control mechanisms may not be operative under such conditions. For instance, elevated circulating Hcy and concurrently reduced SAM occurs in neural tube defect pregnancies, a defect that also appears linked with decreased methylation capacity (50). There is also evidence that clinically relevant levels of Hcy in vitro can reduce SAM levels and thereby induce DNA hypomethylation in vascular smooth muscle cells, and this has been proposed as an important mechanism in the pathogenesis of atherosclerosis (51). Key nutrient deficiencies can also disrupt SAM cycle homeostasis and result in elevated Hcy despite reducing SAM levels (52). Thus, under pathological conditions, it is not uncommon to see increased Hcy in the presence of reduced SAM (Refs. 50–52 and the present study), although the precise reasons for this need additional study.
In summary, arsenic adaptation in human prostate epithelial cells precipitated a variety of profound metabolic changes with potentially diverse effects. On one hand, increased arsenic efflux capacity occurred through increased glutathione production coupled with enhanced ABCC1 expression. These changes are likely favorable to immediate cellular survival in the face of continuous arsenic exposure. On the other hand, the imbalance created in methyl metabolism by the apparent diversion of Hcy to glutathione production and subsequent SAM depletion may have created a hypomethylating environment, a condition potentially associated with altered DNA methylation and potential disrupted expression of genes related to carcinogenesis. Finally, it is clear from the present data that arsenic adaptation is a complex, multifaceted, dynamic, and progressive process in human cells that may favor immediate survival at the cost of long term consequences.
Supplementary Material
Acknowledgments
We thank Drs. Jie Liu, Erik Tokar, and Larry Keefer for careful review of the text, Matthew Bell for help with the figures, Dr. Mukta Webber for supplying the original prostate cell line, and Prof. Doris Kloor for help with AHCY activity assay.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program, National Cancer Institute, Center for the Cancer Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: SAM, S-adenosylmethionine; Hcy, homocysteine; MAT2A, methionine adenosyltransferase 2A; MAT2B, methionine adenosyltransferase 2B; SAH, S-adenosylhomocysteine; AHCY, S-adenosylhomocysteine hydrolase; GCLC, glutamate-cysteine ligase catalytic; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; BHMT, betaine-homocysteine methyltransferase; CBS, cystathionine β-synthase.
References
- 1.Benbrahim-Tallaa, L., and Waalkes, M. P. (2008) Environ. Health Perspect. 116 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Research Council (1999) Arsenic in Drinking Water, 83–101, National Academy Press, Washington, DC
- 3.National Toxicology Program (2004) in National Toxicology Program Report on Carcinogens, 11th Ed., III 18–III 20, United States Department of Health and Human Services, Research Triangle Park, NC
- 4.IARC (2004) IARC Monogr. Eval. Carcinog. Risks Hum. 84 37–267 [PMC free article] [PubMed] [Google Scholar]
- 5.Achanzar, W. E., Brambila, E. M., Diwan, B. A., Webber, M. M., and Waalkes, M. P. (2002) J. Natl. Cancer Inst. 94 1888–1891 [DOI] [PubMed] [Google Scholar]
