Abstract
Transforming growth factor-β (TGF-β) is an important regulator of physiological connective tissue biosynthesis and plays a central role in pathological tissue fibrosis. Previous studies have established that a biologically active lipid mediator, sphingosine 1-phosphate (S1P), mimics some of the profibrotic functions of TGF-β through cross-activation of Smad signaling. Here we report that another product of sphingosine kinase, dihydrosphingosine 1-phosphate (dhS1P), has an opposite role in the regulation of TGF-β signaling. In contrast to S1P, dhS1P inhibits TGF-β-induced Smad2/3 phosphorylation and up-regulation of collagen synthesis. The effects of dhS1P require a lipid phosphatase, PTEN, a key modulator of cell growth and survival. dhS1P stimulates phosphorylation of the C-terminal domain of PTEN and its subsequent translocation into the nucleus. We demonstrate a novel function of nuclear PTEN as a co-factor of the Smad2/3 phosphatase, PPM1A. Complex formation of PTEN with PPM1A does not require the lipid phosphatase activity but depends on phosphorylation of the serine/threonine residues located in the C-terminal domain of PTEN. Upon complex formation with PTEN, PPM1A is protected from degradation induced by the TGF-β signaling. Consequently, overexpression of PTEN abrogates TGF-β-induced Smad2/3 phosphorylation. This study establishes a novel role for nuclear PTEN in the stabilization of PPM1A. PTEN-mediated cross-talk between the sphingolipid and TGF-β signaling pathways may play an important role in physiological and pathological TGF-β signaling.
TGF-β2 is a multifunctional polypeptide growth factor that regulates cell proliferation, functional differentiation, extracellular matrix (ECM) production, cell motility, and apoptosis (1). TGF-β signaling is initiated by ligand binding to a heteromeric complex of transmembrane serine/threonine kinases (type I and type II) and subsequent activation of transcriptional co-regulators, Smad2 and Smad3 (1). Deregulated TGF-β signaling has been implicated in various pathological conditions, including fibrosis and cancer. Depending on the cancer stage, the TGF-β signaling pathway may either suppress or promote tumorigenesis (2). Recent studies suggest that TGF-β may also influence tumor growth through modulation of the tumor microenvironment (3). TGF-β is a potent inducer of the ECM and is required under physiologic conditions, such as wound repair, to induce fibroblasts to produce and contract ECM (4). On the other hand, deregulated TGF-β signaling has long been postulated to underlie pathologic fibrosis, but the specific mechanisms involved in this process have not been fully delineated (5). Recent in vivo and in vitro studies also suggest a novel role for the lipid and protein phosphatase, PTEN, as an inhibitor of fibrosis. Reduced levels of PTEN were found in myofibroblasts in human lung specimens of patients with idiopatic pulmonary fibrosis (6). Additional studies using animal models of fibrosis further supported the antifibrotic role of PTEN (6–8). The specific role of PTEN in regulation of the fibrotic process is not well understood, but studies with cultured cells have indicated that it may function as an antagonist of TGF-β-induced myofibroblast differentiation. Myofibroblasts are the primary effector cells in fibrosis and also play an important role in generating activated tumor stroma (9).
Sphingolipids are ubiquitous structural components of cell membranes, but in recent years their metabolites have been recognized as important biological mediators of various processes (10). Altered sphingolipid signaling has been implicated in the etiology of many disorders, including inflammation, allergies, autoimmune diseases, artheriosclerosis, and cancer (11, 12). Sphingosine kinase (SK) is an evolutionary conserved lipid kinase with two mammalian isoforms, which catalyzes the phosphorylation of sphingosine and dihydrosphingosine to form sphingosine 1-phosphate (S1P) and dhS1P, respectively (10). S1P, which has been widely studied for the past several years, has emerged as an important mediator of a variety of biological processes, including vasculogenesis, cell growth and survival, inflammation, and others (13, 14). The biological actions of S1P are primarily mediated by its binding to the family of specific G-protein-coupled receptors termed Edg or S1P receptors (S1P1–5 receptors). SK is also frequently overexpressed in solid tumors, suggesting an important role for this enzyme during tumorigenesis (15, 16). The oncogenic properties of SK1 have been directly demonstrated in several experimental models of tumorigenesis (17). Furthermore, blockade of S1P via a specific antibody reduced tumor progression in murine models through antitumorigenic and antiangiogenic effects (18). Recent evidence suggests that S1P may also play a role in fibrosis. In mesangial cells, S1P was shown to mimic the effects of TGF-β through cross-activation of Smad signaling. These effects of S1P were mediated through the S1P3 receptor and were dependent on the presence of TGF-βRII (19). In contrast to S1P, dhS1P has not been widely studied, and very little is currently known about its potential biological roles. Recent work from our laboratory suggests that dhS1P is biologically active and may have important biological functions distinct from those of S1P. dhS1P was shown to induce MMP1 (matrix metalloproteinase 1) via activation of ERK/Ets1 signaling in human dermal fibroblasts and to mediate tumor necrosis factor-α induction of MMP1 (20). This function has raised the interesting possibility that dhS1P signaling may functionally interact with the major profibrotic signaling pathway induced by TGF-β.
Since recent studies suggested an involvement of sphingolipid signaling in ECM regulation and a possible cross-talk between the sphingolipid and the TGF-β pathways, this study was undertaken to gain additional insights into this process with the goal to determine the molecular mechanism involved in these interactions. Our findings have established a molecular basis for the functional interaction between SK and the tumor suppressor PTEN, two key regulatory signaling molecules involved in many cellular processes during development and in various pathological conditions. We show that the sphingolipid-PTEN axis regulates TGF-β/Smad signaling in human primary fibroblasts, suggesting that the relative amount of dhS1P versus S1P may be an important determinant of TGF-β signaling during physiological and pathological processes.
EXPERIMENTAL PROCEDURES
Materials—The following antibodies were used: anti-phospho-Smad2, anti-phospho-Smad3, anti-Smad2/3, polyclonal anti-PTEN, anti-phospho-PTEN, anti-phospho-Akt, and anti-Akt from Cell Signaling Technology (Beverly, MA); anti-collagen 1A1 from Southern Biotech (Birmingham, AL); anti-lamin A/C, anti-Smad2/3 (N-19), monoclonal anti-PTEN, anti-TGF-βRI, anti-phospho-Ser from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); and anti-PPM1A from Abcam Inc. (Cambridge, MA). Anti-sphingosine kinase antibody was previously described (20). dhS1P and S1P were from Avanti (Alabaster, AL); pertussis toxin (Ptx) and MG123 were from Calbiochem. PTEN siRNA was purchased from Cell Signaling Technology (Beverly, MA).
