Abstract
σ-1 receptors represent unique binding sites that are capable of interacting with a wide range of compounds to mediate different cellular events. The composition of the ligand binding site of this receptor is unclear, since no NMR or crystal structures are available. Recent studies in our laboratory using radiolabeled photoreactive ligands suggested that the steroid binding domain-like I (SBDLI) (amino acids 91-109) and the steroid binding domain-like II (SBDLII) (amino acids 176-194) regions are involved in forming the ligand binding site(s) (Chen, Y., Hajipour, A. R., Sievert, M. K., Arbabian, M., and Ruoho, A. E. (2007) Biochemistry 46,3532 -3542; Pal, A., Hajipour, A. R., Fontanilla, D., Ramachandran, S., Chu, U. B., Mavlyutov, T., and Ruoho, A. E. (2007) Mol. Pharmacol. 72 ,921 -933). In this report, we have further addressed this issue by utilizing our previously developed sulfhydryl-reactive, cleavable, radioiodinated photocross-linking reagent: methanesulfonothioic acid, S-((4-(4-amino-3-[125I]iodobenzoyl) phenyl)methyl) ester (Guo, L. W., Hajipour, A. R., Gavala, M. L., Arbabian, M., Martemyanov, K. A., Arshavsky, V. Y., and Ruoho, A. E. (2005) Bioconjugate Chem. 16, 685-693). This photoprobe was shown to derivatize the single cysteine residues as mixed disulfides at position 94 in the SBDLI region of the wild type guinea pig σ-1 receptor (Cys94) and at position 190 in the SBDLII region of a mutant guinea pig σ-1 receptor (C94A,V190C), both in a σ-ligand (haloperidol or (+)-pentazocine)-sensitive manner. Significantly, photocross-linking followed by Endo Lys-C cleavage under reducing conditions and intramolecular radiolabel transfer from the SBDLI to the SBDLII region in the wild type receptor and, conversely, from the SBDLII to the SBDLI region in the mutant receptor were observed. These data support a model in which the SBDLI and SBDLII regions are juxtaposed to form, at least in part, a ligand binding site of the σ-1 receptor.
The σ receptors represent unique nonopioid and nonphen-cyclidine binding sites that are distinct from other known neurotransmitters or hormone receptors (4). Initially, Martin et al. (5) proposed the σ receptors as opioid receptors based on the psychomimetic effects of N-allyl-normetazocine (SKF-10047), which could not be explained by μ- or κ-opioid receptors (5). This hypothesis, however, was later corrected when the σ receptors were found to be insensitive to a common opioid receptor antagonist, naloxone (6-8). Two well known subtypes of the σ receptor have been identified, the σ-1 receptor and the σ-2 receptor, which are distinguishable by their ligand binding properties and molecular weights (2, 9). They are ubiquitously expressed in different tissues, such as brain, adrenal gland, testis, ovary, spleen, lung, heart, blood leukocytes, and cancer cells (10-15).
Pharmacological studies have indicated that the σ-1 receptor is able to bind a wide range of compounds, including opiates (7, 16, 17), antipsychotics (18), antidepressants (18), antihistamines (16), phencyclidine-like compounds (19), β-adrenergic receptor ligands (7, 16), serotonergic compounds (16), cocaine and cocaine analogs (1, 20, 21), cholesterol (22), steroids (23-25), and neuropeptides (25), although intriguingly no known natural specific ligands have been discovered. In addition, σ-1 receptor knock-out mice are viable and fertile, showing no overt constitutive phenotype (26). However, the broad tissue distribution as well as the ability to bind different classes of ligands allows the σ-1 receptor to mediate various cellular events, such as modulation of voltage-gated K+ channels (27), calcium release (28), regulation of lipid compartmentalization on the endoplasmic reticulum (29), and “ligand-operated” chaperoning activity at “mitochondrion-associated endoplasmic reticulum membranes” (30). Regulation of cocaine effects (31, 32), neuroprotective effects (33), increase in extracellular acetylcholine levels (34), and inhibition of proliferative responses to mitogens (35) have also been ascribed to the σ-1 receptor. A role in psychostimulant actions has been proposed for the σ-1 receptor, based on methamphetamine binding (36).
The σ-1 receptor has been cloned from different mammalian species, including guinea pigs (37), humans (38, 39), rats (40, 41), and mice (42), and shares about 90% identity and 95% similarity across these species. However, the σ-2 receptor has not been cloned yet. The cloned σ-1 receptor has 223 amino acids and shares 30% identity and 67% similarity with a yeast sterol C8-C7 isomerase (ERG2), which is involved in cholesterol synthesis (43). Interestingly, unlike the yeast sterol isomerase, the σ-1 receptor does not possess any sterol isomerase activity (37) and, in addition, shares no sequence homology with any known mammalian proteins, including the mammalian C8-C7 sterol isomerase. Hydrophobicity analysis indicates that the σ receptor and fungal sterol isomerases are topologically similar, each containing three hydrophobic regions. The first hydrophobic segment was proposed as a transmembrane domain (TMD)2 (amino acids 11-29) (37), and the other two hydrophobic segments (highly conserved regions among the species), were proposed previously as the steroid binding domain-like I (SBDLI) (amino acids 91-109) and steroid binding domain-like II (SBDLII) (amino acids 176-194) regions due to high sequence homology with the steroid binding domain of the ERG2 (1). The model proposed by Aydar et al. (44) suggested that the σ-1 receptor contains two TMDs (amino acids 11-29 and 80-100) with the orientation of both the N and C termini on the same side of the membrane bilayer. A model proposed by Hayashi et al. (30) also contains both the N and C termini at the same side of the membrane bilayer but with an opposite orientation to that proposed previously (44).
Regardless of receptor topology, it has been reported that TMDI, TMDII (including SBDLI regions), and the C terminus of the σ-1 receptor containing the SBDLII region are important for ligand binding (45-47). Mutations of the residues in TMDI (A13T, L28P) and TMDII (A86V) showed ∼60% haloperidol binding compared with the wild type receptor, as reported by Ganapathy et al. (45). Mutations of S98A, Y103F, and L105/106A in the SBDLI region resulted in 70, 16, and 39% of control NE100 binding, respectively (47). It has also been reported that Asp126 and Glu172 are obligatory for σ-1 receptor ligand binding (46). Recently, it has been shown that the regions spanning the amino acids 161-180 and 191-210 at the C-terminal end of the σ-1 receptor are important for cholesterol binding in remodeling lipid rafts (22), and the region of 116-223 is capable of performing protein chaperoning activity (30).
