Abstract
Responses to transforming growth factor β and multiple cytokines involve activation of transforming growth factor β-activated kinase-1 (TAK1) kinase, which activates kinases IκB kinase (IKK) and MKK3/6, leading to the parallel activation of NF-κB and p38 MAPK. Activation of TAK1 by autophosphorylation is known to involve three different TAK1-binding proteins (TABs). Here we report a protein phosphatase subunit known as type 2A phosphatase-interacting protein (TIP) that also acts as a TAB because it co-precipitates with and directly binds to TAK1, enhances TAK1 autophosphorylation at unique sites, and promotes TAK1 phosphorylation of IKKβ and signaling to NF-κB. Mass spectrometry demonstrated that co-expression of TAB4 protein significantly increased phosphorylation of four sites in TAK1, in a linker region between the kinase and TAB2/3 binding domains, and two sites in TAB1. Recombinant GST-TAB4 bound in an overlay assay directly to inactive TAK1 and activated TAK1 but not TAK1 phosphorylated in the linker sites, suggesting a bind and release mechanism. In kinase assays using TAK1 immune complexes, added GST-TAB4 selectively stimulated IKK phosphorylation. TAB4 co-precipitated polyubiquitinated proteins dependent on a Phe-Pro motif that was required to enhance phosphorylation of TAK1. TAB4 mutated at Phe-Pro dominantly interfered with IL-1β activation of NF-κB involving IKK-dependent but not p38 MAPK-dependent signaling. The results show that TAB4 binds TAK1 and polyubiquitin chains to promote specific sites of phosphorylation in TAK1-TAB1, which activates IKK signaling to NF-κB.
Inflammatory cytokines tumor necrosis factor α and IL-1β activate cellular pathways involved in cell proliferation and apoptosis. IL-1β binding to its cognate receptor IL-1R induces recruitment of MyD88, IRAK1, IRAK4, and TRAF6 (1). IRAK4 functions in this complex to phosphorylate IRAK1, thereby triggering release of IRAK1 and TRAF6 into the cytoplasm and subsequent activation of IKK,2 c-Jun N-terminal kinase (JNK), and p38 MAPK (2, 3). The IKK complex consists of IKKα and IKKβ, the two catalytic subunits, and the regulatory subunit IKKγ (known as NEMO), which binds polyubiquitin chains and is ubiquitinated itself (4). It is proposed that polyubiquitin chains act as a scaffold to allow for assembly of a signaling complex (5, 6). Genetic studies have implicated IKKβ and IKKγ in regulating activation of nuclear factor-κB (NF-κB) via phosphorylation of IκB and its subsequent degradation by the 26 S proteasome (7).
Activation of IKK involves two complexes called TRIKA1 and TRIKA2 (8, 9). The first complex, TRIKA1, contains Ubc13/Uev1A (an E2 conjugating enzyme) and TRAF6 (an E3 ligase) (8, 9). The second complex, TRIKA2, contains TAK1, TAB1 (TAK1 activator), and TAB2/3 (ubiquitin-binding proteins) (8, 9). TRAF6 functions with Ubc13/Uev1A to catalyze the addition of polyubiquitin chains to TRAF6 and possibly other proteins via Lys63 linkages in ubiquitin (8–10). TAK1 is activated by TAK1-binding proteins (TABs). TAB1 binds TAK1 and promotes autophosphorylation of the activation loop (11–14). TAB2 and TAB3 activate TAK1 indirectly by binding polyubiquitinated proteins, possibly stabilizing a larger complex (15–19). TAK1 associates with and is inactivated by multiple protein-Ser/Thr phosphatases, including different isoforms of the MPP phosphatases previously called PP2C (20–22) as well as by protein phosphatase 6 (PP6), which dephosphorylates Thr187 in the activation loop of TAK1 (21).
Our interest in PP6 raised a question about inactivation of TAK1. Studies of the TOR pathway in yeast led to the discovery of Tap42, a protein that binds all of the yeast type 2A phosphatases: Sit4 (PP6), Pph3, and Pph21/22 (PP2A) (23). However, Tap42 action on phosphatases is not understood and is controversial. One group claims Tap42 is phosphorylated directly by TOR to increase Tap42 binding to Pph21/22 or Sit4 (24, 25). Another group suggests that Tap42 is restricted from binding to phosphatases by instead binding a protein called Tip41 (Tap42-interacting protein of 41 kDa) and that the Tip41-Tap42 complex is disrupted by TOR phosphorylation of Tip41 (26). Yet a third scenario arose when it was reported that yeast Tip41 interacted with yeast phosphatases Pph21/22 and Pph3 in a two-hybrid assay (27). While our work was in progress another group published that the human orthologue of yeast Tip41 called type 2A phosphatase-interacting protein (TIP) binds human PP2A, PP4, and PP6 (28). Thus, type 2 phosphatases, including PP6, can bind to yeast and human orthologues of both Tap42 (α-4) and Tip41 (TIP). We showed that the human version of Tap42 called α-4 acts as a targeting subunit for PP2A, binding to MEK3 to promote selective dephosphorylation of one of two sites in the activation loop in that way opposing activation of p38 MAPK by cytokines (29).
Here because of the relationship to PP6 we investigated the function of the human TIP protein (NP_690866) relative to TAK1 and discovered properties that qualify this protein as a TAB and led us to call it TAB4. We show that, like TAB1, TAB4 directly binds and activates TAK1 by inducing autophosphorylation of TAK1. The activated TAK1 phosphorylates TAB1 and shows specificity toward the endogenous substrate IKKβ.A Phe-Pro sequence motif found in TAB2/3 is present in the C-terminal region of TAB4. Mutation of Phe254 and Pro255 in TAB4 eliminated binding of polyubiquitinated proteins and activation and phosphorylation of TAK1 and TAB1 without reduction in PP6 binding. These mutations separate multiple functions of TAB4. We propose that TAB4 is a multifunctional protein that promotes the activation of NF-κB using separate domains that bind TAK1 and polyubiquitin chains.
MATERIALS AND METHODS
cDNA Constructs, Plasmids, and Antibodies—Full-length human TAB4 was cloned downstream of GST in pGEX-4T2 using BamHI and EcoRI restriction sites for production of recombinant protein in bacteria. Recombinant GST-TAB4 was used to raise polyclonal antibodies in rabbits. Antibodies were purified using a two-step procedure. 1) GST-conjugated Affi-Gel-15 was used to preclear anti-GST antibodies followed by 2) purification with GST-TAB4-conjugated Affi-Gel-15 using 0.1 m glycine elution-2 m Tris-HCl neutralization as described in the manufacturer's protocol (Bio-Rad). TAB4 was also cloned downstream of HA epitope tag (pKHA vector) and FLAG epitope tag (pcDNA3-FLAG2 vector) using BamHI and EcoRI for expression in mammalian cells. Site-directed mutagenesis of TAB4 was done following the manufacturers' protocol. FLAG-TAK1, HA-TAK1, HA-TAK1(K63W), FLAG-TAB1, and T7-TAB1 were described previously (13). FLAG-Ub vector was a kind gift from Dr. David Wotton at the University of Virginia. Antibodies (dilutions) used in this study are as follows: anti-FLAG (1:3,000) (Sigma-Aldrich), anti-HA (1:5,000) (12CA5), anti-T7 (1:10,000) (Novagen), anti-PP6 (1:5,000) (30), anti-(Thr(P)184/Thr(P)187) TAK1 (1:1,000) and anti-(Ser(P)177/Ser(P)181) IKKβ (1:1,000) (Cell Signaling Technology Inc.), anti-GST and anti-TAB4 (1:5,000) (described above), and anti-(Ser(P)187) MEK3/(Ser(P)207) MEK6 (1:1,000) (Santa Cruz Biotechnology, Inc.).