Plasmids—PTEN WT and G129E in pcDNA 3.1 were kindly provided by Dr. Runzhao Li (Hollings Cancer Center, Medical University of South Carolina). The coding sequence of the PTEN gene was cloned into pCTAP vector (Stratagene), which has two different tags: a streptavidin-binding peptide and a calmodulin binding peptide. PTEN mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Cell Cultures and Transfection—Human dermal fibroblast culture was established from the foreskins of healthy newborns obtained from the delivery suites at the Medical University of South Carolina and in compliance with the Institutional Review Board. Cells between passages 3 and 6 were used for experiments. For experiments, fibroblasts were incubated in serum-free medium for 24 h. Human embryonic kidney 293 (HEK 293) cells were purchased from ATCC and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Transient transfections were carried out using FuGene 6 (Roche Applied Science). Adenoviral infections were conducted as previously described (20).
Immunoblotting—Whole cell protein extracts were prepared according to the manufacturer's recommendations (M-Per; Pierce). Nuclear and cytoplasmic proteins were isolated with a buffer extraction system and centrifugation according to the manufacturer's recommendations (NE-Per; Pierce). Immunoblotting was performed using standard protocols.
Immunoprecipitation—To precipitate nontagged proteins, whole cell extracts (500 μg) were preadsorbed with protein G-Sepharose beads (GE Healthcare) and incubated with 2 μg of appropriate antibodies and then with protein G-Sepharose beads. Streptavidin-coupled agarose beads (Sigma) were used instead of protein G-Sepharose beads for immunoprecipitation of ectopically expressed tagged PTEN. The precipitated proteins were subjected to immunoblotting.
Immunofluorescence—For indirect immunofluorescence, cells were grown on coverslips and fixed in PBS plus 3.7% formaldehyde for 10 min and washed twice with PBS. Cells were then permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature and stained with respective antibodies in PBS plus 1% bovine serum albumin. Antibodies used were as follows: rabbit anti-PTEN or phospho-Smad3 (1:100); mouse anti-Smad (1:100). Secondary antibodies (Invitrogen) were highly cross-absorbed goat antibodies against rabbit IgG or mouse IgG, conjugated to either AlexaFluor 488 or 546 and used at 1:150 dilutions in PBS plus 1% bovine serum albumin. Samples were viewed with an Olympus IX70 microscope with confocal capability (Fluoview 300).
Luciferase Measurements—Cells were seeded at 2 × 105 cells/well in a 6-well plate and grown for 24 h. Transfections were performed using FuGene 6 (Roche Applied Science) according to the manufacturer's instructions with 1 μg of 3TP-Lux or SBE together with a total of 1 μg of the empty vector alone and/or vectors expressing TGF-βRI, Smad3, or PTEN wild type (PTENWT) or mutants as indicated, in the presence and absence of 0.5 μm dhS1P. After a 20-h incubation, luciferase activity was determined using the luciferase assay system (Promega). Total light emission was measured using a luminometer.
RESULTS
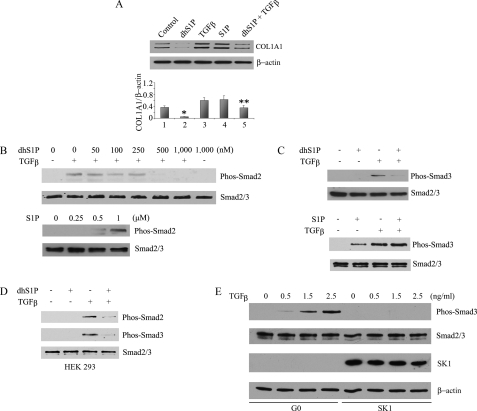
dhS1P and S1P Have Antagonistic Effects on TGF-β/Smad Signaling—We first examined the effects of S1P and dhS1P on the TGF-β induction of collagen type I, the most abundant ECM component. Basal collagen protein levels were significantly reduced in cells treated with dhS1P (Fig. 1A, lane 2) and increased in cells treated with S1P (lane 4) to a level indistinguishable from TGF-β stimulation (lane 3). Furthermore, dhS1P inhibited TGF-β stimulation of collagen production (lane 5). Interestingly, dhS1P treatment inhibited TGF-β induced phosphorylation of Smad2 (Fig. 1B, top) and Smad3 (Fig. 1C, top). Consistent with previous studies, S1P alone stimulated phosphorylation of Smad2 and -3 (Fig. 1, B and C, bottom). The inhibitory effects of dhS1P were not restricted to fibroblasts; dhS1P markedly reduced TGF-β-induced phosphorylation of Smad2 and -3 in HEK 293 cells (Fig. 1D) and mink lung epithelial cells (data not shown). In contrast, TGF-β-induced phosphorylation of Akt was not affected by dhS1P treatment (Fig. S1). Therefore, S1P and dhS1P exert opposite effects on TGF-β signaling at the level upstream or parallel to Smad2/3 activation. Furthermore, the phosphorylation status of TGF-βRI was not affected by dhS1P treatment (Fig. S2). Together, these data suggest that the inhibitory action of dhS1P on TGF-β/Smad signaling occurs downstream from the TGF-β receptors.
FIGURE 1.
DhS1P and S1P have antagonistic effects on collagen production and Smad2/3 signaling. A, the collagen protein level was analyzed by Western blot in fibroblasts incubated with 0.5 μm dhS1P or 1 μm S1P and/or 2.5 ng/ml TGF-β for 24 h. The bar graph shows means ± S.E. of at least three independent experiments. *, dhS1P-treated versus control (p < 0.01); **, TGF-β + dhS1P versus TGF-β alone-treated cells (p < 0.01). B, fibroblasts were incubated with 2.5 ng/ml TGF-β for 30 min with the addition of the indicated concentrations of dhS1P (top) or incubated with S1P (bottom). Phosphorylated Smad2 was detected by Western blot. C, fibroblasts were treated with 2.5 ng/ml TGF-β and either 0.5 μm dhS1P or 1 μm S1P for 30 min. Phosphorylated Smad3 was detected by Western blot. D, HEK 293 cells were treated with 2.5 ng/ml TGF-β and 0.5 μm dhS1P for 30 min. Phosphorylated Smad2 and Smad3 were detected by Western blot. E, overexpression of SK1 prevents TGF-β-induced phosphorylation of Smad3. Fibroblasts were transduced with 50 multiplicity of infection of SK1 or control adenovirus (G0) for 24 h. Next, increasing concentration (0.5–2.5 ng/ml) of TGF-β was added for 30 min. Phosphorylated Smad3, total Smad2/3, and SK1 were detected by Western blot. β-Actin was used as loading control.
Previous studies have shown that overexpression of SK1 in fibroblasts and other cell types results in a predominant increase of intracellular dhS1P compared with S1P (20, 21) (Fig. S3). We next sought to determine whether endogenously produced dhS1P would exert inhibitory effects on TGF-β/Smad signaling. As shown in Fig. 1E, overexpression of SK1 completely abrogated TGF-β-induced phosphorylation of Smad3. It has to be noted that overexpression of SK in foreskin fibroblasts does not lead to appreciable increases of extracellular dhS1P or S1P. Therefore, at present, we cannot exclude the possibility that the dephosphorylation of Smad3 by exogenous dhS1P and endogenous products of SK1 involves different mechanisms.