Previous work from our laboratory showed that a structurally rigid cocaine-based radioiodinated photoaffinity label, methyl-3-(4-azido-3-[125I]iodo-benzoyloxy)-8-methyl-8-azabicyclo[3.2.1]octane-2-carboxylate ([125I]IACoc), specifically photolabeled aspartate 188 of the SBDLII region (amino acids 176-194) as part of the cocaine binding site (1). Recently, we also found that a flexible radioiodinated fenpropimorph-like photoaffinity label, 1-N-(2′,6′-dimethylmorpholino)-3-(4-azido-3-[125I]-iodo-phenyl)propane ([125I]IAF), covalently photolabeled both the SBDLI (amino acids 91-109) and SBDLII (amino acids 176-194) regions of the guinea pig σ-1 receptors in a haloperidol-protectable manner, suggesting that both regions may be involved in forming the ligand binding site(s) (2). Moreover, [127I]IAF showed a single population of binding sites, as determined by competitive radioligand binding against 10 nm (+)-[3H]pentazocine (R2 = 0.992, Kd = 194 ± 27.5 nm). Based on these findings, we proposed two models: a single ligand binding site where the SBDLI and SBDLII regions are juxtaposed to form a single binding site or, alternatively, two ligand binding sites with exactly the same affinity for [125I]IAF in which SBDLI and SBDLII are separate parts of the two closely related ligand binding sites (2). In this paper, we report σ ligand-sensitive mixed disulfide derivatization of the pure, single cysteine containing wild type (Cys94) and mutant (C94A,V190C) guinea pig σ-1 receptors. We have chosen position 190 for cysteine mutation, since this position is near the [125I]IACoc insertion site at Asp188. The mixed disulfide was formed in the dark using a reagent previously reported from our laboratory, which is a sulfhydryl-reactive, cleavable, radioiodinated photocross-linking reagent, methanesulfonothioic acid, S-((4-(4-amino-3-[125I]iodobenzoyl) phenyl)methyl) ester ([125I]IABM) (3). We further show an intramolecular radiolabel transfer between the SBDLI and SBDLII regions of the σ-1 receptor following intramolecular photocross-linking of the derivatized receptors and cleavage with Endo Lys-C and CNBr in a reducing agent-sensitive manner. Thus, this report supports our previously proposed model (2) in which the SBDLI and SBDLII regions are juxtaposed to form, in part, a ligand binding site of the σ-1 receptor.
EXPERIMENTAL PROCEDURES
Materials—The E. coli plasmid pMal-p2X, amylose resin, and all restriction endonucleases were purchased from New England Biolabs (Beverly, MA). Primers for PCR amplification were synthesized by Integrated DNA Technologies (Coralville, IA). Nova Blues chemically competent Escherichia coli and Factor Xa protease were from Novagen (Madison, WI). Radioactive Na125I and (+)-[H3]pentazocine were from PerkinElmer Life Sciences. Endo Lys-C was from Promega (Madison, WI). CNBr and all other chemicals were purchased from σ unless otherwise mentioned.
Expression and Purification of the Wild Type (Cys94) and the Mutant (C94A,V190C) Guinea Pig σ-1 Receptors—The wild type guinea pig σ-1 receptor sequence with a His6 tag on the C terminus was inserted into a pMal-p2X vector (New England Biolabs), as described by Ramachandran et al. (48). This construct was used for the expression of the maltose-binding protein (MBP)-σ-1 receptor fusion protein containing a Factor Xa cleavage site linking the MBP and the σ-1 receptor. The wild type guinea pig σ-1 receptor sequence contains a single cysteine at position 94. For construction of the C94A,V190C mutant σ-1 receptor, cysteine was replaced by alanine at position 94 using QuikChange site-directed mutagenesis (Stratagene). This construct was subsequently used to replace valine with cysteine at position 190 to generate a single cysteine-containing V190C construct. In all cases, the constructs were sequenced to ensure the absence of additional mutations. Sequencing was carried out at the Biotechnology Center (University of Wisconsin, Madison, WI).
Both the wild type (Cys94) and mutant (C94A,V190C) constructs were transformed into BL21(DE3) separately for protein expression and purification as described previously (48). Briefly, the E. coli biomass was collected by centrifugation at 3000 rpm for 30 min, sonicated for 15 min at 4 °C. The lysate was centrifuged at 100,000 × g for 1 h at 4 °C. The pellets were resuspended and extracted for 3 h with stirring at 4 °C in 20 mm Tris, pH 8.0, 0.2 m NaCl, 1 mm 2-mercaptoethanol, and 1 mm EDTA supplemented with a protease inhibitor mixture and 1% Triton X-100 (w/v). The extracts were centrifuged at 100,000 × g for 1 h at 4 °C. The supernatants containing the fusion proteins were then purified on an amylose column and cleaved with Factor Xa at room temperature for 36-48 h. Both the wild type and mutant σ-1 receptors containing a His6 tag at the C terminus were collected on a Ni2+ column and eluted using 0.25 m imidazole in the wash buffer (50 mm sodium phosphate, pH 8.0, 0.3 m NaCl) containing 1% Triton X-100. The proteins eluted from the Ni2+ column were further purified by incubating with an agarose resin coupled with anti-MBP antibody (binding capacity of ∼0.4 mg of MBP/ml) at 4 °C for 18-24 h. After incubation was complete, the mixtures were centrifuged at 4000 rpm at room temperature, and the supernatants containing the pure wild type and mutant guinea pig σ-1 receptors were adjusted to concentrations ranging between 3 and 5 mg/ml and stored at -20 °C with 30% glycerol.
Radiosynthesis of Methanesulfonothioic Acid, [125I]IABM—[125I]IABM was prepared as reported previously with a slight modification (3). Radioactive Na125I (5 mCi) (specific activity = 2200 Ci/mmol) in 14 μl of 0.1 m NaOH was neutralized by adding 14 μl of 0.1 m HCl and then diluted with 50 μl of 0.5 m sodium acetate buffer (pH 5.6). For a standard reaction, 4-methanethiosulfonate methyl-4′-aminobenzophenone (ABM) (3) (10 μl of 25 mg/ml in 0.5 m sodium acetate buffer, pH 5.6) was then added to the reaction. Iodination was initiated by adding 30 μl of chloramine-T (1 mg/ml in water) and continued for 15 min at room temperature. The reaction was terminated by the addition of 100 μl of Na2S2O5 (5 mg/ml in water). The reaction mixture was then extracted three times with 1 ml of ethyl acetate. The pooled extracts were evaporated under N2 to ∼50 μl and streaked on a 0.25-mm-thick silica gel (60/254) plate (10 × 20 cm), which was developed with a solvent system of toluene/methanol (9:1, v/v). The radioactive band that comigrated with authentic [127I]IABM (Rf = 0.54) on the TLC plate was extracted with ethyl acetate (3 × 1 ml), dried under N2 gas, and stored at -20 °C (yield 0.734 mCi, 14.6%). Since the starting ABM (Rf = 0.41) was completely separated from [125I]IABM, the specific radioactivity of [125I]IABM was taken as 2200 Ci/mmol following purification.
Radioligand Binding Assays—The ligand binding properties of the pure WT (Cys94) and mutant (C94A,V190C) σ-1 receptors were determined using methods described previously with slight modifications in times and temperatures (45, 48, 49). The pure receptors (40 ng/well) were incubated with varying concentrations of (+)-[3H]pentazocine at 30 °C for 1 h. Binding was carried out in 50 mm Tris-HCl (pH 8.0) in a total volume of 100 μl. After incubation, the receptors were harvested on a 0.5% polyethyleneimine-treated Whatman GF/B filters using a Brandel Cell Harvester (Brandel, Gaithersburg, MD). Haloperidol (10 μm) was used to determine nonspecific binding. Radioactivity on the filters was detected by liquid scintillation spectrometry using PerkinElmer Life Sciences formula 989 as a scintillation mixture. The values were plotted according to a nonlinear regression curve using the software “Graphpad Prism” version 4.0c (GraphPad Software Inc., San Diego, CA), and the equilibrium dissociation constants, KD, were calculated using the Cheng-Prusoff equation (50).
[125I]IABM Derivatization of the Pure Guinea Pig σ-1 Receptors—Both the pure WT (Cys94) guinea pig σ-1 receptor and the C94A,V190C mutant (10 μg/tube) in 100 μl of incubation buffer (50 mm Tris-HCl, pH 8.0, containing 0.5% (v/v) Triton X-100) were incubated separately with 10 nm [125I]IABM for 6 h at room temperature in the presence or absence of the σ ligands (+)-pentazocine (1 μm) or haloperidol (10 μm). After derivatization to form the mixed disulfide bond with cysteine at position 94 or 190, proteins were then purified with Ni2+ beads and eluted with 200 μl of incubation buffer containing 250 mm imidazole. Half of the eluted protein was used as a dark control, and the other half was used for photocross-linking. To determine the specificity of the mixed disulfide [125I]IABM derivatization, eluted proteins from the Ni2+ beads were treated with SDS-PAGE sample buffer with or without reducing agent (100 mm β-mercaptoethanol) for 8 h at room temperature before separation on 15% SDS-polyacrylamide gels.
