Abstract
The conjugation of polyubiquitin to target proteins acts as a signal that regulates target stability, localization, and function. Several ubiquitin binding domains have been described, and while much is known about ubiquitin binding to the isolated domains, little is known with regard to how the domains interact with polyubiquitin in the context of full-length proteins. Isopeptidase T (IsoT/USP5) is a deubiquitinating enzyme that is largely responsible for the disassembly of unanchored polyubiquitin in the cell. IsoT has four ubiquitin binding domains: a zinc finger domain (ZnF UBP), which binds the proximal ubiquitin, a UBP domain that forms the active site, and two ubiquitin-associated (UBA) domains whose roles are unknown. Here, we show that the UBA domains are involved in binding two different polyubiquitin isoforms, linear and K48-linked. Using isothermal titration calorimetry, we show that IsoT has at least four ubiquitin binding sites for both polyubiquitin isoforms. The thermodynamics of the interactions reveal that the binding is enthalpy-driven. Mutation of the UBA domains suggests that UBA1 and UBA2 domains of IsoT interact with the third and fourth ubiquitins in both polyubiquitin isoforms, respectively. These data suggest that recognition of the polyubiquitin isoforms by IsoT involves considerable conformational mobility in the polyubiquitin ligand, in the enzyme, or in both.
Ubiquitination is a post-translational modification that consists of the covalent attachment of one or more ubiquitin molecules onto target proteins (1). Ubiquitination can alter the localization, function, stability, and interactions of the modified protein (2, 3). Ubiquitination is carried out by the sequential action of a series of enzymes, a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-ligating enzyme (E3). Monoubiquitination, the addition of one ubiquitin molecule to a target protein via an isopeptide bond between the C terminus of ubiquitin and the ε-amino group of a lysine residue on the target protein, is involved in diverse processes including receptor-mediated signaling, maintenance of chromatin, DNA repair, cell cycle, endocytosis, and many others (4, 5). Polyubiquitination occurs when additional ubiquitin molecules are conjugated to any of the seven lysine residues on ubiquitin forming polyubiquitins of different linkages (6–8). It has been shown that these different linkages result in different conformational isoforms (9, 10). These different conformations appear to signal different outcomes, the best understood being protein degradation by the proteasome utilizing K48-linked polyubiquitin chains (6, 8, 11). Polyubiquitin linked through other lysine residues have roles that may be independent of proteasomal degradation (8). In addition to isopeptide-linked polyubiquitin, linear polyubiquitin can be produced from the expression of the proubiquitin gene, a head to tail fusion of three or more ubiquitin monomers, and as recently recognized, by the action of an ubiquitin ligase complex named LUBAC (linear ubiquitin chain assembly complex) (12–15).
The molecular details of ubiquitin recognition have been described for several ubiquitin binding domains (16, 17). These domains contact different surfaces on ubiquitin, although the majority of them interact with ubiquitin through a hydrophobic patch consisting of Leu-8, Ile-44, and Val-70 on ubiquitin (17). One such ubiquitin binding domain is the ubiquitin-associated (UBA)3 domain, which is about 40–50 amino acids long (18). The structure of this domain has been described for several proteins and consists of a three-helix bundle with a hydrophobic surface centered around a conserved (M/L)G(F/Y) motif that is involved in ubiquitin recognition (19–25). In vitro binding studies have shown that isolated UBA domains can bind mono- and polyubiquitin (26). This domain is present in numerous proteins known to function in the ubiquitin proteasome system, including a deubiquitinating enzyme, Isopeptidase T (IsoT) (18, 27).