Cell Culture, Transfection, Immunoprecipitation, and Pulldown Assays—HEK293, HEK293T, and 293IL-1R1 cells were grown in Dulbecco's modified Eagle's medium and 10% fetal bovine serum at 37 °C in a humidified incubator with 5% CO2. HEK293T cells were transfected using Arrest-In (Open Biosystems) as suggested by the manufacturer. In every case, HEK293T cells were seeded into 10-cm dishes at ∼40% confluence the day before transfection. Cells were transfected using ∼5 μg of plasmid for each construct and incubated for 24–48 h before harvesting. Cell extracts were made using a 1% Nonidet P-40 (Igepal CA-630, Sigma) lysis buffer (1% Nonidet P-40, 50 mm MOPS, pH 7.4, 150 mm NaCl, 1 μm microcystin-LR (Alexis Biochemicals), 1 mm sodium orthovanadate, 1 mm sodium fluoride, 1 mm Pefabloc, 1 mg/ml leupeptin, 1 mg/ml pepstatin, 1 mm dithiothreitol). Extracts were immunoprecipitated using either anti-FLAG M2 beads (Sigma-Aldrich), anti-HA beads (Sigma-Aldrich), or anti-TAB4 bound to protein A-agarose (Amersham Biosciences). Immunoprecipitations and pulldowns were done using approximately the same volume of extracts with 10–15 μl of a 50% slurry of anti-FLAG, anti-HA, and microcystin-LR-agarose beads. Immunoprecipitations and pulldowns were incubated at 4 °C for 2 h and then washed with Nonidet P-40 buffer two to three times. Complexes were eluted using 35 μl of 2× SDS sample buffer and boiled for 5 min. Extracts and immunoprecipitates were analyzed by immunoblotting using the antibodies described earlier and the LI-COR Odyssey infrared scanner and software.
Recombinant λ-Phosphatase Treatment—HEK293T cells were transfected with 1) FLAG-TAK1 and T7-TAB1 or 2) FLAG-TAK1 and T7-TAB1 plus HA-TAB4, and extracts were made with RIPA buffer (1% Nonidet P-40, 0.25% sodium deoxycholate, 0.1% SDS, 50 mm MOPS, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 μm microcystin-LR, 1 mm sodium orthovanadate, 1 mm sodium fluoride, 20 mm β-glycerophosphate, 1 mm Pefabloc, 1 mg/ml leupeptin, 1 mg/ml pepstatin, 1 mm dithiothreitol). Immunoprecipitates were collected after 2 h at 4 °C and washed three times with RIPA buffer followed by two washes with 50 mm Tris-HCl, pH 7.5, 0.1 mm EDTA, 2 mm MnCl2, 5 mm dithiothreitol (λ-phosphatase buffer). Immunoprecipitates were taken up in 30 ml of λ-phosphatase buffer and treated with 0, 0.2, or 1 μl of MBP-λ-phosphatase (∼8,330 units/mg) for 2 h at 30 °C. Samples were then analyzed by SDS-PAGE and immunoblotted to detect mobility shifts of TAK1 and/or TAB1.
Far-Western (Overlay) Assay—HEK293T cells were transfected with 1) FLAG-TAK1, 2) FLAG-TAK1 and T7-TAB1, 3) FLAG-TAK1 and T7-TAB1 plus HA-TAB4, or 4) empty vector control. Extracts were made using RIPA buffer and immunoprecipitated using anti-FLAG beads for 2 h at 4 °C. Immunoprecipitates were washed three times with RIPA buffer, eluted with 35 μl of 2× SDS sample buffer, and boiled for 5 min. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose. Membranes were blocked in 1% blocking buffer (1% bovine serum albumin in Tris-buffered saline/Tween 20) for 2 h. Probes were diluted in 1% blocking buffer at a concentration of 5 mg/ml and incubated overnight at 4 °C. Membranes were washed two to three times with 1× phosphate-buffered saline for 1–2 min followed by fixation using 0.5% paraformaldehyde for 30 min at room temperature. Membranes were then rinsed quickly twice with 1× phosphate-buffered saline and quenched using 2% glycine in phosphate-buffered saline for 10 min at room temperature. The membrane was incubated with anti-GST plus secondary antibody and analyzed using the LI-COR Odyssey system and software.
FLAG-Ub Binding Assay—HEK293T cells were transfected with HA-TAB4(wt), HA-TAB4(F254A), or HA-TAB4(FP-AA) or empty vector control along with FLAG-Ub. Extracts were prepared using 1% Nonidet P-40 lysis buffer described above and immunoprecipitated using anti-HA beads for 2 h at 4 °C and washed 2–3 times with Nonidet P-40 buffer. HA-FLAG complexes were eluted using 35 μl 2X SDS sample buffer and boiled for 5 min. Samples were analyzed by SDS-PAGE and immunoblotted with anti-FLAG and anti-HA. Immunoblots were analyzed using the LI-COR Odyssey system and software.
In Vitro TAK1 Kinase Assay with MKK6 or IKKβ—HEK293T cells were transfected with 1) FLAG-TAK1, 2) FLAG-TAK1 and T7-TAB1, or 3) FLAG-TAK1 and T7-TAB1 plus either HA-TAB4(wt), 4) HA-TAB4(F254A), or 5) HA-TAB4(FP-AA). Extracts were prepared using a 1% Nonidet P-40 lysis buffer described above and immunoprecipitated using anti-FLAG beads. Immunoprecipitates were collected after 2 h at 4 °C, washed two to three times with Nonidet P-40 buffer, and then washed twice in 20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 10 mm MgCl2. Immunoprecipitates were taken up in 30 ml of 20 mm Tris-HCl, pH 7.5, 10 mm MgCl2. Kinase assays were done using 1 μg of recombinant His6-MKK6 or GST-IKKβ (Upstate-Chemicon), 5 μl of immunoprecipitate, 2 μl of 5× kinase buffer (50 mm Tris-HCl, pH 7.5, 5 mm dithiothreitol, 25 mm MgCl2), 5 μCi of [γ-32P]ATP at 30 °C for 2 min. Kinase reactions were stopped by adding an equal volume of 2× SDS sample buffer and resolved by SDS-PAGE followed by staining with Gel-Code Blue (Pierce) to visualize bands. Bands corresponding to His6-MKK6 or GST-IKKβ were excised from the stained gel, and 32P was analyzed using a liquid scintillation counter.
NF-κB Luciferase Assays—HEK293T cells were seeded at 30–40% confluence in 12-well dishes and transfected 16 h later with 100 ng of F254A, FP-AA, or empty vector plus 100 ng of NF-κB firefly luciferase and 10 ng of Renilla luciferase vectors or with 100 ng of FLAG-TAB4(wt) or empty vector and HA-TAK1(wt) or HA-TAK1(K63W) plus 100 ng of NF-κB luciferase and 10 ng of Renilla vectors. Cells were incubated for 16 h before changing the medium to serum-free conditions (0.5% fetal bovine serum, Dulbecco's modified Eagle's medium) for the remainder of the experiment. Cells were pretreated with curcumin or SB203580 for 30 min prior to stimulation for 6 h with 10 ng/ml IL-1β or vehicle alone. Cells were harvested and analyzed as described previously (29) to test for luciferase activities in extracts.