Intramolecular Photocross-linking of the Purified, [125I]IABM-derivatized Guinea Pig σ-1 Receptors—Ni2+ bead-purified and specifically [125I]IABM-derivatized receptors (both WT and V190C mutant) were photocross-linked using the methods reported previously (3). Briefly, the derivatized proteins in 1.5-ml ultraclear microcentrifuge tubes (Axygen) were incubated in the dark on ice for 15 min and then illuminated at 350 nm and ∼4 °C in a RPR-100 Rayonet photochemical reactor (Southern New England Ultraviolet) three times (5-min dark intervals on ice after each 10 min of UV light). Similarly, the cross-linked proteins were treated with SDS-PAGE sample buffer with or without reducing agent (100 mm β-mercaptoethanol) for 8 h at room temperature before separation on 15% SDS-polyacrylamide gels.
Endo Lys-C and CNBr Digestion of the Derivatized and Photocross-linked Guinea Pig σ-1 Receptors—Prior to the enzymatic cleavage with Endo Lys-C, both derivatized (dark control) and intramolecularly photocross-linked receptors (both WT and C94A,V190C mutant) were incubated with SDS-PAGE sample buffer with or without reducing agent (100 mm β-mercaptoethanol) for 8 h at room temperature. Proteins were then separated by electrophoresis on a 15% SDS-polyacrylamide gel and visualized by wet gel (unstained) autoradiography. The autoradiogram was used as a template to excise the specifically derivatized photocross-linked as well as the ligand-protected σ-1 receptors from the gel with a razor blade. The gel slices were minced and placed into a 1.5-ml microcentrifuge tube containing 1 ml of water. The tubes were maintained over-night at 4 °C, and the gel slurries were transferred to spin columns (Bio-Rad). The eluted σ-1 receptors in the supernatant extracts were collected by centrifugation at 1600 × g for 5 min and cleaved with Endo Lys-C as reported earlier (2). The proteolytic digestions were terminated by adding SDS-PAGE sample buffer (with or without reducing agent) and separated by 16.5% SDS-Tricine/PAGE. For CNBr digestion, the peptides were again identified by wet gel autoradiography, excised and eluted with water, concentrated (to ∼30 μl), and mixed with 70 μl of CNBr solution (0.15 m in 70% formic acid solution) for 15-18 h at room temperature by tumbling as described previously (2).
RESULTS
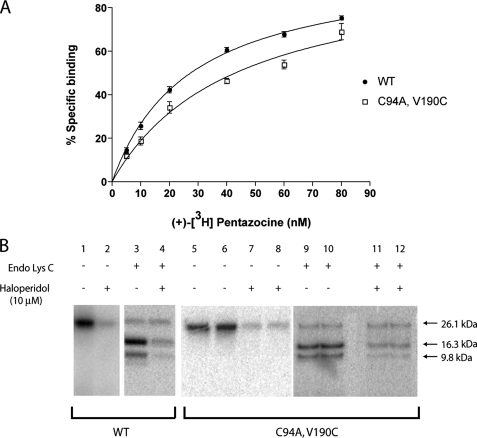
Expression, Purification, and Characterization of the Guinea Pig C94A,V190C Mutant σ-1 Receptor—The C94A,V190C mutant guinea pig σ-1 receptor was expressed and purified in a fashion similar to the WT guinea pig σ-1 receptor. For this purpose, the WT σ-1 receptor was mutated first from cysteine at position 94 to alanine and then further mutated at position 190 to introduce a single cysteine replacing valine. The mutant receptor was expressed in E. coli as an MBP-σ-1 receptor-His6 fusion with a factor Xa cleavage site between MBP and the σ-1 receptor. The fusion protein was purified on an amylose column and cleaved with factor Xa. The mutant σ-1 receptor was then purified using Ni2+ columns to a final concentration of about 3 mg/ml. The purified mutant receptor was characterized based on (+)-[3H]pentazocine binding and photolabeling with [125I]IAF (structure shown in Fig. 1) (2), followed by Endo Lys-C digestion (Fig. 2). The mutant receptor showed binding affinity for (+)-[3H]pentazocine with a Kd value range of 40.3 ± 2.8 nm which was comparable with that of the WT receptor (Kd range of 21.2 ± 2.4 nm) (Fig. 2A). The mutant receptor also showed a specific [125I]IAF photolabeling pattern (Fig. 2B) similar to that reported previously for the WT receptor (2). The specificity of the photolabeling was demonstrated by protection with 10 μm haloperidol (Fig. 2B, compare lanes 5 and 6 with lanes 7 and 8). Following cleavage of the specifically [125I]IAF-photolabeled mutant receptor with Endo Lys-C, both the 16.3- and 9.8-kDa peptide contained specific radiolabel (Fig. 2B, compare lanes 9 and 10 with lanes 11 and 12). Therefore, the purified mutant (C94A,V190C) guinea pig σ-1 receptor was similar to the WT (Cys94) in their ligand binding and photolabeling properties.
FIGURE 1.
Structures of [125I]IACoc) and 125I]IAF). The [125I]IAF photoprobe was used to specifically photolabel the WT and mutant (C94A,V190C) σ-1 receptor in Fig. 2B. The [125I]IACoc photoprobe was used together with [125I]IAF in the experiments reported in Fig. 6.
FIGURE 2.
A, binding of (+)-[3H]pentazocine to the pure guinea pig WT (Cys94) and the mutant (C94A,V190C) σ-1 receptor. Various concentrations of (+)-[3H]pentazocine (0-80 nm) were incubated with pure receptors (40 ng/well) separately for 1 h at 30 °C. Haloperidol (5 μm) was used to determine nonspecific binding. The values were plotted using GraphPad Prism software (version 4.0c) (nonlinear regression curve). (+)-[3H]Pentazocine bound to the WT and mutant receptors with KD values of 21.2 ± 2.4 and 40.3 ± 2.8 nm, respectively, indicating that both the WT and mutant receptors have similar ligand binding properties. Error bars, S.D. values of triplicates from two different experiments. B, PhosphorImager visualization of Endo Lys-C cleavage of the pure guinea pig WT and C94A,V190C mutant σ-1 receptor specifically photolabeled with [125I]IAF (2). The [125I]IAF (structure shown in Fig. 1)-photolabeled pure WT (lanes 1-4) and mutant (C94A,V190C) σ-1 receptor (lanes 5-8) showed a similar Endo Lys-C cleavage pattern as reported previously (2). Specific photolabeling was determined by comparing radiolabel incorporation in the absence of 10 μm haloperidol (lanes 1, 3, 5, 6, 9, and 10) and in the presence of 10 μm haloperidol (lanes 2, 4, 7, 8, 11, and 12). Upon treatment with Endo Lys-C, the specifically [125I]IAF-photolabeled WT and mutant receptors showed radiolabel on both the SBDLI (16.3 kDa) and SBDLII (9.8 kDa) regions (lanes 5 and 6), as previously reported for the WT receptor (2).