Deubiquitinating enzymes (DUBs) are proteases that hydrolyze the isopeptide bond between ubiquitin and target proteins, and between ubiquitins in polyubiquitin. IsoT is a specialized DUB responsible for the disassembly of the majority of unanchored polyubiquitin in vivo (27–29). It is a highly conserved DUB present in organisms ranging from yeast to humans (27–30). Humans have three isoforms: IsoT-S, IsoT-L, and IsoT-3 (28, 31, 32). IsoT-S (hereafter referred as IsoT) and L result from alternative splicing of the human USP5 gene, which leads to a substitution of Ala-629 in IsoT-L by 23 amino acids (Gly-629 to Ser-652) (32). IsoT-3 is encoded by a different gene, USP13, and shares 54.8% identity with IsoT-S (31). Whereas most in vitro studies aimed at understanding the substrate specificity of IsoT have been conducted using human IsoT, very little is known about the substrate specificity of the other two human isoforms (28, 33–36). IsoT disassembles at least four types of polyubiquitin isoforms: K48-, K63-, K29/K6-linked, and linear chains (26, 28). In the case of K48-linked chains, IsoT has been shown to act as an exo-isoamidase that cleaves polyubiquitin from the proximal end of the chain (the end that contains a free C terminus in unanchored chains) (28). Modification or removal of Gly-76 of the proximal ubiquitin in the chain greatly decreases the rate of hydrolysis of K48-linked polyubiquitin.
Primary structure analysis of IsoT reveals that it has four ubiquitin binding domains: a UBP (ubiquitin-specific processing protease) domain, which contains the active site residues, a ZnF UBP domain and two UBA domains (Fig. 1A) (27). The 23-amino acid insertion in IsoT-L is located between the UBP and UBA1 domains. Consistent with the primary structure, kinetic studies using K48-linked chains showed that IsoT has at least four binding sites for ubiquitin, named S1′, S1, S2, and S3 (28). The S1′ site is occupied by the proximal ubiquitin in the chain and contains a pocket that specifically recognizes the intact C terminus of ubiquitin (Fig. 1B). We have previously shown that this site is formed by the ZnF UBP domain of IsoT (37). The S1 site is the active site of the enzyme and is composed of the UBP domain of IsoT containing the characteristic Cys and His boxes (27, 28). Hydrolysis of the in vitro DUB substrate ubiquitin-amidomethyl coumarin (Ub-AMC) occurs at this site and is activated by the binding of ubiquitin to the S1′ site, Fig. 1C (35). Modification of the ubiquitin C terminus or mutation of the ZnF UBP domain of IsoT abolishes the binding of and activation by ubiquitin, suggesting that the activation site is the S1′ site of IsoT (Fig. 3A) (37). The requirement for an intact C terminus in the proximal ubiquitin for efficient hydrolysis of chains and for ubiquitin-mediated activation of Ub-AMC hydrolysis suggests that binding at the S1′ site is required for efficient hydrolysis at the S1 site (37). By definition, the S2-S3 sites are regions of IsoT predicted to bind the remaining distal ubiquitins in the chain.
FIGURE 1.
Ubiquitin binding domains of IsoT. A, IsoT has four ubiquitin binding domains: a ZnF UBP domain, a UBP domain, and two UBA domains. B, putative binding of tetraubiquitin to IsoT. The unanchored proximal ubiquitin occupies the C-terminal specificity pocket formed by the ZnF UBP domain, and the distal ubiquitins occupy the S1, S2, and S3 sites. The active site residues are located in the S1 site. C, Ub-AMC hydrolysis by IsoT is activated by ubiquitin (Ub). Ub-AMC binds to the S1 or active site, and it is hydrolyzed to ubiquitin and AMC. Unanchored ubiquitin binds the S1′ site of IsoT stimulating the hydrolysis of Ub-AMC at the S1 site.
FIGURE 3.
Linear and K48-linked polyubiquitinΔG bind predominantly across the S1-S2 sites. A, binding of monoubiquitin (180 μm in the syringe) in the absence (filled squares) and presence of a 2-fold molar excess linear di-(open squares), tri-(filled circles), and tetraubiquitinΔG(open circles) over C335A IsoT (6 μm in the cell). Monoubiquitin was injected into buffer containing C335A in the absence or presence of polyubiquitinΔG in 18–25 injections of 6 μl each. The solid line corresponds to the fitting of the data points to a single site binding model. B, binding register of linear polyubiquitinΔG. Monoubiquitin binds the S1′ site, and linear polyubiquitinΔG binds across the S1-S3 sites. C, binding of monoubiquitin in the absence (filled squares) and presence of a 2-fold molar excess K48-linked di-(open squares), tri-(filled circles), and tetraubiquitinΔG(open circles). The titration was performed as in A. The solid lines in the absence of K48-linked polyubiquitinΔG correspond to the fitting of the data points to a single site binding model. The solid line in the presence of K48-linked polyubiquitinΔG corresponds to the fitting of the data points to a two independent binding sites model. D, binding register of K48-linked polyubiquitinΔG. Monoubiquitin binds the S1′ site, and a second monoubiquitin can displace the proximal ubiquitin in K48-linked polyubiquitinΔG at the S1 site.