RESULTS
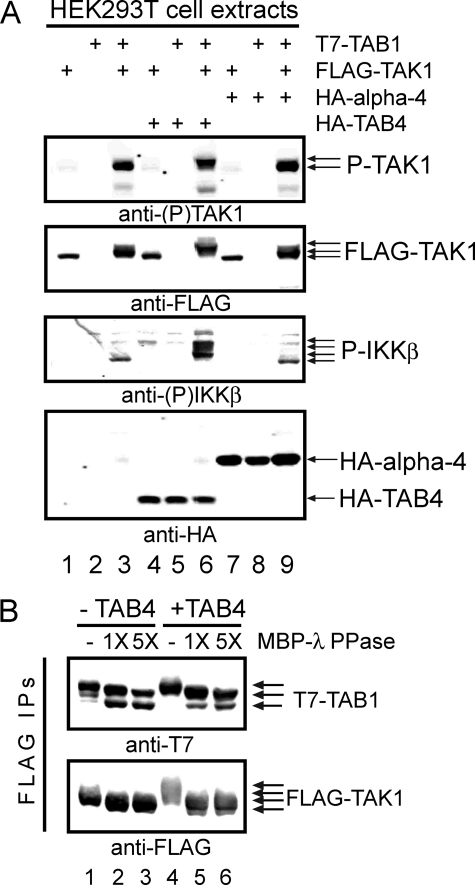
TAB4 Enhances Activation of TAK1-TAB1—The human protein NP_690866 has been reported to bind protein phosphatase catalytic subunits PP2A, PP4, and PP6 (27, 28) and is referred to as TIP. We found that TIP bound type 2 phosphatases in a yeast two-hybrid assay and noted that it showed preferential binding to the C-terminal region of PP6 (residues 177–305) compared with PP2A. Because PP6 dephosphorylates and inactivates TAK1 kinase (21), we tested whether overexpression of this PP6-binding protein (which we called TAB4) would affect TAK1. Activation of TAK1 and its substrate IKKβ were assayed in whole cell extracts by immunoblotting with phosphosite-specific antibodies (Fig. 1A). Neither TAK1 nor IKKβ were phosphorylated in cells transfected with FLAG-TAK1 alone (lane 1) or T7-TAB1 alone (lane 2). However, cells co-expressing FLAG-TAK1 with T7-TAB1 exhibited phospho-TAK1, with retarded migration of the FLAG-TAK1, and increased phosphorylation of endogenous IKKβ (lane 3). Compared with this, co-expression with HA-TAB4 (lane 6) caused more reduced migration of FLAG-TAK1 and greatly increased the phosphorylation of endogenous IKKβ at Ser177/Ser181 with appearance of multiple IKK bands presumably due to other sites of phosphorylation. Co-expression of TAB4 with either FLAG-TAK1 alone (lane 4) or T7-TAB1 alone (lane 5) as controls did not induce phosphorylation of either TAK1 or IKKβ. This shows TAB1 dependence of TAK1 activation by TAB4, the same as for activation of TAK1 by TAB2/3. The effects were specific for TAB4 and not mimicked by overexpression of another PP2A/PP4/PP6-binding protein, α-4 (Fig. 1A). HA-α-4 was expressed with FLAG-TAK1 (lane 7) or T7-TAB1 (lane 8) but produced no increase in phospho-TAK1, no gel shift of FLAG-TAK1, and no detectable phospho-IKKβ. Triple expression of HA-α-4, FLAG-TAK1, and T7-TAB1 (lane 9) was no different from dual expression of FLAG-TAK1 and T7-TAB1 (lane 3). The level of HA-α-4 expressed in these controls was even higher than the levels of HA-TAB4 in parallel samples based on anti-HA immunoblotting (Fig. 2A, lanes 4–6 and 7–9). Together these results showed that TAB4 expression enhanced TAB1-dependent TAK1 phosphorylation and TAK1 activity toward the endogenous substrate IKKβ.
FIGURE 1.
TAK1 phosphorylation and activation by TAB4. A, combinations of FLAG-TAK1, T7-TAB1, HA-TAB4, and HA-α-4 were expressed in HEK293T cells and analyzed by immunoblotting with (top to bottom) 1) anti-(Thr(P)184/Thr(P)187) TAK1, 2) anti-FLAG, 3) anti-(Ser(P)177/Ser(P)181) IKKβ, and 4) anti-HA (bottom panel). Lanes 1–3 show extracts from cells co-expressing 1) FLAG-TAK1, 2) T7-TAB1, or 3) FLAG-TAK1 and T7-TAB1. Lanes 4–6 show extracts from cells co-expressing HA-TAB4 with FLAG-TAK1 and/or T7-TAB1. Lanes 7–9 show extracts from cells co-expressing HA-α-4 with FLAG-TAK1 and/or T7-TAB1. B, phosphatase treatment of FLAG-TAK1 and T7-TAB1. Proteins were co-expressed without (lanes 1–3) and with HA-TAB4 (lanes 4–6) in HEK293T cells, immunoprecipitated using anti-FLAG M2 beads, and immunoblotted with anti-T7 (upper panel) and anti-FLAG (lower panel). Immunoprecipitates (IPs) were treated with vehicle, 1×, or 5× amounts of recombinant MBP-λ-phosphatase (PPase) as indicated. P-, phospho-.
FIGURE 2.
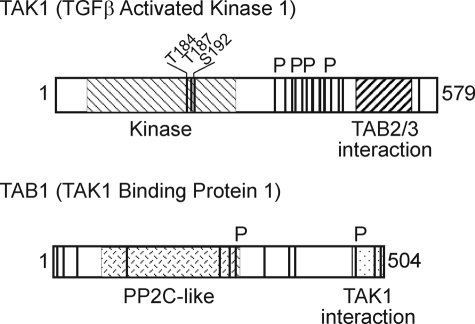
Phosphorylation sites in TAK1 and TAB1. Phosphorylation sites in TAK1 (top) and TAB1 (bottom) are shown as vertical lines, and those sites significantly increased by co-expression of TAB4 are indicated with a P above the boxes. The kinase domain with three sites of activating phosphorylation and the TAB2/3 binding region in TAK1 and the PP2C-like domain and TAK1 binding domain in TAB1 are shown as hatched boxes within the entire sequence. TGFβ, transforming growth factor β.
TAB4 Significantly Increases Multiple Sites of Phosphorylation in TAK1 and TAB1—The reduced mobility of FLAG-TAK1 and T7-TAB1 in SDS-PAGE was due to phosphorylation that was induced by TAB4 and reversed by λ-phosphatase (Fig. 1B). FLAG-TAK1 and T7-TAB1 were expressed in HEK293T cells with and without HA-TAB4, and complexes were immunoprecipitated using anti-FLAG M2 beads. Immunoblotting with anti-FLAG and anti-T7 showed reduced mobility of both TAK1 and TAB1 proteins due to TAB4 co-expression (Fig. 1B; also see Fig. 3A below). Incubation of immunoprecipitates with increasing amounts of recombinant λ-phosphatase-MBP fusion protein dephosphorylated both FLAG-TAK1 and T7-TAB1 as evident from their increased mobility (Fig. 1B). After dephosphorylation by MBP-λ-phosphatase the FLAG-TAK1 and T7-TAB1 had the same mobility whether they originated from cells expressing or not expressing TAB4. This demonstrated that the extra reduced mobility of TAK1 and TAB1 during co-expression with TAB4 was due to phosphorylation. We noted that migration of TAB4 itself in SDS-PAGE was not affected by co-expression with TAK1-TAB1 or incubation with MBP-λ-phosphatase, suggesting that it was not phosphorylated. The results demonstrate that TAB4 increased phosphorylation and kinase activity of TAK1-TAB1 complexes.
FIGURE 3.