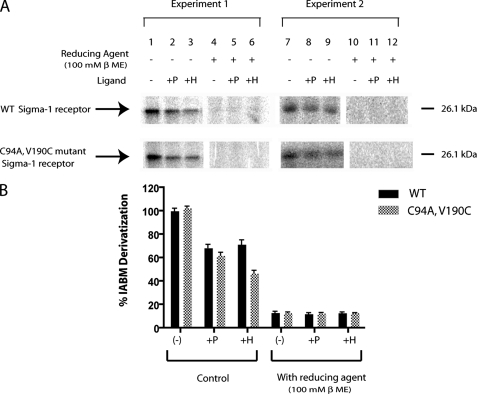
[125I]IABM Derivatization of the Pure WT (Cys94) and the C94A,V190C Mutant Guinea Pig σ-1 Receptors—Both the WT and the C94A,V190C mutant σ-1 receptors contained a single cysteine residue, which was derivatized to form a mixed disulfide under dark conditions with the radioactive cross-linking agent, [125I]IABM (Fig. 3). The derivatizations were highly specific for cysteine, since the mixed disulfide radiolabel was quantitatively removed when treated with 100 mm β-mercaptoethanol (Fig. 3A, compare lanes 1-3 and 7-9 with lanes 4-6 and 10-12, respectively). Interestingly, derivatization was also sensitive to the presence of the σ ligands, (+)-pentazocine and haloperidol (Fig. 3A, compare lanes 1 and 7 with lanes 2 and 8 and lanes 3 and 9, respectively). Densitometric quantitations (n = 3) (Fig. 3B) using the software “Imagej” showed that (+)-pentazocine inhibited the [125I]IABM derivatization of the WT (Cys94) and C94A,V190C mutant by ∼31.4 ± 2.9 and 38 ± 2.6%, respectively, as compared with the control (without ligand; Fig. 3A, lanes 1 and 6), whereas haloperidol inhibited the [125I]IABM derivatization by 28.6 ± 4 and 53.2 ± 2.4%, respectively (Fig. 3B).
FIGURE 3.
A, protection of specific [125I]IABM derivatization of the guinea pig WT (Cys94) and mutant (C94A,V190C) σ-1 receptor by σ receptor ligands. Pure WT (Cys94) and C94A,V190C mutant σ-1 receptors (10 μg/lane, 100-μl volume) were derivatized separately using 10 μl of [125I]IABM in the absence (-) (compare lanes 1 and 6 with lanes 4 and 9, respectively) or the presence of 1 μm (+)-pentazocine (+P) (compare lanes 2 and 7 with lanes 5 and 10, respectively) and 10 μm haloperidol (+H) (compare lanes 3 and 8 with lanes 6 and 12, respectively). After mixed disulfide derivatization for 6 h at room temperature, samples were treated with either no reducing agent (lanes 1-3 and 7-9) or with 100 mm β-mercaptoethanol (lanes 4-6 and 10-12) for 8 h at room temperature and visualized by phosphorimaging after separation on 15% SDS-polyacrylamide gels followed by staining-destaining. B, densitometric quantitation of [125I]IABM derivatization using Imagej software. (+)-Pentazocine (+P) (1 μm) and haloperidol (10 μm) showed the specific [125I]IABM derivatization of the guinea pig WT receptor (Cys94) by 68.6 ± 2.9 and 71.7 ± 3.5%, respectively, and the mutant (C94A,V190C) σ-1 receptor by 62 ± 2.6 and 46.8 ± 2.5%, respectively, as compared with control (no ligand). Error bars, S.D. values from three different experiments (n = 3).
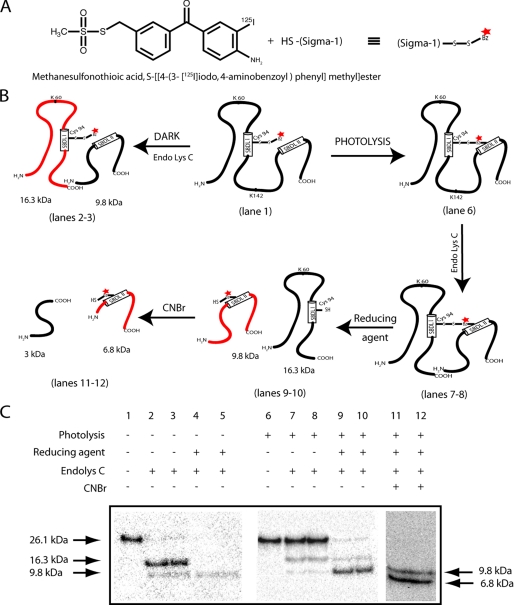
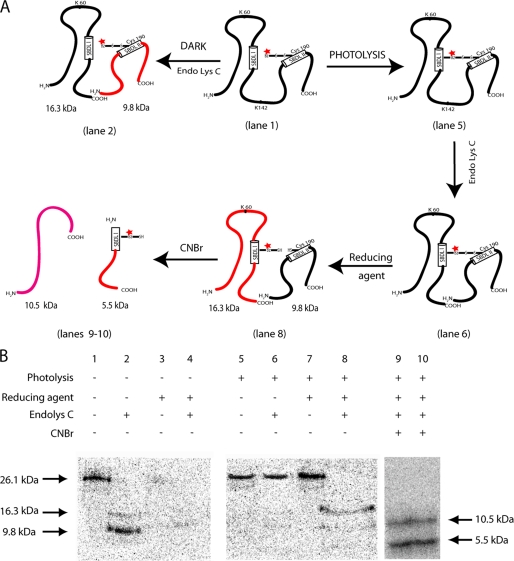
Intramolecular Radiolabel Transfer from SBDLI to the SBDLII Region in the Pure WT σ-1 Receptor—The strategy of the radiolabel transfer assay for the WT σ-1 receptor is diagrammed in Fig. 4, A and B. The mixed disulfide [125I]IABM derivatization of the single cysteine residue at position 94 in the SDBLI region of the WT σ-1 receptor was highly specific as well as sensitive to reducing agent. Under dark conditions, following treatment with Endo Lys-C, the SDS gel-purified, [125I]IABM-derivatized receptor (Fig. 4C, lane 1) produced two peptides with molecular masses of 16.5 and 9.8 kDa (Fig. 4C, lanes 2 and 3), as reported previously (2). Approximately 93% of the total radioactivity was found in the SBDLI-containing 16.3-kDa peptide (Fig. 4C, lanes 2 and 3) and as also depicted in the diagram shown in Fig. 4B. As expected, almost all of the radioactivity was removed by 100 mm β-mercaptoethanol (Fig. 4C, lanes 4 and 5), confirming the cysteine-specific derivatization at position 94. Before photocross-linking, the free excess [125I]IABM was removed from the [125I]IABM-derivatized WT guinea pig σ-1 receptor by purification on Ni2+ beads followed by SDS-PAGE. After photocross-linking and cleavage with Endo Lys-C under nonreducing conditions, the majority of the radioactivity remained at the 26.1 kDa position (see Fig. 4, B and C, lanes 7 and 8), suggesting that the two cleaved peptides were held together by the cross-linker. On the other hand, after Endo Lys-C cleavage of the photocross-linked receptors followed by the disulfide bond reversal under reducing conditions, 90% of the radioactivity was found in the SBDLII-containing 9.8-kDa peptide (Fig. 4C, lanes 9 and 10). In addition, when the 9.8-kDa radiolabeled peptide was further cleaved with CNBr at Met170, the covalently photoinserted radiolabel was located in the SBDLII-containing 6.8-kDa fragment spanning from position 171 to 223 (Fig. 4C, lanes 11 and 12) as reported earlier (2) (see also the diagram in Fig. 4B).
FIGURE 4.