Here we sought to understand the role of the UBA domains of IsoT in recognizing two polyubiquitin isoforms; K48-linked and linear polyubiquitin. These two types of chains are predicted to have different tertiary structures due to the location of the linkage between the ubiquitins (8). Although several studies have focused on determining the binding specificity of isolated ubiquitin binding domains (in particular UBA domains) to polyubiquitin isoforms, very little is known about the roles of each domain in binding polyubiquitin in the context of the respective full-length proteins. Here we show for both K48-linked and linear polyubiquitin, that IsoT has at least four ubiquitin binding sites for the ubiquitin subunits of polyubiquitin, and that binding of both K48-linked and linear polyubiquitin to the S1′ site also requires an intact C terminus in the proximal ubiquitin. Using site-directed mutagenesis, we present evidence that suggests the UBA domains of IsoT interact with the distal ubiquitins in both linear and K48-linked polyubiquitin. Mutation of the UBA1 and UBA2 domains of IsoT suggests that they form the S3 and S2 sites for both polyubiquitin isoforms, respectively. Taken together, we show that all four ubiquitin binding domains of IsoT coordinate the binding of two polyubiquitin isoforms, whereby the ZnF UBP, the UBP, UBA2, and UBA1 domains form the S1′, S1, S2, and S3 sites, respectively.
EXPERIMENTAL PROCEDURES
Expression and Purification of IsoT—To simplify purification, a hexahistidine (His6) tag was introduced at the C terminus of human IsoT. The gene encoding IsoT was PCR-amplified from the pRSIsoT vector, and a His6 tag was introduced in the 3′-end of the coding sequence (28). The amplified DNA was subcloned between the NdeI site and the HindIII site of pRSETB (Invitrogen). Plasmid DNA encoding full-length His6-tagged IsoT (pRSIsoT His6) was mutagenized using the QuikChange Site-directed Mutagenesis kit (Stratagene). The point mutants were confirmed by sequencing and expressed in BL21 (DE3) Escherichia coli cells as described for wild type (WT) IsoT (38). WT, active site, and UBA domain mutant IsoTs were purified using Ni-NTA (nitrilotriacetic acid)-agarose resin (Qiagen) followed by a ubiquitin affinity column as previously described (37, 38). R221A IsoT was purified using Ni-NTA agarose resin only, because binding to ubiquitin was defective.
Ub-AMC Hydrolysis—Enzymatic activity was measured by incubation of 2 μm Ub-AMC with 8 nm purified WT, M643E M711E, or D435A IsoT at 37 °C in 50 mm Tris-HCl, pH 7.5, 1 mm dithiothreitol, 10 μg/ml ovalbumin. The reaction was monitored by following the increase in fluorescence at 440 nm (λex = 340 nm) for 50 s after the addition of IsoT on an AMINCO-BOWMAN Series 2 Luminescence Spectrometer at 37 °C. The activation of Ub-AMC hydrolysis by ubiquitin was measured by the addition of 492 nm ubiquitin to the Ub-AMC reaction and following the increase of fluorescence for 50 s.