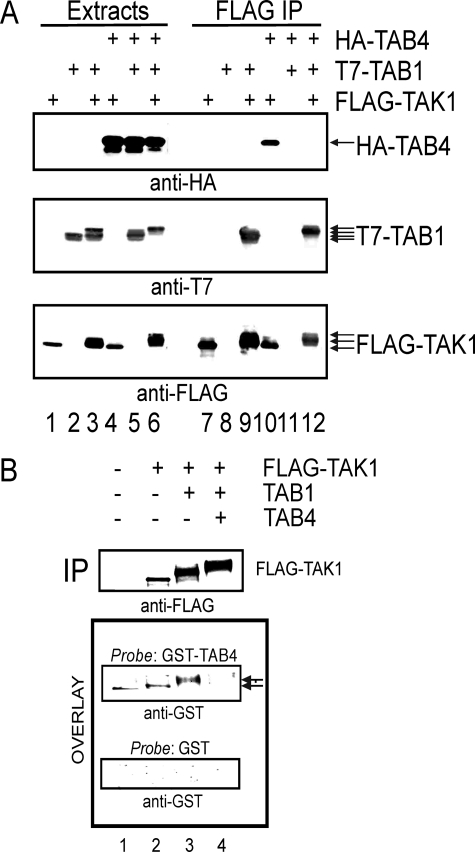
TAB4 associates with and directly binds TAK1. A, FLAG-TAK1 and/or T7-TAB1 were expressed with and without HA-TAB4 in HEK293T cells and immunoprecipitated using anti-FLAG M2 beads. Extracts (lanes 1–6) and immunoprecipitates (IP)(lanes 7–12) were analyzed by immunoblotting with anti-HA, anti-T7, and anti-FLAG (top to bottom). B, empty vector, FLAG-TAK1, FLAG-TAK1 and T7-TAB1, or FLAG-TAK1, T7-TAB1, and HA-TAB4 were expressed in HEK293T cells. Proteins were immunoprecipitated using anti-FLAG M2 beads, resolved by SDS-PAGE, and analyzed by immunoblotting using anti-FLAG (upper panel). A duplicate filter was used in an overlay assay (far-Western) with 5 μg/ml GST-TAB4 or GST as probes and immunoblotted with anti-GST for detection.
Phosphorylation sites in both TAK1 and TAB1 induced by co-expression of TAB4 were mapped by mass spectrometry. Cells were transfected to express TAK1-TAB1 with and without TAB4, FLAG-TAK1 complexes were immunoprecipitated, and the tryptic peptides were analyzed by a linear quadrupole ion trap-Fourier transform mass spectrometer (ThermoScientific) (Table 1). A total of at least 16 phosphorylation sites were identified in FLAG-TAK1, including the well known sites at Thr184, Thr187, and Ser192 in the kinase activation loop (Fig. 2). Phosphorylation of 14 sites in T7-TAB1 were identified. Co-expression of TAB4 induced a significant increase (>5-fold) in phosphorylation of at least four sites in TAK1: 1) residue Thr344 in peptide residues 331–347; 2) peptide 373–386 with residues Ser374, Ser375, and Ser382; 3) peptide 388–398 containing residues Ser389 and Ser393; and 4) peptide 412–429 containing residues Ser412, Thr421, and Ser428. These all mapped to a region C-terminal to the kinase domain (Fig. 2). Likewise co-expression of TAB4 significantly increased at least two sites of phosphorylation in TAB1: 1) peptide 364–386 containing phosphorylated residues Ser273 and Ser278 and 2) peptide 464–477 phosphorylated at residue Ser464 or Ser469. Thus, TAB4 co-expression significantly increased phosphorylation of specific sites in both TAK1 and TAB1.
TABLE 1.
Phosphopeptide analysis of TAK1 and TAB1 by mass spectrometry
Tryptic peptides from FLAG-TAK1-TAB1 complexes were analyzed via nanoflow high pressure liquid chromatography coupled on line to a microelectrospray ionization source on a linear quadrupole ion trap-Fourier transform mass spectrometer (35). triplephos, triply phosphorylated.
| Sites | Peptidesa | Sequenceb |
|---|---|---|
| FLAG-TAK1 | ||
| Thr184 or Thr187 | 173-190 | ICDFGTACDIQp (THMT) NNK |
| Thr184 and Thr187 | ICDFGTACDIQpTHMpTNNK | |
| triplephos (Thr178 or Thr184 or Thr187 or Ser192) | 173-209 | ICDFGppp (TACDIQTHMTNNKGS) AAWMAPEVFEGSNYSEK |
| Thr344 | 331-347 | SDTNMEQVPATNDpTIKR |
| Ser374 or Ser375 | 373-386c | Gp (SS) VESLPPTSEGK |
| Thr382 | GSSVESLPPpTSEGK | |
| Ser389 | 388-398c | MpSADMSEIEAR |
| Ser393 | MSADMpSEIEAR | |
| Thr402 | 399-409 | IVApTAGNGQPR |
| Ser412 and (Thr419 or Thr421) and (Ser427 or Ser428) | 410-429 | RRpSIQDLTVp (TGT) EPGQVp (SS) R |
| Ser412 | 412-429 | pSIQDLTVTGTEPGQVSSR |
| Ser412 and Thr421 | 412-429c | pSIQDLTVTGpTEPGQVSSR |
| Ser412 and Ser428 | pSIQDLTVTGTEPGQVSpSR | |
| Thr440 | 437-450 | MIpTTSGPTSEKPAR |
| Thr444 or Ser445 | MITTSGPp (TS) EKPAR | |
| Thr440 and (Thr444 or Ser445) | MIpTTSGPp (TS) EKPAR | |
| Thr537 | 520-539 | KQELVAELDQDEKDQQNpTSR |
| T7-TAB1 | ||
| Ser7 | 7-35 | pSLLQSEQQPSWTDDLPLCHLSGVGSASNR |
| Ser16 or Thr18 | SLLQSEQQPp (SWT) DDLPLCHLSGVGSASNR | |
| Ser7 and Ser16 | pSLLQSEQQPpSWTDDLPLCHLSGVGSASNR | |
| Ser7 and Thr18 | pSLLQSEQQPSWpTDDLPLCHLSGVGSASNR | |
| Thr44 | 36-55 | SYSADGKGpTESHPPEDSWLK |
| Ser120 | 116-128 | SFLEpSIDDALAEK |
| Ser263 or Thr282 | 263-294 | p (SKPIIAEPEIHGAQPLDGVT) GFLVLMSEGLYK |
| Ser344 | 337-348 | IHSDTFApSGGER |
| Ser373 | 364-386c | NFGYPLGEMpSQPTPSPAPAAGGR |
| Ser378 | NFGYPLGEMSQPTPpSPAPAAGGR | |
| Ser373 and Ser378 | NFGYPLGEMpSQPTPpSPAPAAGGR | |
| Ser396 | 387-402 | VYPVSVPYSpSAQSTSK |
| Ser399, Thr400, or Ser401 | VYPVSVPYSSAQp (STS) K | |
| Ser396 and (Ser399 or Thr400 or Ser401) | VYPVSVPYSpSAQp (STS) K | |
| Ser464 or Ser469 | 464-477 | p (SRPAHS) LPPGEDGR |
| Ser492 | 490-504 | LWpSVDHGEQSVVTAP |
| Ser499 | LWSVDHGEQpSVVTAP |
Peptides with significantly increased phosphorylation (>5-fold) determined by peak area from co-expression of TAB4 are in boldface font.
Listing of TAK1 and TAB1 phosphopeptides detected in single letter code. Confirmed phosphorylation sites (phosphoserine (pS) or phosphothreonine (pT)) are in boldface font. “p()” indicates that a phosphorylation site is located within the parentheses but could not be defined further by the observed fragmentation.
Coelution of more than one phosphorylated form of the peptide collectively contributed to the increased abundance.