A, reaction of methanesulfonothioic acid, [125I]IABM, with the thiol group of the σ-1 receptor. B, schematic diagram of different lanes shown C for the photocross-linking/radiolabel transfer using the pure guinea pig WT (Cys94) σ-1 receptor with [125I]IABM. C, photocross-linking/radiolabel transfer assay of the pure guinea pig WT (Cys94) σ-1 receptor using [125I]IABM. Under dark conditions (lanes 1-5), the gel-purified, [125I]IABM-derivatized receptor (lane 1) produced the SBDLI region-containing 16.5-kDa Endo Lys-C-cleaved peptide as the major radioactive band (lanes 2 and 3) that was sensitive to the reducing agent β-mercaptoethanol (100 mm) (lanes 4 and 5). After photolysis (lanes 6-10), the photocross-linked receptor under nonreducing conditions (lanes 6-8, no β-mercaptoethanol) appeared as the full-length receptor even after the treatment of Endo Lys-C (lane 7-8), whereas under reducing conditions, radiolabel transfer occurred from the SBDLI-containing 16.5-kDa peptide to the SBDLII-containing 9.8-kDa Endo Lys-C-cleaved peptide (compare lanes 2 and 3 with lanes 9 and 10). Further cleavage of this 9.8-kDa radiolabeled peptide with CNBr produced 6.8-kDa peptides (lanes 11 and 12), consistent with the radiolabel incorporation in the SBDLII regions, as reported earlier (2).
Intramolecular Radiolabel Transfer from SBDLII to the SBDLI Region in the C94A,V190C Mutant—If the SBDLI and SBDLII regions are juxtaposed, then the reverse intramolecular radiolabel transfer from SBDLII to SBDLI should occur. Accordingly, the radiolabel transfer assay using the E. coli expressed and purified mutant C94A,V190C guinea pig σ-1 receptor was performed in the same manner as the WT receptor (diagrammed in Fig. 5A). Due to the presence of the single cysteine residue at position 190 in the SDBLII region of the mutated σ-1 receptor, under dark conditions (Fig. 5B, compare lane 1 with lane 3), most of the derivatized radiolabel was found in the 9.8-kDa SBDLII-containing peptides when cleaved with Endo Lys-C (Fig. 5B, compare lane 2 with lane 4, and as depicted in Fig. 5A). After photocross-linking (Fig. 5B, lane 5), Endo Lys-C treatment under nonreducing conditions again showed the full-length, 26.1 kDa radioactive band (Fig. 5B, lane 6), whereas under reducing conditions, the 16.3-kDa SBDLI-containing peptide appeared as the major radioactive band (Fig. 5B, compare lane 7 with lane 8). CNBr digestion at methionine residues (Met2, Met90, and Met93) of the 16.3 kDa band further showed that both the expected 10.5-kDa (amino acids 1-90) and 5.5-kDa (amino acids 94-142) peptides contained radiolabel and the majority of the radioactivity incorporated into the smaller 5.5-kDa fragment (amino acids 94-142) (Fig. 5B, lanes 9 and 10), which contains almost the entire SBDLI region, as reported earlier (2).
FIGURE 5.
A, schematic diagram of the photocross-linking/radiolabel transfer using the pure guinea pig mutant (C94A,V190C) σ-1 receptor with [125I]IABM. B, photocross-linking/radiolabel transfer assay of the pure guinea pig mutant (C94A,V190C) σ-1 receptor using [125I]IABM. Under dark conditions (lanes 1-4), the gel-purified, specific mixed disulfide [125I]IABM-derivatized receptor (compare lane 1 with lane 3) produced the SBDLII region-containing 9.8-kDa Endo Lys-C-cleaved peptide as the major radioactive band that was sensitive to the reducing agent β-mercaptoethanol (100 mm) (compare lane 2 with lane 4). After photolysis (lanes 5-8), the photocross-linked receptor under nonreducing conditions appeared as the full-length receptor even after treatment with Endo Lys-C (compare lane 5 with lane 6). Under reducing conditions, the majority of the radiolabel was transferred from the SBDLII-containing 9.8-kDa peptide to the SBDLI-containing 16.3-kDa Endo Lys-C-cleaved peptide (compare lane 7 with lane 8 for Endo Lys-C cleavage, and compare lane 2 with lane 8 for radiolabel transfer). Further cleavage of this 16.3-kDa radiolabeled peptide with CNBr produced both the expected 10.5- and 5.5-kDa peptides as radiolabel bands, but the majority of the radiolabel was detected in the smaller 5.5-kDa peptide (lanes 9 and 10), consistent with the radiolabel photoincorporation into the SBDLI regions.
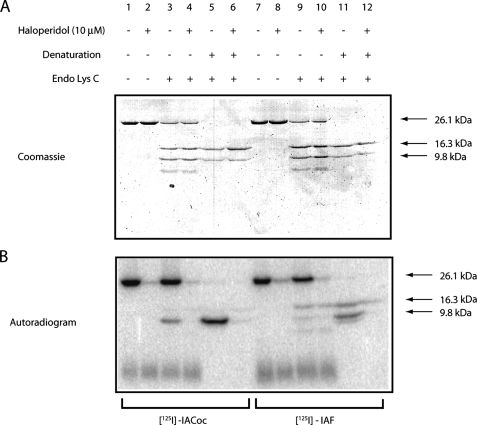
Protection of Endo Lys-C Cleavage of the Native, Ligand-photolabeled σ-1 Receptor—As previously reported (2), both the photoaffinity ligands [125I]IACoc and [125I]IAF (structures shown in Fig. 1) specifically covalently photolabeled the pure WT guinea pig σ-1 receptor binding site(s) in the SBDLI and SBDLII regions. We hypothesized that the presence of the covalently attached ligand binding site-occupying photoprobes in the SBDLI and SBDLII domains of the pure σ-1 receptor would protect against Endo Lys-C cleavage at Lys142. Accordingly, the photolabeled pure guinea pig σ-1 receptor was cleaved with Endo Lys-C under native (i.e. in 0.5% Triton X-100) and denatured conditions (i.e. SDS gel-eluted). The Coomassie staining pattern of the native specifically photolabeled σ-1 receptor is shown in Fig. 6A (lanes 1 and 2 for [125I]IACoc-labeled and lanes 7 and 8 for [125I]IAF-labeled), and the corresponding autoradiogram is shown in Fig. 6B (lanes 1 and 2 for [125I]IACoc-labeled and lanes 7 and 8 for [125I]IAF-labeled). Following Endo Lys-C cleavage of the native receptor, the nonphotoincorporated population of σ-1 receptor molecules was readily cleaved into the expected 16.3- and 9.8-kDa fragments (Fig. 6A, lanes 3 and 4 and lanes 9 and 10). However, the σ-1 receptor molecules that were specifically photolabeled by the photoprobes were not as readily cleaved (Fig. 6B, lanes 3 and 4 for [125I]IA-Coc and lanes 9 and 10 for [125I]IAF). This result occurred because a fraction of the σ-1 receptor molecules were specifically covalently labeled with the photoprobes, and the covalent positioning of the photoprobes in the SBDLI/SBDLII binding site protected against Endo Lys-C cleavage at Lys142. To further support these conclusions, when the native specifically photolabeled receptors were first denatured with SDS prior to Endo Lys-C cleavage, then a more robust cleavage of the σ-1 receptors to the 16.3- and 9.8-kDa fragments occurred (Fig. 6A, lanes 5 and 6 and lanes 11 and 12). Now the denatured photolabeled σ-1 receptor molecules were also cleaved to reveal the peptides of specific derivatization; i.e. for [125I]IACoc, radiolabel was seen exclusively on the 9.8-kDa fragment (Fig. 6B, lanes 5 and 6), whereas with [125I]IAF, specific labeling was distributed between the 16.3- and 9.8-kDa fragments (Fig. 6B, lanes 11 and 12). This result is further consistent with juxtaposition of the SBDLI and SBDLII regions of the σ-1 receptor binding site.
FIGURE 6.