Subcloning, Expression, and Purification of Linear Polyubiquitin—To create linear diubiquitin, the ubiquitin coding sequence was excised from a pRSET ubiquitin Csp6I cloning vector by digestion with NdeI and Csp6I. This vector contains a single Csp6I site at the 3′-end of the ubiquitin coding sequence. The excised ubiquitin coding sequence was ligated into a NdeI-digested pRSET ubiquitin vector. Tri- and tetraubiquitin were created similarly by ligating the excised ubiquitin coding sequence from pRSET ubiquitin Csp6I vector to a NdeI-digested pRSET diubiquitin and triubiquitin vectors, respectively. Linear polyubiquitin lacking the C-terminal glycine (polyubiquitinΔG) was made by first mutating the pRSET ubiquitin vector using the QuikChange Site-directed Mutagenesis kit to delete Gly-76, creating pRSET ubiquitinΔG. The ubiquitin coding sequence from pRSET ubiquitin Csp6I was ligated to NdeI-digested pRSET ubiquitinΔG yielding linear diubiquitinΔG. Linear tri- and tetraubiquitinΔG were subcloned similarly to WT chains except that the ubiquitin coding sequence was ligated into a NdeI-digested pRSET diubiquitinΔG and triubiquitinΔG vector, respectively. All newly created vectors were verified by sequencing. Linear polyubiquitin was expressed in BL21(DE3) cells and purified as previously described for the yeast ubiquitin proprotein (39).
Synthesis and Purification of K48- and K63-linked Polyubiquitin—WT K48 and K63-linked diubiquitin were synthesized by conjugating WT ubiquitin as described by Cook et al. and Komander et al., respectively (40, 41). K48-linked polyubiquitinΔG was synthesized using the same method described by Raasi and Pickart (42). WT, K48C, D77 ubiquitin, and ubiquitinΔG were expressed in bacteria and purified as previously described (42). K48-linked diubiquitinΔG was prepared by conjugating K48C ubiquitin to ubiquitinΔG using recombinant E2–25K and E1 (42). To create K48-linked triubiquitinΔG, the distal ubiquitin in K48-linked diubiquitinΔG was deblocked by alkylation with ethyleminine. The third ubiquitin was added by conjugation of K48C ubiquitin to alkylated K48-linked diubiquitinΔG using recombinant E2–25K and E1. To synthesize K48-linked tetraubiquitinΔG, K48-linked diubiquitin D77 was made as described by Raasi and Pickart (42), followed by deblocking the proximal ubiquitin by removal of Asp-77 with UCH-L3. The C-terminally deblocked K48-linked diubiquitin was conjugated to alkylated K48-linked diubiquitinΔG to create K48-linked tetraubiquitinΔG. All the chains were purified to near homogeneity by FPLC as previously described (42).
Isothermal Titration Calorimetry (ITC)—WT, mutant IsoT, monoubiquitin, and polyubiquitin chains were dialyzed extensively against 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10 mm β-mercaptoethanol. ITC measurements were carried out in triplicate on a MicroCal VP-ITC instrument at 30 °C. Monoubiquitin or polyubiquitin was injected into 1.8 ml of buffer containing WT or mutant IsoT. Binding constants were calculated by fitting the data using Origin 7.0 (OriginLab Corp.). WT and mutant IsoT concentrations were determined using an ε280 of 86,290 cm-1 m-1 (43). Ubiquitin concentrations were determined by HPLC using a ubiquitin standard.
RESULTS
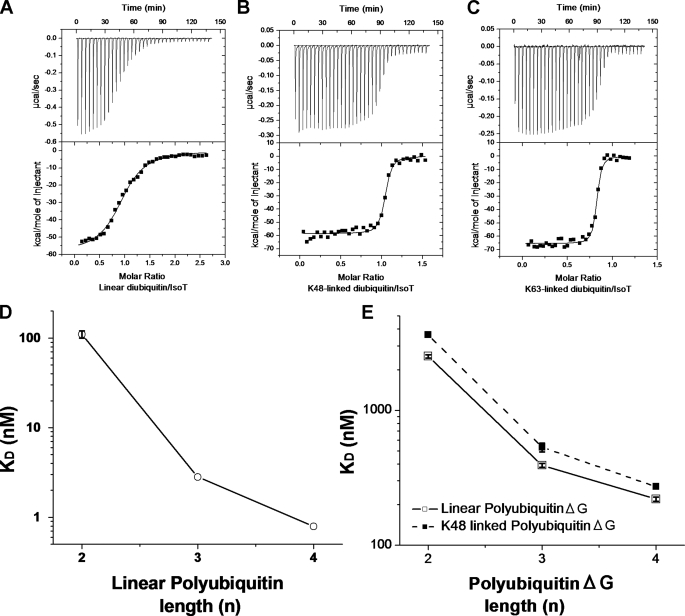
Binding of Polyubiquitin—To address the role of the UBA domains in the binding of polyubiquitin isoforms, we first characterized the binding of polyubiquitin to IsoT. For these experiments, we mutated the active site cysteine of IsoT, Cys-335, to alanine to prevent the hydrolysis of the chains. The binding of linear, K48-, and K63-linked diubiquitin to C335A IsoT, as measured by ITC, revealed that although IsoT can hydrolyze these substrates, the enzyme binds them with different affinities (supplemental Table S1 and Fig. 2, A–C). The binding of K48- and K63-linked diubiquitin was too tight to accurately measure by ITC. However, we are able to estimate that IsoT binds K48- and K63-linked diubiquitin with at least 50-fold tighter affinity (KD ≤ 2 nm) than linear diubiquitin (KD = 110 ± 20 nm). The binding affinities for linear triubiquitin and tetraubiquitin sharply increased compared with linear diubiquitin, resulting in binding affinities too tight, KD ≤ 2 nm, to accurately measure by ITC (supplemental Table S2 and Fig. 2D). These binding data suggest that IsoT has multiple ubiquitin binding sites for linear polyubiquitin.