- 6.Brambila, E. M., Achanzar, W. E., Qu, W., Webber, M. M., and Waalkes, M. P. (2002) Toxicol. Appl. Pharmacol. 183 99–107 [PubMed] [Google Scholar]
- 7.Leslie, E. M., Haimeur, A., and Waalkes, M. P. (2004) J. Biol. Chem. 279 32700–32708 [DOI] [PubMed] [Google Scholar]
- 8.Liu, J., Chen, H., Miller, D. S., Saavedra, J. E., Keefer, L. K., Johnson, D. R., Klaassen, C. D., and Waalkes, M. P. (2001) Mol. Pharmacol. 60 302–309 [DOI] [PubMed] [Google Scholar]
- 9.Thomas, D. J., Styblo, M., and Lin, S. (2001) Toxicol. Appl. Pharmacol. 176 127–144 [DOI] [PubMed] [Google Scholar]
- 10.Kligerman, A. D., Doerr, C. L., Tennant, A. H., Harrington-Brock, K., Allen, J. W., Winkfield, E., Poorman-Allen, P., Kundu, B., Funasaka, K., Roop, B. C., Mass, M. J., and DeMarini, D. M. (2003) Environ. Mol. Mutagen. 42 192–205 [DOI] [PubMed] [Google Scholar]
- 11.Mass, M. J., Tennant, A., Roop, B. C., Cullen, W. R., Styblo, M., Thomas, D. J., and Kligerman, A. D. (2001) Chem. Res. Toxicol. 14 355–361 [DOI] [PubMed] [Google Scholar]
- 12.Loenen, W. A. (2006) Biochem. Soc. Trans. 34 330–333 [DOI] [PubMed] [Google Scholar]
- 13.Razin, A., and Kantor, B. (2005) Prog. Mol. Subcell. Biol. 38 151–167 [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich, M. (2006) Curr. Top. Microbiol. Immunol. 310 251–274 [DOI] [PubMed] [Google Scholar]
- 15.Benbrahim-Tallaa, L., Waterland, R. A., Styblo, M., Achanzar, W. E., Webber, M. M., and Waalkes, M. P. (2005) Toxicol. Appl. Pharmacol. 206 288–298 [DOI] [PubMed] [Google Scholar]
- 16.Zhao, C. Q., Young, M. R., Diwan, B. A., Coogan, T. P., and Waalkes, M. P. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10907–10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichard, J. F., Schnekenburger, M., and Puga, A. (2007) Biochem. Biophys. Res. Commun. 352 188–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie, Y., Trouba, K. J., Liu, J., Waalkes, M. P., and Germolec, D. R. (2004) Environ. Health Perspect. 112 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, H., Li, S., Liu, J., Diwan, B. A., Barrett, J. C., and Waalkes, M. P. (2004) Carcinogenesis 25 1779–1786 [DOI] [PubMed] [Google Scholar]
- 20.Waalkes, M. P., Liu, J., Chen, H., Xie, Y., Achanzar, W. E., Zhou, Y. S., Cheng, M. L., and Diwan, B. A. (2004) J. Natl. Cancer Inst. 96 466–474 [DOI] [PubMed] [Google Scholar]
- 21.Okoji, R. S., Yu, R. C., Maronpot, R. R., and Froines, J. R. (2002) Carcinogenesis 23 777–785 [DOI] [PubMed] [Google Scholar]
- 22.Pastore, A., Federici, G., Bertini, E., and Piemonte, F. (2003) Clin. Chim. Acta 333 19–39 [DOI] [PubMed] [Google Scholar]
- 23.Kojima, C., Qu, W., Waalkes, M. P., Himeno, S., and Sakurai, T. (2006) Toxicol. Sci. 91 70–81 [DOI] [PubMed] [Google Scholar]
- 24.Davison, K., Cote, S., Mader, S., and Miller, W. H. (2003) Leukemia 17 931–940 [DOI] [PubMed] [Google Scholar]
- 25.Lu, S. C. (2000) Int. J. Biochem. Cell Biol. 32 391–395 [DOI] [PubMed] [Google Scholar]
- 26.Bello, D., Webber, M. M., Kleinman, H. K., Wartinger, D. D., and Rhim, J. S. (1997) Carcinogenesis 18 1215–1223 [DOI] [PubMed] [Google Scholar]
- 27.Webber, M. M., Bello, D., Kleinman, H. K., and Hoffman, M. P. (1997) Carcinogenesis 18 1225–1231 [DOI] [PubMed] [Google Scholar]
- 28.Kloor, D., Kurz, J., Fuchs, S., Faust, B., and Osswald, H. (1996) Kidney Blood Press Res. 19 100–108 [DOI] [PubMed] [Google Scholar]