Binding of TAB4 to TAK1 Prevented by Phosphorylation of Linker Region—TAB4 associates with TAK1 by direct binding, and phosphorylation of the TAK1 linker region interferes with the interaction (Fig. 3). FLAG-TAK1 co-precipitated HA-TAB4 when expressed without TAB1, showing that unphosphorylated TAK1 stably associates with TAB4 (Fig. 3A, lane 10). However, FLAG immunoprecipitates of activated TAK1-TAB1 complexes did not contain HA-TAB4 (Fig. 3, lane 12). Thus, TAB4 induced phosphorylation of co-expressed TAK1 and TAB1 but did not remain bound to the complex. We propose that TAB4 acts as a “hit-and-run” activator. TAB4 binds to unphosphorylated TAK1 and to TAK1-TAB1 complexes because TAB1 was required for induced phosphorylation (see Fig. 1, lanes 4 versus lanes 6), but TAB4 did not co-precipitate with the active, phosphorylated TAK1. This is in contrast to a “hit-and-hold” mechanism in which TAB1 remains bound to TAK1 after inducing kinase autophosphorylation. FLAG-TAK1 co-precipitated T7-TAB1 whether or not HA-TAB4 was co-expressed (Fig. 3A, lanes 9 and 12), confirming a stable association between these proteins that was not lost upon phosphorylation. We also examined co-precipitation of the endogenous TAK1 and TAB4 in 293IL-1R1 cells and mouse dermal fibroblasts. Precipitation of TAK1 from unstimulated cells did not yield detectable TAB4; however, 5 min after activation of the cells with IL-1 (5 ng/ml) TAB4 could be detected by immunoblotting of the immunoprecipitates (see the supplemental figure). At 15 min after stimulation TAB4 was no longer detected in the TAK1 immunoprecipitates. The results support a transient, IL-1-stimulated association of TAB4 with TAK1.
A protein overlay assay showed direct protein-protein binding of GST-TAB4 to FLAG-TAK1 (Fig. 3B). Different phosphorylated forms of FLAG-TAK1 were produced by transfecting HEK293T cells with 1) empty vector as blank control, 2) FLAG-TAK1 alone, 3) FLAG-TAK1 and T7-TAB1, or 4) FLAG-TAK1, T7-TAB1, and HA-TAB4. FLAG immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-FLAG to show the reduced migration due to phosphorylation as well as the relative loading of the FLAG-TAK1 protein in the different samples (Fig. 3B, upper panel). The proteins on this filter were probed with purified GST-TAB4 protein or GST protein as control and stained with anti-GST antibodies to detect bound probe proteins (Fig. 3B, lower panels). The GST-TAB4 bound to unphosphorylated FLAG-TAK1 (lane 2) and the slower migrating phospho-TAK1 (lane 3) formed by co-expression with TAB1. However, there was no GST-TAB4 binding to the more highly phosphorylated TAK1 formed by co-expression with both TAB1 and HA-TAB4 even though this sample (lane 4) had the highest amount of FLAG-TAK1. These results reinforce the lack of co-immunoprecipitation between the most highly phosphorylated form of TAK1 and HA-TAB4 (see Fig. 3A, lane 12). GST as a control probe showed no binding to any forms of FLAG-TAK1, demonstrating the specificity of GST-TAB4 binding. These results show that TAB4 binds directly to TAK1, supporting its assignment as an authentic TAB, but TAB4 does not bind to TAK1-TAB1 complexes when TAK1 is phosphorylated in the linker region.
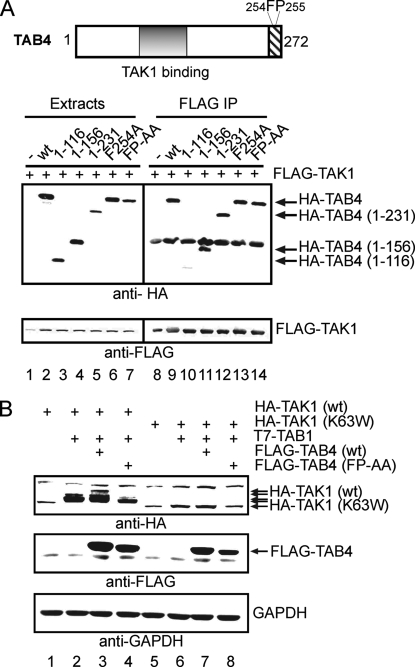
Deletion and point mutants of HA-TAB4 were co-expressed with TAK1 to map TAK1 binding to a central region of the TAB4 protein (Fig. 4A). FLAG-TAK1 co-precipitated all the HA-TAB4 proteins (lanes 9 and 11–14) except HA-TAB4-(1–116) (lane 10). Whole cell extracts were immunoblotted to show that the expression levels of FLAG-TAK1 and several HA-TAB4 proteins were similar (lanes 1–7). The results indicated that the central region of TAB4 consisting of residues 116–156 was necessary for association with TAK1. It is important to note that truncation of the C terminus of TAB4 up to residue 156 did not reduce binding to FLAG-TAK1 (lane 11). Truncation of TAB4 residues 177–272 reportedly eliminates binding to phosphatases (28), suggesting that TAB4 requires separate regions for binding to TAK1 and to PP6.
FIGURE 4.
TAB4 activation of TAK1-TAB1 is dependent on TAK1 kinase activity. A, the diagram shows two domains in TAB4: a TAK1 binding domain (residues 116–156) and a putative ubiquitin binding domain motif (residues 241–272) with residues Phe254 and Pro255. FLAG-TAK1 was expressed in HEK293T cells with empty vector or one of the following deletion or point mutants of HA-TAB4 (wt, 1–116, 1–156, 1–231, F254A, or FP-AA), and proteins were immunoprecipitated with anti-FLAG beads. Extracts (lanes 1–7) and immunoprecipitates (IP)(lanes 8–14) were analyzed by immunoblotting with anti-HA (top) and anti-FLAG (bottom). B, HA-TAK1(wt) or HA-TAK1(K63W) were expressed in 293IL-1R1 cells alone or with T7-TAB1 and either FLAG-TAB4(wt) or FLAG-TAB4(FP-AA) or empty vector. Extracts were immunoblotted (top to bottom) with anti-HA, anti-FLAG, and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control.
TAB4 Requires a Phe-Pro Motif to Stimulate TAK1 Autophosphorylation and Bind Polyubiquitin—TAB4 induced phosphorylation of wild type TAK1 protein but not a kinasedead mutant, TAK1(K63W), consistent with TAB4 activating TAK1 autophosphorylation (Fig. 4B). Phosphorylation of wild type HA-TAK1 was stimulated by TAB1 in 293IL-1R1 cells based on the reduced migration in SDS-PAGE in comparison with HA-TAK1 expressed alone (lane 2 versus lane 1). In contrast, kinase-dead HA-TAK1(K63W) was not phosphorylated with or without co-expression of TAB1 (lanes 5 and 6). Phosphorylation of HA-TAK1 was increased by FLAG-TAB4 with the characteristic extra shift in migration (lane 3 versus lane 2). However, FLAG-TAB4 did not induce any reduced mobility due to phosphorylation of HA-TAK1(K63W) co-expressed with T7-TAB1 (lane 7), showing that the kinase activity of TAK1 was required. This makes it unlikely that TAB4 was activating or recruiting some other kinase that phosphorylated TAK1. TAB4 has a Phe-Pro (FP) sequence, a motif known to be important for TAB2/3 function and association with polyubiquitin chains (16). Expression of mutated FLAG-TAB4(FP-AA) did not induce phosphorylation of TAK1 (Fig. 4B, lane 4) even though the expression levels of mutated and wild type TAB4 proteins were comparable in these experiments (lanes 3 and 4). The results show two important points. 1) TAB4 induction of TAK1 phosphorylation requires TAK1 activity, indicative of autophosphorylation, and 2) residues Phe254 and Pro255 in TAB4 are required to induce this autophosphorylation of TAK1.