Length of the Polyubiquitin Binding Site for Linear and K48-linked PolyubiquitinΔG—Because the KD for the binding of polyubiquitin to IsoT was too low to be accurately determined by ITC, we took advantage of the fact that deletion of the C-terminal glycine of the proximal ubiquitin greatly reduces its affinity for the S1′ site. Thus, we generated mutant polyubiquitin lacking Gly-76 in the proximal ubiquitin (polyubiquitinΔG) to decrease their binding affinities for IsoT. Given that IsoT exhibits different binding affinities for linear and K48-linked diubiquitin, and that these two types of polyubiquitin are predicted to have different three-dimensional structures, we focused on determining the binding properties of IsoT for linear and K48-linked chains of different length. As predicted, linear and K48-linked polyubiquitinΔG bind IsoT with lower affinity than WT chains (Fig. 2, D and E and supplemental Tables S2 and S3). The binding affinity increases about 6-fold upon lengthening of the chain from linear diubiquitinΔG (2510 ± 70 nm) to linear triubiquitinΔG (390 ± 10 nm), and less than 2-fold from linear triubiquitinΔG to linear tetraubiquitinΔG (219 ± 8 nm), suggesting that IsoT has at least three ubiquitin binding sites for linear polyubiquitinΔG. While K48-linked diubiquitin binds to C335A IsoT with greater affinity than linear diubiquitin, K48-linked polyubiquitinΔG chains bound with similar affinity as linear polyubiquitinΔG (Fig. 2E and supplemental Table S3). The simplest explanation for the similar binding affinities of the different polyubiquitinΔG isoforms is that both chains bind in a similar manner to the S1-S3 sites. However, we were concerned that there may be differences in the binding register occupied by the chains; i.e. while both linear and K48-linked diubiquitin bind across the S1′-S1 sites because of the requirement for an intact C terminus for the binding to the S1′ site, the binding register for linear or K48-linked polyubiquitinΔG is not defined by this experiment.
FIGURE 2.
Binding of linear, K48-, and K63-linked polyubiquitin. A, binding of linear diubiquitin (91 μm in the syringe) to C335A IsoT (2 μm in the cell) as measured by ITC (as described under “Experimental Procedures”). B, binding of K48-linked diubiquitin (40 μm in the syringe) to C335A IsoT (1.5 μm in the cell). C, binding of K63-linked diubiquitin (30 μm in the syringe) to C335A IsoT (1.5 μm in the cell). Linear, K48- or K63-linked diubiquitin were injected into 1.8 ml of buffer containing C335A IsoT in 35 injections of 3 μl each. The solid line corresponds to the fitting of the data points in A–C (filled squares) to a single site binding model. D, C335A IsoT binding affinities for linear polyubiquitin chains of increasing length. Error bars correspond to the S.D. of three measurements. E, binding affinities of linear (open squares) and K48-linked (filled squares) polyubiquitinΔG by C335A IsoT. Error bars correspond to the S.D. of three measurements.