A, Coomassie staining pattern of Endo Lys-C cleavage of the pure guinea pig σ-1 receptor photolabeled with [125I]IACoc and [125I]IAF under native (0.5% Triton X-100) and denatured (SDS gel-eluted) conditions. B, phosphorimaging visualization of the gels shown in Fig. 6A. Both [125I]IACoc-photolabeled (lanes 1-6) and [125I]IAF-photolabeled (lanes 7-12) pure guinea pig σ-1 receptor showed specific derivatization, as determined by comparing the photolabeling in the absence of 10 μm haloperidol (lanes 1, 3, 5, 7, 9, and 11) and in the presence of 10 μm haloperidol (lanes 2, 4, 6, 8, 10, and 12). Under native conditions, the specifically photolabeled receptor using both of the photoprobes showed resistance to the cleavage at Lys142 with Endo Lys-C (lanes 3 and 4 for the [125I]IACoc-labeled receptor and lanes 9 and 10 for the [125I]IAF-labeled receptor). The Coomassie staining pattern (Fig. 6A) showed that the majority of the receptors that were not photolabeled were cleaved by Endo Lys-C. On the other hand, under denatured conditions (SDS gel-eluted), the specifically photolabeled receptor showed no resistance to Endo Lys-C cleavage as visualized by phosphorimaging and Coomassie staining (lanes 5 and 6 for [125I]IACoc-labeled receptor and lanes 11 and 12 for [125I]IAF-labeled receptor), as reported earlier (2).
DISCUSSION
For the past few decades, the σ-1 receptor has been persistently enigmatic. The ability to bind different classes of ligands allows this receptor to mediate various cellular events. As a result, it is extremely important to obtain structural information regarding the ligand binding site(s). The fact that neither any atomic structure nor any homolog is available for the σ-1 receptor, necessitates biochemical approaches to investigate the binding site(s). Thus, we utilized a photocross-linking/radiolabel transfer approach, which is a powerful technique to investigate intramolecular domain-domain interaction(s) or intermolecular protein-protein interactions. Location of the radiolabeled “donor” and “receiver” regions of the σ-1 receptor were determined by taking advantage of Endo Lys-C-sensitive cleavage at Lys142 and chemical cleavage with cyanogen bromide at positions 2, 90, 93, and 170. In recent years, our laboratory has successfully applied this technique to identify protein-protein interactions (3, 51-54).
Recent reports from our laboratory showed that the highly conserved hydrophobic regions of the σ-1 receptor SBDLI (amino acids 91-109) and SBDLII (amino acids 176-194) comprise at least parts of the ligand binding site(s), as shown by photolabeling with two highly specific radioactive photoprobes [125I]IAF and [125I]IACoc (1, 2). The [125I]IACoc photoprobe specifically photolabeled the σ-1 receptor at Asp188, which is in the SBDLII regions, whereas the [125I]IAF-mediated photolabel incorporation was found in the peptides containing both the SBDLI and SBDLII regions. From these [125I]IAF photolabeling data, we hypothesized that the SBDLI and SBDLII regions are at least partly involved in constituting the σ-1 receptor ligand binding site(s), and possibly these regions are in close proximity to each other.
Since the cloned wild type guinea pig σ-1 receptor contains only a single cysteine residue at position 94 (in the SBDLI region), we utilized this opportunity to test the hypothesis of the intramolecular juxtaposition of the SBDLI and SBDLII regions. We derivatized the single cysteine at position 94 via the MTS group of the radioactive benzophenone cross-linking agent, [125I]IABM. This cross-linking agent has a photoactive carbonyl carbon atom, which is ∼8 Å from the mixed disulfide bond. After photocross-linking followed by Endo Lys-C cleavage, the appearance of full-length receptor under nonreducing conditions indicated that the 16.5- and 9.8-kDa peptides generated by Endo Lys-C cleavage remained tethered together via the mixed disulfide bond (Fig. 4C, lanes 7 and 8). Intermolecular cross-linking was not observed, since a dimer was not obtained under any of the conditions reported in this paper (e.g. see Fig. 4C, lanes 7 and 8). On the other hand, Endo Lys-C cleavage, followed by treatment with reducing agent (100 mm β-mercaptoethanol), showed that almost all of the radiolabel was transferred to the SBDLII-containing 9.8-kDa peptide (Fig. 4C, lanes 9 and 10). Further cleavage of the 9.8-kDa peptide with CNBr also indicated that the radioactive photolabel was incorporated into the SBDLII-containing 6.8-kDa peptides (Fig. 4C, lanes 11 and 12). These results suggest that the SBDLI and SBDLII regions are likely to be juxtaposed within an approximate distance of 8 Å.
To explore the possibility of juxtaposition of the SBDLII with the SBDLI further, we generated a mutant of the guinea pig σ-1 receptor in which an alanine residue replaced the cysteine at position 94 and a cysteine residue was introduced at valine 190. Since the specific [125I]IACoc photolabel insertion in the guinea pig σ-1 receptor occurred at the Asp188 (1), we chose position 190 for replacement with cysteine. This E. coli expressed and purified C94A,V190C mutant σ-1 receptor was further characterized by determining the Kd value for (+)-[3H]pentazocine binding and Endo Lys-C cleavage pattern after photolabeling with [125I]IAF (Fig. 2). The mutant receptor showed similar ligand binding as well as [125I]IAF photolabeling compared with the WT receptor.
In a similar fashion to the derivatization at position 94 in the WT receptor, we used [125I]IABM to derivatize the position 190 cysteine in the SBDLII region of the purified mutant receptor (C94A,V190C) as a mixed disulfide and proved the specificity of the derivatization by the same methods described for the WT receptor. In contrast to the data from position 94, ∼68% of the total radioactivity after photocross-linking was transferred to the larger 16.5-kDa peptide following Endo Lys-C cleavage as depicted in the diagram (Fig. 5A). Interestingly, the CNBr digestion of this 16.5-kDa peptide produced two radioactive bands with molecular masses of 10.5 and 5.5 kDa. The larger 10.5 kDa band consists of the putative TMDI as well as a portion of putative TMDII, whereas the lower 5.5 kDa band contained the SBDLI region. This result leads to the conclusion that the radiolabeled [125I]IABM, which derivatized the cysteine at position 190, was intramolecularly transferred to the SBDLI region as well as to the putative TMDI and/or a portion of putative TMDII following photolysis, reduction, and Endo Lys-C cleavage (Fig. 5B). These data further indicate that TMDI may be also closely juxtaposed to SBDLI and SBDLII and are consistent with the mutagenesis experiments previously reported by Ganapathy et al. (45).
Furthermore, partial protection of [125I]IABM derivatization by the well known σ ligands (+)-pentazocine and haloperidol from both positions 94 and 190 (Fig. 3) supports the conclusion that these residues are located within a portion of the ligand binding site(s) or conformationally connected to this region, since the presence of these ligands rendered cysteine at positions 94 and 190 less accessible to the [125I]IABM. In addition, the SBDLI and SBDLII regions comprise at least a portion of the ligand binding site of the σ-1 receptors, since the native [125I]IACoc or [125I]IAF photolabeled receptors were found to be not readily cleaved by Endo Lys-C (Fig. 6). This result indicated that the ligand-bound receptors could be structurally different from the non-ligand-bound receptors and comparatively less sensitive to Endo Lys-C cleavage. Since the Lys142 position is straddled by the SBDLI and SBDLII regions, this cleavage site was protected from cleavage by Endo Lys-C when the binding site was occupied by the ligand photoprobe. Use of limited proteolysis in the presence of substrates or small molecules to query binding sites and other structural properties of proteins has been previously utilized extensively with great success (55-58).