TAB4 residues Phe254 and Pro255 that are necessary for activation of TAK1 are involved in binding to polyubiquitinated proteins (Fig. 5). HA-TAB4 co-precipitated proteins conjugated with FLAG-Ub, and the mutant TAB4(FP-AA) precipitated much less of these ubiquitinated proteins compared with wild type TAB4 (Fig. 5A). Recovery of the different HA-tagged TAB4 proteins was assayed by anti-HA immunoblotting (Fig. 5B). Normalized to the amount of HA-tagged protein there was >60% reduction in the amount of polyubiquitinated proteins bound to HA-TAB4(FP-AA) compared with wild type TAB4. This control also showed that the FLAG-Ub-conjugated proteins recovered by anti-HA immunoprecipitation in Fig. 5A were bound to TAB4 and not ubiquitinated forms of the HA-TAB4 protein itself that would have appeared in the upper part of the anti-HA immunoblot in Fig. 5B. These results show that TAB4 has a polyubiquitin binding activity that requires Phe254 and Pro255, the same residues required for activation of TAK1 autophosphorylation.
FIGURE 5.
TAB4 co-precipitation with polyubiquitinated proteins. FLAG-Ub and empty vector (lane 1) or HA-TAB4(wt) (lane 2), HA-TAB4(F254A) (lane 3), or HA-TAB4(FP-AA) (lane 4) were expressed in HEK293T cells, and extracts were immunoprecipitated with anti-HA beads. Immunoprecipitates (IP) were analyzed by immunoblotting with anti-FLAG (A) and anti-HA (B).
TAB4 Effects on TAK1 in Vitro Phosphorylation of IKKβ and MKK6—Kinase assays of FLAG-TAK1 immune complexes used [32P]ATP plus either recombinant His6-MKK6 or GST-IKKβ as substrates (Fig. 6). Following reaction the proteins were resolved by SDS-PAGE and stained with Coomassie (Fig. 6A), and substrates were excised from the gel and quantitated for 32P incorporation by scintillation counting (Fig. 6B). FLAG-TAK1 alone exhibited little kinase activity with either His6-MKK6 or GST-IKKβ (columns 1 and 6), whereas FLAG-TAK1 co-expressed with T7-TAB1 showed about the same level of activity with both protein substrates (columns 2 and 7) consistent with TAB1 induced autophosphorylation and activation of TAK1. Expression of HA-TAB4 with FLAG-TAK1 plus T7-TAB1 further increased kinase-specific activity 3-fold toward both substrates (column 3 versus column 2 and column 8 versus column 7). Expression of either HA-TAB4(F254A) or HA-TAB4(FP-AA) increased TAK1 kinase activity toward His6-MKK6 more than expression of wild type TAB4 (columns 4 and 5). However, either mutated form of TAB4 did not increase but blocked TAK1 activity with GST-IKKβ as substrate (columns 9 and 10). Thus, wild type TAB4 activated TAK1 kinase in vitro using either of two different substrates. However, TAB4 mutated in the FP motif inhibited TAK1 in vitro phosphorylation of IKKβ but not MKK6.
FIGURE 6.
In vitro TAK1 kinase assay with recombinant MKK6 and IKKβ as substrates. A, FLAG-TAK1 and T7-TAB1 plus either HA-TAB4(wt), HA-TAB4(F254A), or HA-TAB4(FP-AA) were expressed in HEK293T cells as indicated. TAK1 complexes were immunoprecipitated using anti-FLAG M2 beads for in vitro kinase assays with [32P]ATP. Following reaction proteins were resolved by SDS-PAGE and stained with Coomassie Blue. Lanes 1–5 are assays using recombinant His6-MKK6, and lanes 6–10 are assays using recombinant GST-IKKβ as substrate. B, substrates were excised from the gel, and radioactivity was determined by scintillation counting and corrected by subtracting cpm in blank samples of substrate without added TAK1.
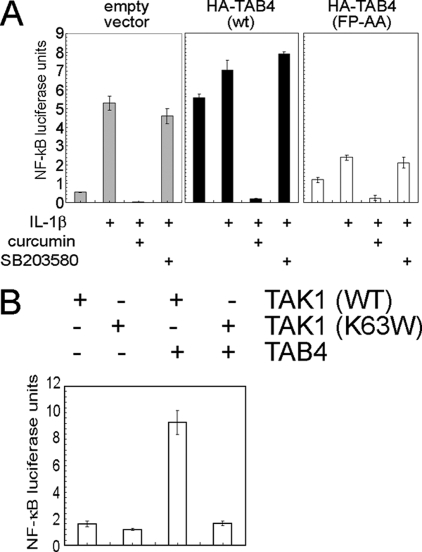
TAB4 Regulates TAK1 Activation of NF-κB via IKK in Living Cells—TAB4 mutated in the FP motif also interfered with IKK-dependent NF-κB activation in response to IL-1β in living cells (Fig. 7A). IL-1β activated an NF-κB-dependent luciferase reporter >5-fold in control cells, and this response was inhibited completely by pretreatment with 100 μm curcumin (used to inhibit IKK) but was unaffected by 20 μm SB203580 (a p38 MAPK inhibitor), showing that the response primarily involved signaling by IKK rather than p38 MAPK (Fig. 7A, left panel). Basal NF-κB activity was increased 6-fold in cells overexpressing wild type HA-TAB4 compared with control; this response was equivalent to the response of the cells to IL-1β. Addition of IL-1β elicited little further increase in NF-κB activity, suggesting that TAB4 expression had fully activated the pathway (Fig. 7A, center panel). The NF-κB activation by TAB4 in these cells was inhibited by curcumin but not by SB203580, showing that the response primarily involved IKK signaling. Expression of the TAB4(FP-AA) mutant modestly increased basal NF-κB activity and dramatically blunted activation by IL-1β (Fig. 7A, right panel). This activity was inhibited by curcumin but not by SB203580. The expression levels of HA-TAB4(wt) and TAB4(FP-AA) proteins were similar in all these experiments (not shown). As an additional control we observed equivalent expression of luciferase from an AP-1-dependent promoter in cells expressing wild type TAB4 or the TAB4(FP-AA) mutant (data not shown). We concluded that in living cells TAB4 overexpression activated TAK1 reaction with IKK to increase NF-κB activity, whereas the FP-mutated form of TAB4 interfered with TAK1 reaction with IKK, thereby preventing stimulation of NF-κB in response to IL-1β. Furthermore we demonstrated that TAB4 activation of NF-κB required TAK1 kinase activity (Fig. 7B). Co-expression of FLAG-TAB4 with wild type HA-TAK1 activated NF-κB 5-fold compared with FLAG-TAB4 plus kinase-dead HA-TAK1(K63W). Single expression of HA-TAK1 wild type or K63W did not activate NF-κB in this assay. Together the results indicate that TAB4 promotes TAK1 phosphorylation of IKK in the NF-κB pathway.
FIGURE 7.
A, HEK293T cells were transfected with luciferase reporter plasmid containing an NF-κB promoter, Renilla reporter plasmid, and either HA-TAB4(wt) or HA-TAB4(FP-AA) or empty vector; grown in serum-free conditions; and pretreated with kinase inhibitors curcumin (100 μm) or SB203580 (20 μm) prior to stimulation with IL-1β (10 ng/ml) for 6 h. Cell extracts were prepared and assayed for luciferase and Renilla activity using firefly reagent (Promega) and coelenterazine as substrates, respectively. Cells containing empty vector (light gray bars, left), HA-TAB4(wt) (black bars, center), or HA-TAB4(FP-AA) (white bars, right) were analyzed for luciferase/Renilla activity. Values are luciferase units normalized to Renilla and plotted as mean ± S.D. (n = 3). B, HEK293T cells were transfected with luciferase reporter plasmid containing an NF-κB-dependent promoter, Renilla reporter plasmid, and HA-TAB4(wt) or empty vector and HA-TAK1(wt) or HA-TAK1(K63W). Extracts were prepared and assayed as described above. The histogram shows NF-κB luciferase units normalized against Renilla units (mean ± S.D., n = 3).