PolyubiquitinΔG Binds across the S1-S3 Sites—To determine the binding register for linear and K48-linked polyubiquitinΔG, we took advantage of the fact that IsoT binds ubiquitin tightly. WT IsoT binds one ubiquitin at the S1′ site with a binding constant of ∼600 nm (supplemental Table S4 and Fig. S1A). Surprisingly, C335A IsoT binds two ubiquitins with a KD ∼ 20–30 nm, (supplemental Table S4 and Fig. S1A), one at the S1′ site as shown for WT IsoT and another at an undefined site. The second site appears to be the S1 site, as covalent attachment of ubiquitin aldehyde (Ubal) to the S1 site of IsoT competes for the binding of one ubiquitin and significantly increases the binding affinity at the S1′ site (KD = 6.8 ± 0.9 nm, supplemental Table S4 and Fig. S1A). Further, ubiquitinΔG does not bind if Ubal is present (arguing again that the second site is the S1 site, supplemental Fig. S1B).
If polyubiquitinΔG occupies the S1-S3 sites of IsoT, then monoubiquitin should bind only one site, the S1′ site, when polyubiquitinΔG is prebound to C335A IsoT. Thus, we titrated monoubiquitin into C335A IsoT that was prebound to linear or K48-linked polyubiquitinΔG of increasing lengths. As shown in Fig. 3A, titration of monoubiquitin into IsoT prebound to either linear di-, tri-, or tetraubiquitinΔG results in the binding of only one monoubiquitin with a dissociation constant of about 100 nm (supplemental Table S5). Titration of ubiquitinΔG (which cannot bind to the S1′ site) into C335A prebound to linear triubiquitinΔG resulted in no detectable binding (data not shown). These data suggest that the monoubiquitin binds in the S1′ site, whereas linear polyubiquitinΔG simultaneously binds across the S1-S3 sites (Fig. 3B).
Unlike linear polyubiquitinΔG, titration of monoubiquitin into preformed C335A IsoT-K48-linked polyubiquitinΔG complex resulted in the binding of two monoubiquitin molecules (Fig. 3C and supplemental Table S6). One monoubiquitin bound with a KD of about 60 nm, whereas the second ubiquitin bound with a KD depending on the length of the polyubiquitin in the preformed complex (70 ± 10 nm, 380 ± 60 nm, and 310 ± 50 nm for K48-linked di-, tri-, and tetraubiquitinΔG, respectively). Because the apparent KD of the second ubiquitin increases from K48-linked diubiquitinΔG to tri- and tetraubiquitinΔG this suggests that the second monoubiquitin is competing with polyubiquitin for binding to the S1 site of IsoT (Fig. 3D). The thermodynamic parameters support this explanation because they reveal that the binding of the second ubiquitin at the S1 site is entirely entropy-driven (ΔH ∼ 0, ΔS ∼ 30 cal mol-1 K-1, see supplemental Table S6), consistent with the idea that there is no net increase in the number of ubiquitin subunits bound upon adding ubiquitin to IsoT preloaded with polyubiquitinΔG (44). Similarly there is only a modest increase in entropy meaning that most of the chain remains constrained (bound) upon addition of ubiquitin. Thus, S1′ is always occupied indicating there is only one predominant binding register (S1-S3), and the proximal ubiquitin in K48-linked polyubiquitinΔG is displaced by the binding of monoubiquitin to the S1 site (Fig. 3D).