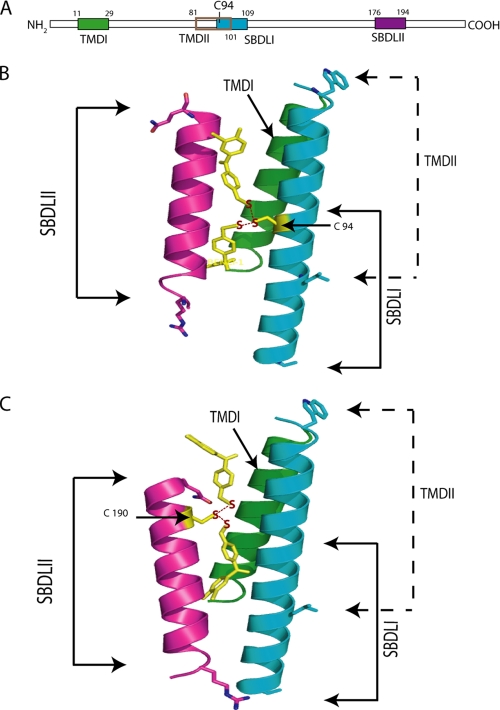
In conclusion, the radiolabel transfer experiments between the SBDLI and SBDLII regions are summarized as models in Fig. 7. The helical structures for SBDLI and SBDLII are considered based on the high order predicted for these regions by the PONDR program (available on the World Wide Web) and the hydrophobicity prediction from the TMPred program (available on the World Wide Web) (2), and the assumption for the helical structures of these regions also agrees with a model reported by Palmer et al. (22). The benzophenones were oriented 180° from the center of the Cβ atom of the cysteine in the helix to demonstrate the possible 20 Å reach on either side of the targeted helix. Both models illustrate that the SBDLI and SBDLII regions are juxtaposed to form at least part of the guinea pig σ-1 receptor ligand binding site(s) and may be important to mediate the proposed functions, such as lipid raft remodeling (22) or ligand-operated chaperoning (30), and support the model proposed earlier (2). There also remains the possibility that the TMDI or a part of the TMDII (residues 80-89) outside the SBDLI region is also in close apposition to the SBDLII domain and could contribute residues to the ligand binding site(s), as previously reported (45).
FIGURE 7.
Summary models of the [125I]IABM radiolabel transfer assays between the SBDLI and SBDLII regions of the pure guinea pig σ-1 receptor. A linear diagram of the σ-1 receptor (A) is color-coded to match the putative helical TMDI, TMDII/SBDLI, and SBDLII regions shown in B and C. The [125I]IABM radiolabel transfer from cysteine at position 94 (B) and 190 (C) is shown schematically. The specified sequences were assumed to be α-helical structures and built in a standard α-helix using the Molecular Modeling program Sybyl from Tripos International (St. Louis, MO). The helices were manipulated to orient them in agreement with the known chemistry and their predicted position in the membrane. The benzophenones were oriented 180° from the center of the Cβ atom of the cysteine in the helix to illustrate the 20-Å reach on either side of the targeted helix.
Acknowledgments
We thank Dr. Kenneth A. Satyshur for helping to create the models.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 MH065503 (to A. E. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TMD, transmembrane domain; SBDLI, steroid binding domain-like I; SBDLII, steroid binding domain-like II; [125I]IACoc, methyl-3-(4-azido-3-[125I]iodo-benzoyloxy)-8-methyl-8-azabicyclo[3.2.1]octane-2-carboxylate; [125I]IAF, 1-N-(2′,6′-dimethylmorpholino)-3-(4-azido-3-[125I]iodo-phenyl)propane; [125I]IAMBM, S-[[4-(4-amino-3-[125I]iodobenzoyl) phenyl] methyl] ester; Endo, endoproteinase; MBP, maltose-binding protein; ABM, 4-methanethiosulfonate methyl-4′-aminobenzophenone; WT, wild type; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1.Chen, Y., Hajipour, A. R., Sievert, M. K., Arbabian, M., and Ruoho, A. E. (2007) Biochemistry 463532 -3542 [DOI] [PubMed] [Google Scholar]
- 2.Pal, A., Hajipour, A. R., Fontanilla, D., Ramachandran, S., Chu, U. B., Mavlyutov, T., and Ruoho, A. E. (2007) Mol. Pharmacol. 72921 -933 [DOI] [PubMed] [Google Scholar]
- 3.Guo, L. W., Hajipour, A. R., Gavala, M. L., Arbabian, M., Martemyanov, K. A., Arshavsky, V. Y., and Ruoho, A. E. (2005) Bioconjugate Chem. 16685 -693 [DOI] [PubMed] [Google Scholar]
- 4.Quirion, R., Bowen, W. D., Itzhak, Y., Junien, J. L., Musacchio, J. M., Rothman, R. B., Su, T. P., Tam, S. W., and Taylor, D. P. (1992) Trends Pharmacol. Sci. 13 85-86 [DOI] [PubMed] [Google Scholar]
- 5.Martin, W. R., Eades, C. G., Thompson, J. A., Huppler, R. E., and Gilbert, P. E. (1976) J. Pharmacol. Exp. Ther. 197517 -532 [PubMed] [Google Scholar]
- 6.Iwamoto, E. T. (1981) J. Pharmacol. Exp. Ther. 217451 -460 [PubMed] [Google Scholar]
- 7.Su, T. P. (1982) J. Pharmacol. Exp. Ther. 223284 -290 [PubMed] [Google Scholar]
- 8.Vaupel, D. B. (1983) Eur. J. Pharmacol. 92269 -274 [DOI] [PubMed] [Google Scholar]
- 9.Hellewell, S. B., Bruce, A., Feinstein, G., Orringer, J., Williams, W., and Bowen, W. D. (1994) Eur. J. Pharmacol. 2689 -18 [DOI] [PubMed] [Google Scholar]
- 10.Walker, J. M., Bowen, W. D., Walker, F. O., Matsumoto, R. R., De Costa, B., and Rice, K. C. (1990) Pharmacol. Rev. 42355 -402 [PubMed] [Google Scholar]
- 11.Seth, P., Leibach, F. H., and Ganapathy, V. (1997) Biochem. Biophys. Res. Commun. 241535 -540 [DOI] [PubMed] [Google Scholar]
- 12.Bowen, W. D. (2000) Pharm. Acta Helv. 74211 -218 [DOI] [PubMed] [Google Scholar]
- 13.Bermack, J. E., and Debonnel, G. (2005) J. Pharmacol. Sci. 97317 -336 [DOI] [PubMed] [Google Scholar]
- 14.Aydar, E., Palmer, C. P., and Djamgoz, M. B. (2004) Cancer Res. 645029 -5035 [DOI] [PubMed] [Google Scholar]
- 15.Wheeler, K. T., Wang, L. M., Wallen, C. A., Childers, S. R., Cline, J. M., Keng, P. C., and Mach, R. H. (2000) Br. J. Cancer 821223 -1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tam, S. W. (1983) Proc. Natl. Acad. Sci. U. S. A. 806703 -6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber, E., Sonders, M., Quarum, M., McLean, S., Pou, S., and Keana, J. F. (1986) Proc. Natl. Acad. Sci. U. S. A. 838784 -8788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell, B. G., Scherz, M. W., Keana, J. F., and Weber, E. (1989) J. Neurosci. 93380 -3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Contreras, P. C., Contreras, M. L., O'Donohue, T. L., and Lair, C. C. (1988) Synapse 2 240-243 [DOI] [PubMed] [Google Scholar]