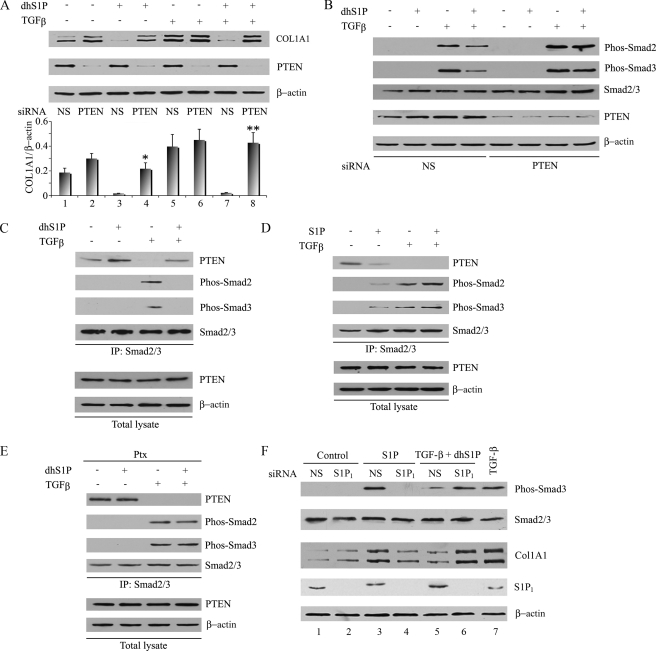
PTEN Mediates Inhibitory Effects of dhS1P—Recent studies have implicated PTEN in mediating an S1P-induced antimigratory response in embryonic fibroblasts through the S1P2 receptor (22). Since PTEN has also been shown to negatively regulate TGF-β induction of collagen (6), we explored the intriguing possibility that PTEN may be a link between dhS1P/S1P and TGF-β signaling pathways. Blockade of PTEN using siRNA resulted in a modest induction of basal and TGF-β-induced collagen levels (Fig. 2A, lanes 2 and 6). Strikingly, the inhibitory effects of dhS1P on basal (compare lanes 3 and 4) and TGF-β-induced (compare lanes 7 and 8) collagen production were completely reversed in the absence of PTEN. Consistent with the effect on collagen expression, blockade of PTEN prevented the inhibitory effects of dhS1P on Smad2/3 phosphorylation (Fig. 2B). Therefore, the antagonistic effect of dhS1P on TGF-β signaling requires PTEN function. We next examined whether PTEN can directly modulate the phosphorylation status of Smad2/3.
FIGURE 2.
PTEN mediates inhibitory effects of dhS1P. A and B, depletion of endogenous PTEN prevents collagen down-regulation and Smad2/3 dephosphorylation by dhS1P. Fibroblasts were transfected with 30 nm PTEN siRNA or nonsilencing (NS) siRNA for 24 h and then serum-starved overnight. Cells were treated with 2.5 ng/ml TGF-β with the addition of 0.5 μm dhS1P for 30 min. Collagen 1A1 (A) and phosphorylated Smad2 and -3 (B) were detected by Western blot. The bar graph shows means ± S.E. of at least three independent experiments. *, dhS1P plus PTEN siRNA-treated versus dhS1P plus NS siRNA-treated cells (p < 0.01). **, TGF-β plus dhS1P and PTEN siRNA-treated versus TGF-β plus dhS1P and NS siRNA-treated cells (p < 0.001). C and D, dhS1P enhances whereas S1P diminishes binding of endogenous Smad2/3 and PTEN. Cells were treated with 2.5 ng/ml TGF-β with the addition of 0.5 μm dhS1P (C) or S1P (D) for 30 min. Smad2/3 was immunoprecipitated from cell extracts with anti-Smad2/3 antibody, followed by immunoblotting with PTEN, phospho-Smad2, and phospho-Smad3 antibodies. E, Ptx inhibits formation of Smad-PTEN complexes. Cells were pretreated with 100 ng/ml Pxt for 24 h and then treated with 0.5 μm dhS1P plus 2.5 ng/ml TGF-β for 30 min. Smad2/3 was immunoprecipitated from cell extracts, followed by immunoblotting with the indicated antibodies. F, depletion of endogenous S1P1 abrogates effects of S1P and dhS1P. Cells were transfected with 30 nm S1P1 siRNA or nonsilencing siRNA for 24 h and then serum-starved overnight. Cells were treated with 1 μm S1P or 2.5 ng/ml TGF-β plus 0.5 μm dhS1P for 30 min to measure phospho-Smad3 or 24 h to measure collagen. Phosphorylated Smad3 and COL1A1 protein were detected by immunoblotting. Cells treated with TGF-β only served as control (lane 7).
To explore the potential mechanism of PTEN action on Smad2/3, we first used co-immunoprecipitation to examine whether PTEN interacts with Smad2/3 under our experimental conditions. Formation of PTEN-Smad complexes was observed in unstimulated dermal fibroblasts (Fig. 2C, lane 1), and the amount of Smad2/3 associated with PTEN was further increased in cells treated for 30 min with dhS1P (Fig. 2C, lane 2). TGF-β treatment induced phosphorylation of Smad2 and -3 and concomitantly reduced the PTEN-Smad2/3 complex formation (Fig. 2C, lane 3). However, costimulation with TGF-β and dhS1P reversed the TGF-β effect (lane 4). Conversely, stimulation with S1P decreased association of PTEN with Smad2/3, mimicking the TGF-β effect (Fig. 2D). Together, these data indicate that PTEN-Smad2/3 complex formation is inversely correlated with TGF-β stimulation and Smad2/3 phosphorylation and that dhS1P and S1P regulate binding of PTEN to Smad2/3 in a manner consistent with their respective inhibitory and stimulatory effects on TGF-β signaling.
To examine whether the effects of dhS1P on TGF-β signaling involve G-protein-coupled receptor-mediated mechanisms, the experiment was performed in cells treated with PTX, an inhibitor of the Gαi/o subunit. As shown in Fig. 2E, PTX treatment did not affect formation of the Smad-PTEN complexes in unstimulated cells but prevented further increase in complex formation in response to dhS1P. TGF-β-induced dissociation of Smad-PTEN complexes was not affected by PTX; however, the action of dhS1P in restoring Smad-PTEN complex formation was abolished under this condition, thus suggesting the involvement of G-protein-coupled receptors in the actions of dhS1P. Human foreskin fibroblasts express S1P1–3 high affinity receptors for S1P. Previous studies have shown that dhS1P efficiently competes with S1P for binding to the S1P1 receptor (23). To examine which of the S1P receptors mediates the effects of sphingolipids on TGF-β signaling, we used specific siRNA to individually reduce the expression of endogenous S1P receptors. As shown in Fig. 2F, depletion of the S1P1 receptor abolished S1P stimulation of phospho-Smad3 and abrogated up-regulation of collagen (Fig. 2F, compare lanes 3 and 4). Likewise, inhibitory effects of dhS1P on TGF-β signaling were reversed (lanes 5 and 6). Depletion of either the S1P2 or S1P3 receptor had no effect (Fig. S4). These results indicate that S1P and dhS1P compete for the same receptor, S1P1, for their effects on Smad signaling.