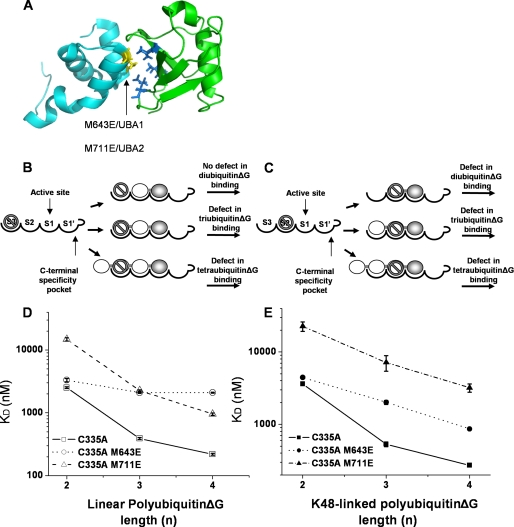
The UBA Domains of IsoT Form the S2 and S3 Sites—We know from previous work that the ZnF UBP domain forms the S1′ site of IsoT and that the S1 site is formed by the UBP domain (37, 45). Given that IsoT binds to four ubiquitin subunits in polyubiquitin, we predict that the UBA domains are involved in forming the S2 and S3 sites. To test the role of the UBA domains in forming the S2 and S3 sites, we mutated conserved UBA residues implicated in the recognition of ubiquitin by UBA domains (Fig. 4A) (17, 19, 21, 22, 24, 46). Affirming the soundness of this approach, we note that the UBA domains of IsoT are nearly identical in structure to all other UBA domains (PDB accession codes 2DAG for UBA1 and 2DAK for UBA2) (17). Because the interaction between ubiquitin and UBA domains is predominantly hydrophobic, Met-643 from UBA1 and Met-711 from UBA2 were mutated to glutamic acid to introduce a charged residue that should disrupt the interaction. We individually tested the role of each of the UBA domains of IsoT by measuring the binding of linear and K48-linked di-, tri- and tetraubiquitinΔG to C335A/M643E IsoT and C335A/M711E IsoT. If we mutate the UBA domain of IsoT that forms the S3 site, the prediction is that this mutant IsoT would be defective in binding tri- and tetraubiquitinΔG, but not diubiquitinΔG (Fig. 4B). Likewise, if the UBA domain of IsoT that forms the S2 site is mutated, it should be defective in di-, tri-, and tetraubiquitinΔG binding (Fig. 4C).
FIGURE 4.
UBA1 and UBA2 form the S3 and S2 sites of IsoT. A, structure of the DSK2 UBA domain (cyan) bound to ubiquitin (green). The PDB accession code is 1WR1. A conserved residue in the UBA domain that contacts the hydrophobic patch of ubiquitin (blue) was mutated to glutamic acid in each UBA domain. This residue corresponds to Met-643 in UBA1 and Met-711 in UBA2 of IsoT (yellow). B and C, testing the role of the UBA domains in forming the S2 and S3 sites. Mutation of the S3 site should lead to defects in the binding to tri- and tetraubiquitinΔG but not to diubiquitinΔG. Defects in the S2 site should lead to a defect in di-, tri-, and tetraubiquitinΔG binding. D, binding of linear polyubiquitinΔG to C335A, C335A/M643E and C335A/M711E IsoT (open squares, circles, and triangles, respectively). Error bars correspond to the S.D. of three measurements. E, binding of K48-linked polyubiquitinΔG to C335A, C335A/M643E, and C335A/M711E IsoT (filled squares, circles, and triangles, respectively). Error bars correspond to the S.D. of three measurements.
As shown in Fig. 4D and supplemental Tables S3 and S7, upon mutation of UBA1 (C335A/M643E) IsoT binds to linear diubiquitinΔG with a similar affinity as C335A IsoT (3300 ± 200 nm and 2510 ± 70 nm, respectively). In contrast, it binds to both linear tri- and tetraubiquitinΔG with ∼5- and 10-fold weaker affinity. Likewise, C335A/M643E binding to K48-linked diubiquitinΔG was not defective, but was defective for K48-linked tri- and tetraubiquitinΔG (Fig. 4E and supplemental Tables S3 and S7). These data are consistent with UBA1 contacting the ubiquitin subunits positioned at the S3 site in both polyubiquitin isoforms. Although the C335A/M643E IsoT was also deficient in the binding K48-linked tri- and tetraubiquitinΔG, the magnitude of the defect was smaller, 4- and 3-fold, respectively (Fig. 4D and supplemental Tables S3 and S7). These data suggest that residue Met-643 of UBA1 makes different contacts with the ubiquitin positioned at the S3 site for each polyubiquitin isoforms. Unlike the UBA1 domain mutant, the UBA2 mutation (C335A/M711E) exhibited a decrease in the binding affinity of all chain lengths tested; di-, tri-, and tetraubiquitinΔG, consistent with a role for UBA2 in forming the S2 site of IsoT (Fig. 4, D and E and supplemental Tables S3 and S7). Also, unlike the UBA1 mutation, C335A/M711E IsoT exhibited greater defects in binding K48-linked chains in comparison to linear chains. C335A/M711E IsoT bound linear di-, tri-, and tetraubiquitinΔG with ∼6-, 6-, and 4-fold defects, respectively. In comparison, it bound K48-linked di-, tri-, and tetraubiquitinΔG with ∼6-, 13-, and 12-fold weaker affinity, respectively. These data demonstrate a role for the UBA domains in binding the distal ubiquitins in linear and K48-linked polyubiquitin.