- 20.Kahoun, J. R., and Ruoho, A. E. (1992) Proc. Natl. Acad. Sci. U. S. A. 891393 -1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharkey, J., Glen, K. A., Wolfe, S., and Kuhar, M. J. (1988) Eur. J. Pharmacol. 149171 -174 [DOI] [PubMed] [Google Scholar]
- 22.Palmer, C. P., Mahen, R., Schnell, E., Djamgoz, M. B., and Aydar, E. (2007) Cancer Res. 6711166 -11175 [DOI] [PubMed] [Google Scholar]
- 23.Su, T. P., London, E. D., and Jaffe, J. H. (1988) Science 240219 -221 [DOI] [PubMed] [Google Scholar]
- 24.Monnet, F. P., Mahe, V., Robel, P., and Baulieu, E. E. (1995) Proc. Natl. Acad. Sci. U. S. A. 923774 -3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonard, B. E. (2004) Pharmacopsychiatry 37Suppl. 3 , 166-170 [Google Scholar]
- 26.Langa, F., Codony, X., Tovar, V., Lavado, A., Gimenez, E., Cozar, P., Cantero, M., Dordal, A., Hernandez, E., Perez, R., Monroy, X., Zamanillo, D., Guitart, X., and Montoliu, L. (2003) Eur. J. Neurosci. 182188 -2196 [DOI] [PubMed] [Google Scholar]
- 27.Wilke, R. A., Mehta, R. P., Lupardus, P. J., Chen, Y., Ruoho, A. E., and Jackson, M. B. (1999) J. Biol. Chem. 27418387 -18392 [DOI] [PubMed] [Google Scholar]
- 28.Hayashi, T., and Su, T. P. (2001) Proc. Natl. Acad. Sci. U. S. A
- 29.Hayashi, T., and Su, T. P. (2003) J. Pharmacol. Exp. Ther. 306718 -725 [DOI] [PubMed] [Google Scholar]
- 30.Hayashi, T., and Su, T. P. (2007) Cell 131596 -610 [DOI] [PubMed] [Google Scholar]
- 31.McCracken, K. A., Bowen, W. D., de Costa, B. R., and Matsumoto, R. R. (1999) Eur. J. Pharmacol. 370225 -232 [DOI] [PubMed] [Google Scholar]
- 32.McCracken, K. A., Bowen, W. D., and Matsumoto, R. R. (1999) Eur. J. Pharmacol. 365 35-38 [DOI] [PubMed] [Google Scholar]
- 33.Lysko, P. G., Gagnon, R. C., Yue, T. L., Gu, J. L., and Feuerstein, G. (1992) Stroke 23 414-419 [DOI] [PubMed] [Google Scholar]
- 34.Matsuno, K., Matsunaga, K., Senda, T., and Mita, S. (1993) J. Pharmacol. Exp. Ther. 265851 -859 [PubMed] [Google Scholar]
- 35.Paul, R., Lavastre, S., Floutard, D., Floutard, R., Canat, X., Casellas, P., Le Fur, G., and Breliere, J. C. (1994) J. Neuroimmunol. 52183 -192 [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, E. C., McCracken, K. A., Liu, Y., Pouw, B., and Matsumoto, R. R. (2005) Neuropharmacology 49 638-645 [DOI] [PubMed] [Google Scholar]
- 37.Hanner, M., Moebius, F. F., Flandorfer, A., Knaus, H. G., Striessnig, J., Kempner, E., and Glossmann, H. (1996) Proc. Natl. Acad. Sci. U. S. A. 938072 -8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kekuda, R., Prasad, P. D., Fei, Y. J., Leibach, F. H., and Ganapathy, V. (1996) Biochem. Biophys. Res. Commun. 229553 -558 [DOI] [PubMed] [Google Scholar]
- 39.Prasad, P. D., Li, H. W., Fei, Y. J., Ganapathy, M. E., Fujita, T., Plumley, L. H., Yang-Feng, T. L., Leibach, F. H., and Ganapathy, V. (1998) J. Neurochem. 70 443-451 [DOI] [PubMed] [Google Scholar]
- 40.Mei, J., and Pasternak, G. W. (2001) Biochem. Pharmacol. 62349 -355 [DOI] [PubMed] [Google Scholar]
- 41.Seth, P., Fei, Y. J., Li, H. W., Huang, W., Leibach, F. H., and Ganapathy, V. (1998) J. Neurochem. 70 922-931 [DOI] [PubMed] [Google Scholar]
- 42.Pan, Y. X., Mei, J., Xu, J., Wan, B. L., Zuckerman, A., and Pasternak, G. W. (1998) J. Neurochem. 702279 -2285 [DOI] [PubMed] [Google Scholar]
- 43.Moebius, F. F., Reiter, R. J., Hanner, M., and Glossmann, H. (1997) Br. J. Pharmacol. 121 1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aydar, E., Palmer, C. P., Klyachko, V. A., and Jackson, M. B. (2002) Neuron 34 399-410 [DOI] [PubMed] [Google Scholar]
- 45.Ganapathy, M. E., Prasad, P. D., Huang, W., Seth, P., Leibach, F. H., and Ganapathy, V. (1999) J. Pharmacol. Exp. Ther. 289251 -260 [PubMed] [Google Scholar]
- 46.Seth, P., Ganapathy, M. E., Conway, S. J., Bridges, C. D., Smith, S. B., Casellas, P., and Ganapathy, V. (2001) Biochim. Biophys. Acta 154059 -67 [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto, H., Miura, R., Yamamoto, T., Shinohara, K., Watanabe, M., Okuyama, S., Nakazato, A., and Nukada, T. (1999) FEBS Lett. 44519 -22 [DOI] [PubMed] [Google Scholar]
- 48.Ramachandran, S., Lu, H., Prabhu, U., and Ruoho, A. E. (2007) Protein Expression Purif. 51 283-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torrence-Campbell, C., and Bowen, W. D. (1996) Eur. J. Pharmacol. 304201 -210 [DOI] [PubMed] [Google Scholar]
- 50.Cheng, Y., and Prusoff, W. H. (1973) Biochem. Pharmacol. 223099 -3108 [DOI] [PubMed] [Google Scholar]
- 51.Guo, L. W., Muradov, H., Hajipour, A. R., Sievert, M. K., Artemyev, N. O., and Ruoho, A. E. (2006) J. Biol. Chem. 28115412 -15422 [DOI] [PubMed] [Google Scholar]
- 52.Guo, L. W., Grant, J. E., Hajipour, A. R., Muradov, H., Arbabian, M., Artemyev, N. O., and Ruoho, A. E. (2005) J. Biol. Chem. 28012585 -12592 [DOI] [PubMed] [Google Scholar]
- 53.Liu, Y., Arshavsky, V. Y., and Ruoho, A. E. (1999) Biochem. J. 337281 -288 [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, Y., Arshavsky, V. Y., and Ruoho, A. E. (1996) J. Biol. Chem. 27126900 -26907 [DOI] [PubMed] [Google Scholar]
- 55.Esteban, B., Carrascal, M., Abian, J., and Lamparter, T. (2005) Biochemistry 44 450-461 [DOI] [PubMed] [Google Scholar]
- 56.de Laureto, P. P., Tosatto, L., Frare, E., Marin, O., Uversky, V. N., and Fontana, A. (2006) Biochemistry 4511523 -11531 [DOI] [PubMed] [Google Scholar]
- 57.Dokudovskaya, S., Williams, R., Devos, D., Sali, A., Chait, B. T., and Rout, M. P. (2006) Structure 14 653-660 [DOI] [PubMed] [Google Scholar]
- 58.Guo, L. W., Assadi-Porter, F. M., Grant, J. E., Wu, H., Markley, J. L., and Ruoho, A. E. (2007) Protein Expression Purif. 51187 -197 [DOI] [PubMed] [Google Scholar]