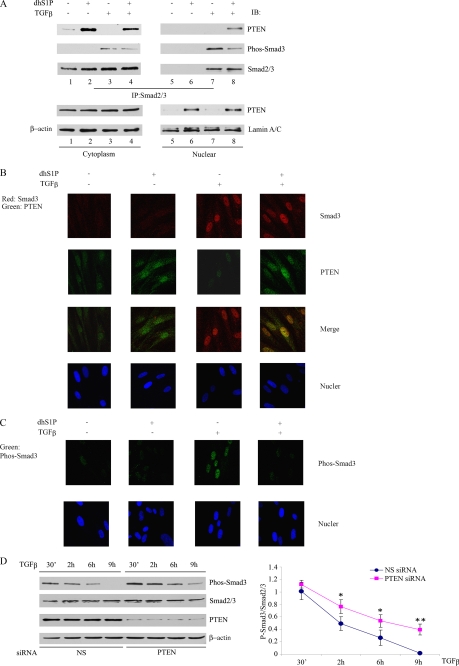
We next examined whether PTEN association with Smad2/3 can be correlated with transcription activity. Consistent with the known properties of Smad2/3, in unstimulated cells, Smad2/3 was unphosphorylated and located in the cytoplasm (Fig. 3A, IP panels, compare lanes 1 and 5). As expected, after TGF-β treatment, phosphorylated Smad2/3 was enriched in the nucleus, with a minor proportion of phosphorylated Smad remaining in the cytoplasm (Fig. 3A, IP panels, compare lanes 3 and 7). Similarly, PTEN was present primarily in the cytoplasm in unstimulated cells with just above background level in the nuclear fraction (Fig. 3A, bottom, compare lanes 1 and 5). PTEN-Smad complexes that were rapidly formed in response to dhS1P treatment were also detected in the cytoplasm (Fig. 3A, IP panels, lane 2). Interestingly, dhS1P treatment resulted in a nuclear translocation of a fraction of PTEN molecules, suggesting that dhS1P may be involved in regulating the subcellular distribution of PTEN (Fig. 3A, bottom, compare lanes 5 and 6). This fraction was not associated with Smad2/3 with or without dhS1P (Fig. 3A, IP panels, lanes 5 and 6), since Smad2/3 is not present in the nucleus without TGF-β stimulation. The simultaneous addition of dhS1P and TGF-β resulted in the appearance of PTEN-Smad complexes in the nuclear fraction (Fig. 3A, IP panels, lane 8). Interestingly, upon the appearance of the PTEN-Smad2/3 complex, the amounts of phopspho-Smad3 in the nuclear fraction were significantly reduced (Fig. 3A, IP panels, lanes 7 and 8), indicating that PTEN may promote dephosphorylation of Smad2/3 in the nucleus.
FIGURE 3.
PTEN is required for Smad3 dephosphorylation in the nucleus. A, cytoplasmic and nuclear fractions were isolated from fibroblasts treated for 30 min with 2.5 ng/ml TGF-β and 0.5 μm dhS1P alone or in combination. Binding of PTEN to phosphorylated and total Smad2/3 were examined by immunoprecipitation (IP)/Western blot (IB). Distribution of PTEN in cytoplasmic and nuclear fractions was examined by immunoblotting. Lamin A/C and β-actin were used as control for nuclear and cytoplasmic fractions, respectively. B, immunofluorescence imaging of Smad3 (red) and PTEN (green) in cells treated individually or in combination with 0.5 μm dhS1P and 2.5 ng/ml TGF-β for 30 min. Note the co-localization of Smad3 and PTEN in cells treated by a combination of TGF-β and dhS1P. C, subcellular distribution of phosphorylated Smad3 (green) under experimental conditions described in B. Note the significantly reduced presence of nuclear phospho-Smad3 in cells treated with TGF-β and dhS1P. D, depletion of endogenous PTEN prolongs TGF-β-induced phosphorylation of Smad3. Cells were transfected with 30 nm PTEN siRNA or nonsilencing siRNA for 24 h and then starved overnight and incubated with 2.5 ng/ml TGF-β for the indicated times. The lower panel presents average kinetics of phospho-Smad3 from three independent experiments. *, p < 0.05; **, p < 0.01.
To verify the results described above, we next used immunofluorescence to further analyze the subcellular localization of PTEN and Smad complexes. In unstimulated cells, PTEN was diffusely detected in the cytoplasm as well as in the nucleus at low levels (Fig. 3B, PTEN). dhS1P treatment increased the level of nuclear localization, consistent with the biochemical data (compare with Fig. 3A). TGF-β resulted in nuclear translocation of endogenous Smad3. The simultaneous addition of dhS1P and TGF-β led to co-localization of Smad3 and PTEN in the nucleus. As expected, phosphorylated Smad3 was present in the nucleus in fibroblasts treated with TGF-β alone, but there was a significant reduction of nuclear phospho-Smad3 in cells treated with a combination of TGF-β and dhS1P (Fig. 3C). These results suggest that dhS1P promotes PTEN nuclear localization, which in turn inactivates, via dephosphorylation, nuclear Smad3. This is consistent with recent studies demonstrating that Smad dephosphorylation occurs in the nucleus (24).
To examine the effects of PTEN on the kinetics of endogenous Smad phosphorylation, we utilized an siRNA approach to deplete endogenous PTEN. Reduced levels of PTEN resulted in a prolonged presence of phosphorylated Smad3 (Fig. 3D), consistent with the notion that PTEN enhances dephosphorylation of nuclear Smad3.
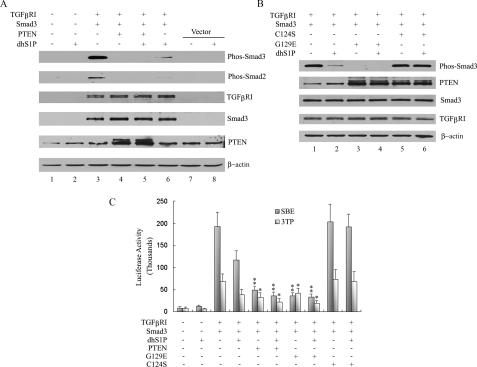
PTEN Mediates Dephosphorylation of Smad2/3 in a Lipid Phosphatase-independent Manner—PTEN has dual phosphatase activity: a well characterized lipid phosphatase and a less well defined protein phosphatase activity (25). To gain further insights into the mechanism governing PTEN-mediated Smad2/3 dephosphorylation, PTEN mutants deficient in lipid and protein phosphatase activity were used in biochemical and functional assays. Constitutively activated TGF-βRI (TGF-βRI 204D) was co-expressed together with Smad3 and PTEN in HEK 293 cells. Smad3 and endogenous Smad2 are phosphorylated in the presence of TGF-βRI 204D (Fig. 4A, lane 3). Co-transfection of PTEN resulted in a complete inhibition of Smad2/3 phosphorylation (lane 4). Consistent with previous results, dhS1P treatment also strongly inhibited phosphorylation of Smad2/3 in the absence of ectopic PTEN (lane 6).
FIGURE 4.