DISCUSSION
Inflammatory cytokines, chemokines, and Toll-like receptor agonists such as lipopolysaccharide activate TAK1 downstream of receptors by promoting the binding of TABs that stimulate TAK1 autophosphorylation (11, 12, 14). TAB1 is the primary TAK1 activator; it also is a substrate for TAK1 (11, 12, 14). TAB2 and TAB3 are unrelated in sequence to TAB1 and bind to a different region of TAK1 near the C terminus. Activation of TAK1 by TAB2/3 requires their interaction with polyubiquitin chains (15–19). The exact mechanisms for TAK1 action are not understood, but polyubiquitin is though to act as a scaffold to assemble different kinase complexes, potentially conferring specificity for upstream activators as well as downstream substrates. Here we show another protein that qualifies as a new TAB because 1) it co-precipitates with and binds directly to TAK1; 2) it stimulates TAK1 autophosphorylation at multiple sites outside the kinase domain, enhancing phosphorylation above the levels induced by TAB1; and 3) like TAB2/3 it has an FP sequence motif that is required for association with polyubiquitinated proteins, activation of TAK1, and activation of IKK and NF-κB in living cells.
Multiple protein phosphatases regulate TAK1 activity, but the interplay among them is still not understood (20–22). Different individual PP2C isoforms associate with TAK1 and inactivate the kinase by dephosphorylation of residues in the activation loop (20–22). Okadaic acid, which does not inhibit PP2C phosphatases, greatly enhances phosphorylation and activation of TAK1 in response to stimulation (21), implicating the PPP family protein-Ser/Thr phosphatases in the control of TAK1. This led to our discovery that PP6 associates with TAK1 and dephosphorylates Thr187 in the activation loop (21). We suspected that PP6 was targeted to TAK1 by a regulatory subunit; however, we have not detected any of the SAPS-related subunits (30) associated with immunoprecipitated TAK1.3 Alternatively other regulatory subunits bind PP6 and other type 2A phosphatases. Yeast Tip41 has been implicated in TOR regulation of type 2A phosphatases, and the mammalian orthologue of yeast Tip41, called TIP, is a smaller protein of 32 kDa. Both the yeast and mammalian TIP proteins directly bind to type 2A protein phosphatases, including PP6 (27, 28). We carried out yeast two-hybrid assays and found relatively specific binding of TIP to PP6 compared with PP2A and found that the interaction involved the C-terminal region of PP6 not otherwise known for regulatory subunit association. Furthermore we produced a specific antibody and co-immunoprecipitated the endogenous TIP (TAB4) protein with endogenous PP6 from cell extracts. Results from another group showed that TIP inhibits PP2A phosphatase activity in vitro and enhances phosphorylation of an ataxia telangiectasia mutated kinase substrate (28).
Association of TIP with PP6 suggested to us a possible role in targeting PP6 to TAK1; however, in testing this idea we found that the protein activates TAK1 by physical association and increases phosphorylation of both TAK1 and TAB1 at multiple novel sites. Therefore we refer to TIP as TAB4. Phosphopeptide analysis by mass spectrometry confirmed that co-expression significantly increased (>5-fold) phosphorylation of four sites in TAK1 and two sites in TAB1 over and above levels of phosphorylation achieved with TAK1-TAB1 co-expression. The sites in TAK1 are in a linker region, C-terminal to the TAK1 kinase domain, and N-terminal to the domain that binds TAB2/3. Increased phosphorylation of these sites in TAK1 had differential effects on TABs; binding to TAB1 appeared to be unaffected in terms of co-precipitation, but binding of TAB4 to TAK1 was eliminated. This was seen both by co-precipitation from cell extracts and, more stringently, in an overlay assay of direct TAK1-TAB4 protein-protein interaction. Therefore, TAB4 binds to TAK1 to activate the kinase involving phosphorylation, but then TAB4 is released from the phosphorylated TAK1. We nicknamed this as a hit-and-run activation distinct from TAB1 action that involves persistent binding to activated TAK1. In assays with unstimulated 293IL-1R1 cells or mouse dermal fibroblasts we did not see co-precipitation of endogenous TAB4 with endogenous TAK1; however, at 5 min following IL-1β stimulation there was co-precipitation, which was no longer evident at 15 min after IL-1β stimulation, supporting our proposed mechanism (see the supplemental figure). Apparently the four sites of phosphorylation in the TAK1 linker region, residues 330–430, drastically reduce affinity for TAB4. We speculate that this is the region of TAK1 that binds TAB4 or at least regulates TAB4 binding.
We propose that TAB4 functions similarly to the TAB2/3 proteins to mediate activation of TAK1 through a polyubiquitin-dependent mechanism. The receptor-activated TRAFF proteins are polyubiquitinated, and these chains are thought to act as scaffolds to assemble multiprotein complexes for signaling. The TAB2/3 proteins depend on a Phe-Pro motif for polyubiquitin binding in activation of TAK1 (16). Likewise we found that residues Phe254 and Pro255 near the C terminus of TAB4 were required for stimulation of TAK1 phosphorylation and activation. We imagine the TABs act as adaptors to mediate TAK1 recruitment to polyubiquitin scaffolds. Mutation of the FP residues in TAB4 reduced co-precipitation of ubiquitinated proteins by more than half without affecting binding to PP6 (not shown). We speculate that polyubiquitin and PP6 are probably mutually exclusive TAB4 binding partners. Mutation of the FP motif disabled TAB4 stimulation of TAK1 phosphorylation of endogenous IKKβ in transfected cells and eliminated specific phosphorylation of IKKβ by TAK1-TAB1 immune complexes in an in vitro assay. Thus, without binding polyubiquitin TAB4 cannot stimulate activation of TAK1. Moreover the FP mutant TAB4 acted as a dominant negative interfering protein and blocked IL-1β stimulation of NF-κB via IKK. Thus, Phe254 and Pro255 are required for TAB4 to direct substrate specificity of TAK1 to IKKβ. We propose that TAB4 binds to TAK1 and to polyubiquitin chains to assemble complexes where TAK1 becomes phosphorylated in the linker sites. Without polyubiquitin association we speculate that TAB4 binds and holds TAK1 without an increase in phosphorylation of the linker sites, forming a complex that has reduced activity with IKK. The phosphosites in the TAK1 linker region induced by TAB4 cause release of the TAB4, might also mediate specificity for IKK, and enhance binding of IKK to TAK1, a hypothesis that can be tested in future experiments.
Expression of NF-κB and NF-κB-dependent genes occurs in inflammatory disease and cancer, and pharmaceutical intervention may be therapeutic. Mutational activation of the p52/p100 gene NFKB2 is prevalent in certain T-cell lymphomas, myelomas, and B-cell lymphomas (31–33). Amplification of the c-Rel gene, another subunit in the NF-κB family, has been detected in some B-cell lymphomas (31–33). Dysregulation of IκB is evident in some cases of Hodgkin lymphoma (31–33). Genes dependent on NF-κB activity such as c-myc, cyclin D1, MMP2, vascular endothelial growth factor, tumor necrosis factor α, IL-1, and cellular inhibitor of apoptosis proteins are involved in human cancers (31–33). Inhibition of NF-κBin tumor cell lines using a “super-repressor” form of IκB that is degradation-resistant leads to increased apoptotic cell death (31–33). We propose that TAB4 promotes NF-κB activity via IKK, and this may be critical for cell survival because depletion of TAB4 by small interfering RNA in HeLa or HEK293 cells produced extensive cell death within 48 h (not shown). Side by side this was a more drastic response than RNA interference knock-down of α-4, a dominant antiapoptotic factor in cells (29, 34). The FP-mutated version of TAB4 potently interferes with signaling from TAK1 to IKK and effectively blocked NF-κB activation by IL-1β. TAB4 offers a potential target for chemical modulation of NF-κB for therapeutic benefit.