- 29.Welsh, J., and McClelland, M. (1990) Nucleic Acids Res. 18 7213–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peinado, M. A., Malkhosyan, S., Velazquez, A., and Perucho, M. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 10065–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalgo, M. L., Liang, G., Spruck, C. H., III, Zingg, J. M., Rideout, W. M., III, and Jones, P. A. (1997) Cancer Res. 57 594–599 [PubMed] [Google Scholar]
- 32.Lertratanangkoon, K., Orkiszewski, R. S., and Scimeca, J. M. (1996) Cancer Res. 56 995–1005 [PubMed] [Google Scholar]
- 33.Lertratanangkoon, K., Wu, C. J., Savaraj, N., and Thomas, M. L. (1997) Cancer Lett. 120 149–156 [DOI] [PubMed] [Google Scholar]
- 34.James, S. J., Melnyk, S., Pogribna, M., Pogribny, I. P., and Caudill, M. A. (2002) J. Nutr. 132 2361S–2366S [DOI] [PubMed] [Google Scholar]
- 35.Glick, J. M., Ross, S., and Leboy, P. S. (1975) Nucleic Acids Res. 2 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crooks, P. A., Tribe, M. J., and Pinney, R. J. (1984) J. Pharm. Pharmacol. 36 85–89 [DOI] [PubMed] [Google Scholar]
- 37.Ueland, P. M. (1982) Pharmacol. Rev. 34 223–253 [PubMed] [Google Scholar]
- 38.Brodie, A. E., and Reed, D. J. (1985) Arch. Biochem. Biophys. 240 621–626 [DOI] [PubMed] [Google Scholar]
- 39.Park, E. I., and Garrow, T. A. (1999) J. Biol. Chem. 274 7816–7824 [DOI] [PubMed] [Google Scholar]
- 40.Martinov, M. V., Vitvitsky, V. M., Mosharov, E. V., Banerjee, R., and Ataullakhanov, F. I. (2000) J. Theor. Biol. 204 521–532 [DOI] [PubMed] [Google Scholar]
- 41.Brosnan, J. T., Jacobs, R. L., Stead, L. M., and Brosnan, M. E. (2004) Acta Biochim. Pol. 51 405–413 [PubMed] [Google Scholar]
- 42.Townsend, D. M., Tew, K. D., and Tapiero, H. (2003) Biomed. Pharmacother. 57 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savic-Radojevic, A., Mimic-Oka, J., Pljesa-Ercegovac, M., Opacic, M., Dragicevic, D., Kravic, T., Djokic, M., Micic, S., and Simic, T. (2007) Eur. Urol. 52 470–477 [DOI] [PubMed] [Google Scholar]
- 44.Romach, E. H., Zhao, C. Q., Del Razo, L. M., Cebrian, M. E., and Waalkes, M. P. (2000) Toxicol. Sci. 54 500–508 [DOI] [PubMed] [Google Scholar]
- 45.Ramirez, T., Garcia-Montalvo, V., Wise, C., Cea-Olivares, R., Poirier, L. A., and Herrera, L. A. (2003) Mutat. Res. 528 61–74 [DOI] [PubMed] [Google Scholar]
- 46.Wainfan, E., and Poirier, L. A. (1992) Cancer Res. 52 2071S–2077S [PubMed] [Google Scholar]
- 47.Xie, Y., Liu, J., Benbrahim-Tallaa, L., Ward, J. M., Logsdon, D., Diwan, B. A., and Waalkes, M. P. (2007) Toxicology 236 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serman, A., Vlahovic, M., Serman, L., and Bulic-Jakus, F. (2006) Coll. Antropol. 30 665–671 [PubMed] [Google Scholar]
- 49.Das, P. M., and Singal, R. (2004) J. Clin. Oncol. 22 4632–4642 [DOI] [PubMed] [Google Scholar]
- 50.Zhao, W., Mosley, B. S., Cleves, M. A., Melnyk, S., James, S. J., and Hobbs, C. A. (2006) Birth Defects Res. A. Clin. Mol. Teratol. 76 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yideng, J., Jianzhong, Z., Ying, H., Juan, S., Jinge, Z., Shenglan, W., Xiaoqun, H., and Shuren, W. (2007) DNA Cell Biol. 26 603–611 [DOI] [PubMed] [Google Scholar]
- 52.Guerra-Shinohara, E. M., Morita, O. E., Peres, S., Pagliusi, R. A., Sampaio Neto, L. F., D'Almeida, V., Irazusta, S. P., Allen, R. H., and Stabler, S. P. (2004) Am. J. Clin. Nutr. 80 1312–1321 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.