PTEN mediates dephosphorylation of Smad2/3 in a lipid phosphatase-independent manner. A, HEK 293 cells were co-transfected with the indicated combination of TGF-βRI 204D, Smad3, and PTENWT for 24 h and, where indicated, treated with 0. 5 μm dhS1P for 30 min. Expression levels of phospho-Smad3, phospho-Smad2, TGF-βRI, Smad3, and PTEN were assessed by immunoblotting. B, HEK 293 cells were cotransfected with the indicated combination of TGF-βRI 204D, Smad3, PTEN C124S, and PTEN G129E for 24 h and then treated with 0.5 μm dhS1P. Phospho-Smad3, PTEN, Smad3, and TGF-βRI were detected by immunoblotting. C, HEK 293 cells were transiently transfected with TGF-βRI 204D, Smad3, PTENWT, PTEN C124S, PTEN G129RE, and 3TP-Lux or SBE-Lux for 24 h and then incubated with 0.5 μm dhS1P for 30 min. Cells were harvested, and luciferase activity was measured. Activity is expressed as relative luminometer units (n ≥ 3). *, p < 0.05; **, p < 0.01.
To test involvement of PTEN phosphatase activity in the dephosphorylation of Smad3, cells were cotransfected with the PTEN G129E mutant that lacks lipid phosphatase activity or the PTEN C124S mutant that lacks total phosphatase (lipid and protein) activity (26). Similar to wild-type PTEN, co-transfection with the PTEN G129E mutant completely abrogated phosphorylation of Smad3 (Fig. 4B, lane 3). In contrast, the PTEN C124S mutant did not inhibit Smad3 phosphorylation (lane 5). Significantly, overexpression of the PTEN C124S mutant abolished the inhibitory effect of dhS1P on Smad3 phosphorylation (compare lanes 4 and 6), suggesting a dominant negative effect. We next investigated the functional effects of PTEN and its mutated forms on Smad3 signaling using two different Smad3-responsive promoters, 3TPLux and SBE. Cells were co-transfected with TGF-βRI 204D together with Smad3 and either PTEN or an empty vector. Potent activation of both promoters was observed in the presence of the empty vector, whereas either dhS1P treatment or co-expression of PTEN significantly inhibited promoter activity (Fig. 4C). Co-expression of the PTEN G129E mutant also exerted an inhibitory effect similar to wild-type PTEN, whereas the PTEN C124S mutant had no effect on TGF-β stimulation but blocked the dhS1P-mediated inhibitory effect, consistent with the effect on Smad2/3 phosphorylation (Fig. 4B).
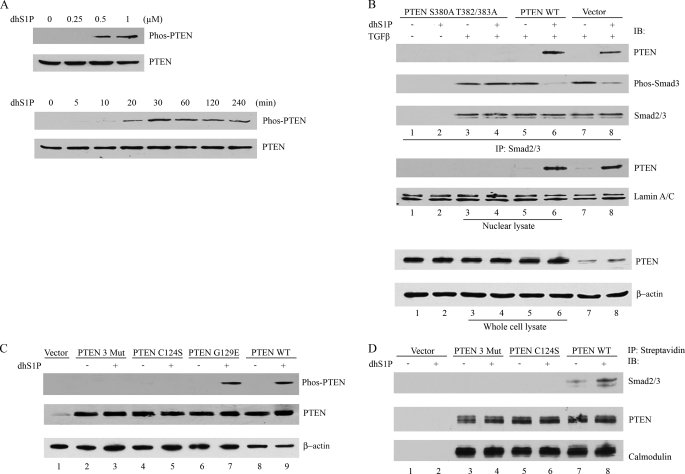
PTEN protein stability and selected biological function are regulated by the phosphorylation of three residues (Ser380, Thr382, and Thr383) located in its C-terminal domain (27). We next determined whether these phosphorylation sites are involved in mediating PTEN effects in response to dhS1P treatment. Stimulation of dermal fibroblasts with dhS1P induced phosphorylation of S380/T382/T383 in a dose- and time-dependent manner (Fig. 5A), whereas S1P did not have this effect (data not shown). To further examine the role of phosphorylation at these sites, we introduced three alanine substitutions into the above Ser/Thr sites. Mutated PTEN lost its ability to dephosphorylate Smad3 (Fig. 5B, compare lanes 4 and 6). C-terminal mutations also abolished the ability of PTEN to translocate to the nucleus in response to dhS1P (middle). We concluded from these data that phosphorylation of the C terminus is required for the dhS1P-mediated nuclear translocation of PTEN and subsequent dephosphorylation of Smad2/3. Therefore, we also examined the ability of dhS1P to induce phosphorylation of C124S and G129E PTEN mutants. Phosphorylation of the lipid phosphatase G129E mutant was similar to that of wild type, whereas the double phosphatase mutant, C124S, was not phosphorylated, suggesting that the conformation required for the dual phosphatase activity is necessary for the C-terminal phosphorylation of PTEN by dhS1P (Fig. 5C). Together, these data suggested that the inability of PTEN mutants to dephosphorylate Smad3 might be caused by their failure to translocate to the nucleus. In addition, complex formation between Smad and PTEN may also depend on the phosphorylation status of PTEN. To verify this possibility, we used co-immunoprecipitation. In contrast to wild type PTEN, the C124S mutant and the triple phosphorylation site mutant were not able to interact with Smad2/3 either at the basal level or in response to dhS1P (Fig. 5, B (top) and D). We therefore concluded that the C-terminal phosphorylation events are required for PTEN activity in response to dhS1P and that the mechanism of dhS1P action on Smad2/3 activation is through modulation of the phosphorylation state of PTEN.
FIGURE 5.
C-terminal phosphorylation of PTEN mediates dhS1P-induced PTEN nuclear translocation and dephosphorylation of Smad3. A, dhS1P stimulates C-terminal phosphorylation of PTEN in a dose-dependent (top) and time-dependent (bottom) manner. C-terminal phosphorylation of PTEN was detected by immunoblotting. B, HEK 293 cells were transfected with the phosphorylation-deficient PTEN mutant, PTENWT, or vector only for 24 h and then incubated with 0.5 μm dhS1P and 2.5 ng/ml TGF-β for 30 min. The nuclear fraction was immunoprecipitated (IP) by Smad2/3 and then probed for PTEN, phospho-Smad3, and Smad2/3 by Western blot (IB). PTEN was also tested in nuclear lysate and whole cell lysate. Lamin A/C and β-actin served as controls for nuclear and cytoplasmic fractions, respectively. C, HEK 293 cells were transfected with the phosphorylation-deficient PTEN mutant (Mut3), PTEN C124S, PTEN G129E, and PTENWT and incubated with 0.5 μm dhS1P for 30 min. C-terminal phosphorylation of PTEN was detected by Western blotting. D, C-terminal phosphorylation (Mut3) and C124S PTEN mutants do not associate with Smad2/3. The PTEN proteins contained calmodulin and streptavidin tags from the pCTAP vector (Stratagene). Streptavidin-coupled agarose beads were used to pull down wild-type and mutated PTEN. Smad2/3 and PTEN were detected by immunoblotting. Immunoblotting with calmodulin served as control for equal loading.