Supplementary Material
Acknowledgments
We thank Abbi Daugherty for technical assistance and Dr. David Wotton (University of Virginia) for the FLAG-Ub plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants CA77584 (to D. L. B.), GM068812 (to J. N.-T.), and GM-37537 (to D. F. H.) from the United States Public Health Service. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
Footnotes
The abbreviations used are: IKK, IκB kinase; MAPK, mitogen-activated protein kinase; TAK1, transforming growth factor β-activated kinase-1; TAB, TAK1-binding protein; TIP, type 2A phosphatase-interacting protein; GST, glutathione S-transferase; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; PP, protein phosphatase; HA, hemagglutinin; MOPS, 4-morpholinepropanesulfonic acid; MBP, myelin basic protein; wt, wild type; FP-AA, F254A/P255A; NF-κB, nuclear factor-κB.
T. D. Prickett, J. Ninomiya-Tsuji, P. Broglie, and D. L. Brautigan, unpublished results.
References
- 1.Martin, M. U., and Wesche, H. (2002) Biochim. Biophys. Acta 1592 265–280 [DOI] [PubMed] [Google Scholar]
- 2.Sakurai, H., Shigemori, N., Hasegawa, K., and Sugita, T. (1998) Biochem. Biophys. Res. Commun. 243 545–549 [DOI] [PubMed] [Google Scholar]
- 3.Shirakabe, K., Yamaguchi, K., Shibuya, H., Irie, K., Matsuda, S., Moriguchi, T., Gotoh, Y., Matsumoto, K., and Nishida, E. (1997) J. Biol. Chem. 272 8141–8144 [DOI] [PubMed] [Google Scholar]
- 4.Pomerantz, J. L., and Baltimore, D. (2002) Mol. Cell 10 693–695 [DOI] [PubMed] [Google Scholar]
- 5.Crosetto, N., Bienko, M., and Dikic, I. (2006) Mol. Cancer Res. 4 899–904 [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay, D., and Riezman, H. (2007) Science 315 201–205 [DOI] [PubMed] [Google Scholar]
- 7.Perkins, N. D. (2007) Nat. Rev. Mol. Cell Biol. 8 49–62 [DOI] [PubMed] [Google Scholar]
- 8.Wang, C., Deng, L., Hong, M., Akkaraju, G. R., Inoue, J., and Chen, Z. J. (2001) Nature 412 346–351 [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z. J., Bhoj, V., and Seth, R. B. (2006) Cell Death Differ. 13 687–692 [DOI] [PubMed] [Google Scholar]
- 10.Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C., and Chen, Z. J. (2000) Cell 103 351–361 [DOI] [PubMed] [Google Scholar]
- 11.Shibuya, H., Yamaguchi, K., Shirakabe, K., Tonegawa, A., Gotoh, Y., Ueno, N., Irie, K., Nishida, E., and Matsumoto, K. (1996) Science 272 1179–1182 [DOI] [PubMed] [Google Scholar]
- 12.Sakurai, H., Miyoshi, H., Mizukami, J., and Sugita, T. (2000) FEBS Lett. 474 141–145 [DOI] [PubMed] [Google Scholar]
- 13.Ninomiya-Tsuji, J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z., and Matsumoto, K. (1999) Nature 398 252–256 [DOI] [PubMed] [Google Scholar]
- 14.Kishimoto, K., Matsumoto, K., and Ninomiya-Tsuji, J. (2000) J. Biol. Chem. 275 7359–7364 [DOI] [PubMed] [Google Scholar]
- 15.Takaesu, G., Kishida, S., Hiyama, A., Yamaguchi, K., Shibuya, H., Irie, K., Ninomiya-Tsuji, J., and Matsumoto, K. (2000) Mol. Cell 5 649–658 [DOI] [PubMed] [Google Scholar]
- 16.Kishida, S., Sanjo, H., Akira, S., Matsumoto, K., and Ninomiya-Tsuji, J. (2005) Genes Cells 10 447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanayama, A., Seth, R. B., Sun, L., Ea, C. K., Hong, M., Shaito, A., Chiu, Y. H., Deng, L., and Chen, Z. J. (2004) Mol. Cell 15 535–548 [DOI] [PubMed] [Google Scholar]
- 18.Besse, A., Lamothe, B., Campos, A. D., Webster, W. K., Maddineni, U., Lin, S. C., Wu, H., and Darnay, B. G. (2007) J. Biol. Chem. 282 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung, P. C., Nebreda, A. R., and Cohen, P. (2004) Biochem. J. 378 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, M. G., Katsura, K., Nomiyama, H., Komaki, K., Ninomiya-Tsuji, J., Matsumoto, K., Kobayashi, T., and Tamura, S. (2003) J. Biol. Chem. 278 12013–12021 [DOI] [PubMed] [Google Scholar]
- 21.Kajino, T., Ren, H., Iemura, S., Natsume, T., Stefansson, B., Brautigan, D. L., Matsumoto, K., and Ninomiya-Tsuji, J. (2006) J. Biol. Chem. 281 39891–39896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanada, M., Ninomiya-Tsuji, J., Komaki, K., Ohnishi, M., Katsura, K., Kanamaru, R., Matsumoto, K., and Tamura, S. (2001) J. Biol. Chem. 276 5753–5759 [DOI] [PubMed] [Google Scholar]
- 23.Di Como, C. J., and Arndt, K. T. (1996) Genes Dev. 10 1904–1916 [DOI] [PubMed] [Google Scholar]
- 24.Duvel, K., and Broach, J. R. (2004) Curr. Top. Microbiol. Immunol. 279 19–38 [DOI] [PubMed] [Google Scholar]
- 25.Jiang, Y., and Broach, J. R. (1999) EMBO J. 18 2782–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacinto, E., Guo, B., Arndt, K. T., Schmelzle, T., and Hall, M. N. (2001) Mol. Cell 8 1017–1026 [DOI] [PubMed] [Google Scholar]
- 27.Gingras, A. C., Caballero, M., Zarske, M., Sanchez, A., Hazbun, T. R., Fields, S., Sonenberg, N., Hafen, E., Raught, B., and Aebersold, R. (2005) Mol. Cell. Proteomics 4 1725–1740 [DOI] [PubMed] [Google Scholar]
- 28.McConnell, J. L., Gomez, R. J., McCorvey, L. R., Law, B. K., and Wadzinski, B. E. (2007) Oncogene 26 6021–6030 [DOI] [PubMed] [Google Scholar]
- 29.Prickett, T. D., and Brautigan, D. L. (2007) Mol. Cell. Biol. 27 4217–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefansson, B., and Brautigan, D. L. (2006) J. Biol. Chem. 281 22624–22634 [DOI] [PubMed] [Google Scholar]
- 31.Courtois, G., and Gilmore, T. D. (2006) Oncogene 25 6831–6843 [DOI] [PubMed] [Google Scholar]
- 32.Basseres, D. S., and Baldwin, A. S. (2006) Oncogene 25 6817–6830 [DOI] [PubMed] [Google Scholar]
- 33.Inoue, J., Gohda, J., Akiyama, T., and Semba, K. (2007) Cancer Sci. 98 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong, M., Fox, C. J., Mu, J., Solt, L., Xu, A., Cinalli, R. M., Birnbaum, M. J., Lindsten, T., and Thompson, C. B. (2004) Science 306 695–698 [DOI] [PubMed] [Google Scholar]
- 35.Schroeder, M. J., Webb, D. J., Shabanowitz, J., Horwitz, A. F., and Hunt, D. F. (2005) J. Proteome Res. 4 1832–1841 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.