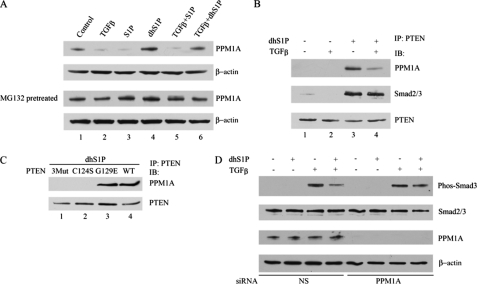
PTEN Facilitates Interaction of Smad2/3 with PPM1A—Since complex formation of PTEN with Smad2/3 leads to Smad2/3 dephosphorylation, it raises the possibility that PTEN is a novel Smad2/3 phosphatase. However, this possibility seems unlikely, since PTEN mutants that lost the ability to translocate to the nucleus and interact with the phospho-Smad2/3 have dominant negative effects (Fig. 4). This observation suggests that PTEN is an essential co-factor in a dephosphorylation complex. Therefore, we explored the possibility that the action of PTEN is mediated through the recently described Smad2/3 phosphatase, PPM1A. Expression of PPM1A protein was rapidly down-regulated in response to TGF-β or S1P treatment, whereas dhS1P treatment increased basal expression of PPM1A and prevented its TGF-β-induced down-regulation (Fig. 6A). Rapid loss of PPM1A in response to TGF-β or S1P suggested proteosome-mediated degradation. In support of this possibility, pretreatment with proteosomal inhibitor MG132 reversed the effects of TGF-β or S1P (Fig. 6A). We next examined the potential interaction between PTEN and PPM1A. Complexes of PTEN and PPM1A were not detectable in untreated cells but were markedly increased in cells treated with dhS1P and to a lesser extent in cells treated with a combination of dhS1P and TGF-β (Fig. 6B). Importantly, the dhS1P-induced PTEN-PPM1A complex also contains Smad2/3 (Fig. 6B), consistent with the dephosphorylation function of the PTEN-PPM1A complex. In agreement with earlier data, the C124S mutant and the triple phosphorylation site mutant were not able to interact with PPM1A either at the basal level or in response to dhS1P (Fig. 6C). To further verify the dependence of the effects of PTEN on PPM1A, we depleted endogenous PPM1A using siRNA. As shown in Fig. 6D, in the absence of PPM1A, dhS1P did not affect TGF-β-induced Smad3 phosphorylation, suggesting that PPM1A is required for the inhibitory effects of PTEN on TGF-β/Smad signaling. These results suggest a novel function of PTEN as a co-factor of the protein phosphatase PPM1A.
FIGURE 6.
Nuclear PTEN interacts with PPM1A and prevents its degradation by TGF-β. A, TGF-β reduces whereas dhS1P enhances PPM1A protein levels. Fibroblasts were incubated for 30 min with 2.5 ng/ml TGF-β and 0.5 μm dhS1P alone or in combination. PPM1A was detected by Western blot. The lower panel shows PPM1A levels under the same conditions in cells pretreated with 20 μm MG132 for 1 h. B, fibroblasts were treated for 30 min with 2.5 ng/ml TGF-β and 0.5 μm dhS1P alone or in combination. Binding of PTEN to PPM1A was examined by immunoprecipitation (IP)/Western blot (IB). C, HEK 293 cells were transfected with the phosphorylation-deficient PTEN mutant (Mut3), PTEN C124S, PTEN G129E, and PTENWT and incubated with 0.5 μm dhS1P for 30 min. Binding of PTEN to PPM1A was examined by immunoprecipitation/Western blot. D, depletion of endogenous PPM1A prevents inhibitory effects of PTEN on TGF-β-induced phosphorylation of Smad3. Cells were transfected with 30 nm PPM1A siRNA or nonsilencing siRNA for 24 h and then starved overnight and incubated for 30 min with 2.5 ng/ml TGF-β and 0.5 μm dhS1P alone or in combination. Smad3 and PPM1A were detected by Western blotting.
DISCUSSION
The present study has delineated the molecular basis for the cross-talk between the sphingolipid and TGF-β signaling pathways. Treatment of cells with S1P or its structural analog, FTY720, has previously been shown to reproduce many of the profibrotic effects of TGF-β, including Smad phosphorylation and myofibroblast differentiation (19, 28, 29). Herein we show for the first time that a closely related product of SK, dhS1P, is an antagonist of TGF-β/Smad signaling. We demonstrate that dhS1P induces dephosphorylation of Smad2/3 through a PTEN-mediated stabilization of the Smad2/3 phosphatase, PPM1A. Dephosphorylation of Smad2/3 is a key mechanism of inactivating TGF-β signaling, and the role of PTEN as a facilitator of this process is consistent with its recently described role as antagonist of the profibrotic and antitumorigenic effects of TGF-β.
The tumor suppressor PTEN is one of the most common targets for mutation in human cancers and is also deregulated in several other human diseases (30). The role of PTEN in tumorigenesis as a negative regulator of the PI3-K/Akt pathway is well documented. PTEN encodes a lipid phosphatase that dephosphorylates phosphatidylinositol 3,4,5-trisphosphate, thus inactivating downstream signaling. Phosphatidylinositol 3,4,5-trisphosphate is generated in response to a variety of stimuli and regulates a wide range of cellular processes, including proliferation, survival, growth, and motility (30). More recent evidence indicates that PTEN, through protein-protein interaction involving its C-terminal domain, has cellular functions that do not depend on its lipid phosphatase activity. The C-terminal domain was shown to inhibit cellular transformation by binding to an oncogenic, nucleolar protein MSP58 (58-kDa microspherule protein), and that interaction required intact Thr366 (31). A recently described ability of PTEN to maintain chromosomal integrity also required binding of the C-terminal domain of PTEN to a centromeric protein, CENP-C (32). Recent studies have suggested that PTEN may also play an important role as a negative regulator of the TGF-β signaling pathway through direct interaction with Smad2/3. Specifically, PTEN was shown to inhibit TGF-β-induced motility and invasion of epithelial cells, including the keratinocyte cell line HaCaT and U87MG glioma cells (33). Another group has reported a role for PTEN as a negative regulator of fibrosis (6). The authors demonstrated that pharmacologic inhibition of PTEN resulted in worsened manifestation in an experimental model of pulmonary fibrosis. Furthermore, fibroblasts derived from pten-/- mice showed elevated collagen and α-SMA levels, suggesting that PTEN negatively regulates myofibroblast differentiation. In addition, pten-/- cells lost responsiveness to TGF-β stimulation, which was attributed to televated Smad7 expression in these cells (6).
In the present study, we utilized primary human fibroblasts to delineate the molecular mechanism that underlies the inhibitory effects of PTEN on TGF-β signaling. Consistent with the previous report, we demonstrate that PTEN forms cytoplasmic complexes with Smad2/3. In contrast to HaCaT cells, in which TGF-β stimulation promoted formation of such complexes (33), in fibroblasts, TGF-β has an opposite effect by dissociating existing complexes. PTEN did not interfere with nuclear translocation of Smad2/3 in either fibroblasts or HaCaT cells, indicating that the inhibition of TGF-β signaling by PTEN does not occur through sequestration of Smad2/3 in the cytoplasm. We show that the inhibitory function of PTEN depends on its nuclear localization and interaction with a phosphatase, PPM1A/PP2Cα. Smad2/3 was recently shown to be a target of this phosphatase, which resides in the nucleus and promotes dephosphorylation and nuclear export of Smad2/3 (24). Nuclear PTEN forms complexes with PPM1A and Smad2/3. Importantly, in the presence of PTEN PPM1A is protected from degradation in response to TGF-β signaling, resulting in rapid dephosphorylation of Smad2/3. Biological effects of TGF-β are also mediated through non-Smad dependent signaling pathways. Since known PPM1A targets include other signaling molecules, such as JNK and p38 MAPK (34), which also contribute to the various biological effects of TGF-β, it is possible that PTEN may suppress TGF-β signaling at multiple levels. Further studies are needed to explore these possibilities.
Antagonistic effects of PTEN on TGF-β signaling described in this study depended on the translocation of PTEN into the nucleus. PTEN lacks a typical nuclear localization signal, and several mechanisms regulating nuclear import have been described to date in various experimental models, including Ca2+-dependent MVP (major vault protein)-mediated import (35), Ran-mediated import that required the N-terminal nuclear localization domain of PTEN (36), or a passive transfer by diffusion (37). A recent study has also suggested that monoubiquitination of the Lys289 residue enhances nuclear localization of PTEN (38). However, the mechanism governing nuclear localization of PTEN needs further clarification, and surprisingly little is known about external signals modulating this process. We demonstrate that in fibroblasts nuclear translocation of PTEN requires phosphorylation of the serine/threonine residues in the tail region, which could be induced in these cells by treatment with dhS1P. Previous study has implicated CK2 (casein kinase 2) in constitutive phosphorylation of PTEN in MCF-7 cells (39), but the nature of the kinase mediating dhS1P effects remains to be elucidated. Our observation is consistent with the behavior of endometrial stromal cells, in which increased nuclear localization of PTEN occurred in response to estradiol-mediated tail phosphorylation (40). However, in glioblastoma U87MG cells, mutation of the same serine/threonine residues led to an increased nuclear localization (36). These contradictory observations may reflect different mechanisms in cells of different origin or a difference between the primary and cancer cells. It has to be noted that, unlike the majority of cell lines studied to date in which PTEN is constitutively phosphorylated (30), quiescent fibroblasts contain mainly nonphosphorylated forms of PTEN. It has been reported that dephosphorylation of the tail residues results in decreased protein stability (27); however, in that form, PTEN is presumably more biologically active due to increased colocalization with the cell membranes (30). Since most of the studies of PTEN were performed with cancer cell lines, it needs to be verified that similar mechanisms are also applicable to fibroblasts and other primary cells.
In conclusion, this study describes a novel function of dhS1P as an antagonist of TGF-β signaling. Both S1P and dhS1P are generated in response to activation of SK (20); however, the relative ratio in which they are produced may depend on other factors. SK and its product S1P are induced by TGF-β and mediate some of the profibrogenic responses in fibroblasts (41). On the other hand, overexpression of SK alone preferentially leads to generation of dhS1P (20, 21), resulting in up-regulation of MMP1 (20) and, as shown in this study, repression of Smad2/3 signaling. Unexpectedly, the antagonistic effects of S1P and dhS1P on Smad2/3 signaling were mediated through the same receptor, S1P1. Furthermore, in contrast to stimulation of Erk1/2 phosphorylation, which occurs with similar kinetics in response to S1P and dhS1P (20), only dhS1P triggered phosphorylation of PTEN, thus suggesting that additional determinants of the receptor status could contribute to the activation of distinct signaling pathways by these agonists (42). The mechanism responsible for the distinct effects of S1P and dhS1P may involve novel pathways downstream of the S1P1 receptor. Although future studies will focus on further elucidation of the mechanisms underlying this phenomenon, the current study provides strong support for the biological role of dhS1P. More importantly, the antagonistic role of dhS1P in TGF-β signaling opens new possibilities to control pathological effects of TGF-β via modulation of the G protein-coupled receptor signaling and may lead to development of new therapeutic approaches for treatment of fibrotic diseases.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants P60 AR049459 and R01 AR44883 (to M. T.), R01 CA109860 (to T. H.), and P01 CA78582. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: TGF-β, transforming growth factor-β; TGF-βRI and TGF-βRII, TGF-β recptor I and II, respectively; S1P, sphingosine 1-phosphate; dhS1P, dihydrosphingosine 1-phosphate; ECM, extracellular matrix; SK, sphingosine kinase; Ptx, pertussis toxin; PTEN, phosphatase and tensin homologue deleted on chromosome 10; HEK 293, human embryonic kidney cells; siRNA, small interfering RNA; WT, wild type.
References
- 1.Massague, J. (1998) Annu. Rev. Biochem. 67 753-791 [DOI] [PubMed] [Google Scholar]
- 2.Bierie, B., and Moses, H. L. (2006) Nat. Rev. Cancer 6 506-520 [DOI] [PubMed] [Google Scholar]
- 3.Bhowmick, N. A., Chytil, A., Plieth, D., Gorska, A. E., Dumont, N., Shappell, S., Washington, M. K., Neilson, E. G., and Moses, H. L. (2004) Science 303 848-851 [DOI] [PubMed] [Google Scholar]
- 4.Faler, B. J., Macsata, R. A., Plummer, D., Mishra, L., and Sidawy, A. N. (2006) Perspect. Vasc. Surg. Endovasc. Ther. 18 55-62 [DOI] [PubMed] [Google Scholar]
- 5.Verrecchia, F., and Mauviel, A. (2007) World J. Gastroenterol. 13 3056-3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White, E. S., Atrasz, R. G., Hu, B., Phan, S. H., Stambolic, V., Mak, T. W., Hogaboam, C. M., Flaherty, K. R., Martinez, F. J., Kontos, C. D., and Toews, G. B. (2006) Am. J. Respir. Crit. Care Med. 173 112-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahimainathan, L., Das, F., Venkatesan, B., and Choudhury, G. G. (2006) Diabetes 55 2115-2125 [DOI] [PubMed] [Google Scholar]
- 8.Sato, W., Horie, Y., Kataoka, E., Ohshima, S., Dohmen, T., Iizuka, M., Sasaki, J., Sasaki, T., Hamada, K., Kishimoto, H., Suzuki, A., and Watanabe, S. (2006) Hepatol. Res. 34 256-265 [DOI] [PubMed] [Google Scholar]
- 9.Desmouliere, A., Guyot, C., and Gabbiani, G. (2004) Int. J. Dev. Biol. 48 509-517 [DOI] [PubMed] [Google Scholar]
- 10.Spiegel, S., and Milstien, S. (2007) J. Biol. Chem. 282 2125-2129 [DOI] [PubMed] [Google Scholar]
- 11.El Alwani, M., Wu, B. X., Obeid, L. M., and Hannun, Y. A. (2006) Pharmacol. Ther. 112 171-183 [DOI] [PubMed] [Google Scholar]
- 12.Gardell, S. E., Dubin, A. E., and Chun, J. (2006) Trends Mol. Med. 12 65-75 [DOI] [PubMed] [Google Scholar]
- 13.Spiegel, S., and Milstien, S. (2003) Nat. Rev. Mol. Cell. Biol. 4 397-407 [DOI] [PubMed] [Google Scholar]
- 14.Saba, J. D., and Hla, T. (2004) Circ. Res. 94 724-734 [DOI] [PubMed] [Google Scholar]
- 15.French, K. J., Schrecengost, R. S., Lee, B. D., Zhuang, Y., Smith, S. N., Eberly, J. L., Yun, J. K., and Smith, C. D. (2003) Cancer Res. 63 5962-5969 [PubMed] [Google Scholar]
- 16.Ogretmen, B., and Hannun, Y. A. (2004) Nat. Rev. Cancer 4 604-616 [DOI] [PubMed] [Google Scholar]
- 17.Xia, P., Gamble, J. R., Wang, L., Pitson, S. M., Moretti, P. A., Wattenberg, B. W., D'Andrea, R. J., and Vadas, M. A. (2000) Curr. Biol. 10 1527-1530 [DOI] [PubMed] [Google Scholar]
- 18.Visentin, B., Vekich, J. A., Sibbald, B. J., Cavalli, A. L., Moreno, K. M., Matteo, R. G., Garland, W. A., Lu, Y., Yu, S., Hall, H. S., Kundra, V., Mills, G. B., and Sabbadini, R. A. (2006) Cancer Cell 9 225-238 [DOI] [PubMed] [Google Scholar]
- 19.Xin, C., Ren, S., Kleuser, B., Shabahang, S., Eberhardt, W., Radeke, H., Schafer-Korting, M., Pfeilschifter, J., and Huwiler, A. (2004) J. Biol. Chem. 279 35255-35262 [DOI] [PubMed] [Google Scholar]
- 20.Bu, S., Yamanaka, M., Pei, H., Bielawska, A., Bielawski, J., Hannun, Y. A., Obeid, L., and Trojanowska, M. (2006) FASEB J. 20 184-186 [DOI] [PubMed] [Google Scholar]
- 21.Berdyshev, E. V., Gorshkova, I. A., Usatyuk, P., Zhao, Y., Saatian, B., Hubbard, W., and Natarajan, V. (2006) Cell. Signal. 18 1779-1792 [DOI] [PubMed] [Google Scholar]
- 22.Sanchez, T., Thangada, S., Wu, M. T., Kontos, C. D., Wu, D., Wu, H., and Hla, T. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 4312-4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Brocklyn, J. R., Lee, M. J., Menzeleev, R., Olivera, A., Edsall, L., Cuvillier, O., Thomas, D. M., Coopman, P. J., Thangada, S., Liu, C. H., Hla, T., and Spiegel, S. (1998) J. Cell Biol. 142 229-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, X., Duan, X., Liang, Y. Y., Su, Y., Wrighton, K. H., Long, J., Hu, M., Davis, C. M., Wang, J., Brunicardi, F. C., Shi, Y., Chen, Y. G., Meng, A., and Feng, X. H. (2006) Cell 125 915-928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maehama, T., Taylor, G. S., and Dixon, J. E. (2001) Annu. Rev. Biochem. 70 247-279 [DOI] [PubMed] [Google Scholar]
- 26.Tang, Y., and Eng, C. (2006) Cancer Res. 66 736-742 [DOI] [PubMed] [Google Scholar]
- 27.Vazquez, F., Ramaswamy, S., Nakamura, N., and Sellers, W. R. (2000) Mol. Cell. Biol. 20 5010-5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller, C. D., Rivera Gil, P., Tolle, M., van der Giet, M., Chun, J., Radeke, H. H., Schafer-Korting, M., and Kleuser, B. (2007) Am. J. Pathol. 170 281-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kono, Y., Nishiuma, T., Nishimura, Y., Kotani, Y., Okada, T., Nakamura, S., and Yokoyama, M. (2007) Am. J. Respir. Cell Mol. Biol. 37 395-404 [DOI] [PubMed] [Google Scholar]
- 30.Leslie, N. R., and Downes, C. P. (2004) Biochem. J. 382 1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okumura, K., Zhao, M., Depinho, R. A., Furnari, F. B., and Cavenee, W. K. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2703-2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen, W. H., Balajee, A. S., Wang, J., Wu, H., Eng, C., Pandolfi, P. P., and Yin, Y. (2007) Cell 128 157-170 [DOI] [PubMed] [Google Scholar]
- 33.Hjelmeland, A. B., Hjelmeland, M. D., Shi, Q., Hart, J. L., Bigner, D. D., Wang, X. F., Kontos, C. D., and Rich, J. N. (2005) Cancer Res. 65 11276-11281 [DOI] [PubMed] [Google Scholar]
- 34.Takekawa, M., Maeda, T., and Saito, H. (1998) EMBO J. 17 4744-4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minaguchi, T., Waite, K. A., and Eng, C. (2006) Cancer Res. 66 11677-11682 [DOI] [PubMed] [Google Scholar]
- 36.Gil, A., Andres-Pons, A., Fernandez, E., Valiente, M., Torres, J., Cervera, J., and Pulido, R. (2006) Mol. Biol. Cell 17 4002-4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, F., Wagner, S., Campbell, R. B., Nickerson, J. A., Schiffer, C. A., and Ross, A. H. (2005) J. Cell. Biochem. 96 221-234 [DOI] [PubMed] [Google Scholar]
- 38.Trotman, L. C., Wang, X., Alimonti, A., Chen, Z., Teruya-Feldstein, J., Yang, H., Pavletich, N. P., Carver, B. S., Cordon-Cardo, C., Erdjument-Bromage, H., Tempst, P., Chi, S. G., Kim, H. J., Misteli, T., Jiang, X., and Pandolfi, P. P. (2007) Cell 128 141-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres, J., and Pulido, R. (2001) J. Biol. Chem. 276 993-998 [DOI] [PubMed] [Google Scholar]
- 40.Guzeloglu-Kayisli, O., Kayisli, U. A., Al-Rejjal, R., Zheng, W., Luleci, G., and Arici, A. (2003) J. Clin. Endocrinol. Metab. 88 5017-5026 [DOI] [PubMed] [Google Scholar]
- 41.Yamanaka, M., Shegogue, D., Pei, H., Bu, S., Bielawska, A., Bielawski, J., Pettus, B., Hannun, Y. A., Obeid, L., and Trojanowska, M. (2004) J. Biol. Chem. 279 53994-54001 [DOI] [PubMed] [Google Scholar]
- 42.Maudsley, S., Martin, B., and Luttrell, L. M. (2005) J. Pharmacol. Exp. Ther. 314 485-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.