The UBA Domains of IsoT Do Not Contribute to the S1′ and/or S1 Site—Although the UBA domains have a role in forming the S2-S3 sites, it does not exclude a role for the UBA domains in contacting the ubiquitins at the S1′-S1 sites. To test whether the UBA domains contact the ubiquitins at the S1′ and/or S1 site, we examined the effects of mutating both UBA domains on the hydrolysis of Ub-AMC in the presence or absence of ubiquitin. Simultaneous mutation of both UBA domains (M643E/M711E IsoT) did not significantly alter Ub-AMC hydrolysis, suggesting that the UBA domains of IsoT are not forming the S1 site of IsoT (Fig. 5A). In contrast, mutation of residue Asp-435 to alanine in the UBP domain causes a defect in the hydrolysis of Ub-AMC (Fig. 5A). Based on the structure of other UBP domains, Asp-435 is predicted to form a hydrogen bond with the backbone amide group of Leu-73 of ubiquitin (47, 48). To test if the UBA domains of IsoT contribute to the S1′ site, we measured Ub-AMC hydrolysis in the presence or absence of monoubiquitin (35, 37). The mutant UBA domain IsoT did not show a defect in activation by ubiquitin, suggesting that these two domains are not part of the S1′ site. The UBP domain mutant did not exhibit measurable Ub-AMC hydrolysis even in the presence of free ubiquitin (Fig. 5A).
FIGURE 5.
The UBA domains do not form the S1′-S1 sites. A, hydrolysis of Ub-AMC and ubiquitin modulation of activity by WT, M643E/M711E, and D453A IsoT. WT and mutant IsoT, 8 nm, were incubated with 2 μm Ub-AMC for 50 s in the absence (white bars) and presence of 492 nm ubiquitin (gray bars). Error bars correspond to the S.D. of three measurements. B, binding of monoubiquitin to C335A IsoT (filled squares), C335A/M643E/M711E IsoT (open squares), R221A./C335A IsoT (filled circles), and C335A/D435A IsoT (open triangles). The concentration of ubiquitin in the syringe for the C335A IsoT (2 μm in the cell), C335A/M643E/M711E IsoT (1.57 μm in the cell), and C335A/D435A IsoT (2 μm in the cell) was 91 μm and for R221A/C335A IsoT (2 μm) was 154 μm. The titration was done as in Fig. 2. The solid lines correspond to the fitting of the data points to a single site binding model.
Further, C335A/M643E/M711E IsoT binds two monoubiquitin molecules with similar affinity as C335A IsoT (30 ± 2 nm) (Fig. 5B and supplemental Table S4). In contrast, C335A/D435A IsoT binds ubiquitin with a stoichiometry of one instead of two as determined for C335A IsoT (Fig. 5B and supplemental Table S4). This is consistent with a model in which the defect in activity observed for D435A IsoT is due to a defect in the binding of ubiquitin to the S1 site. An active site mutant having an additional mutation in the ZnF UBP domain (R221A/C335A IsoT), which attenuates monoubiquitin binding at the S1′ site, shows no detectable ubiquitin binding (Fig. 5B). This suggests that binding the S1′ site is required for monoubiquitin binding to the S1 site. These data indicate that binding of ubiquitin to the S1′ site is coupled to binding of ubiquitin to the S1 site in C335A IsoT.
We also found that linear diubiquitinΔG interacts with C335A/D435A IsoT with approximately a 2.4-fold defect in affinity (supplemental Table S8). These data clearly indicate a role for the S1 site in the binding of diubiquitinΔG, and that the defect observed is due to a disruption in the S1-ubiquitin interaction. This defect is not as strong as that which we observe with the S1′ mutation, with regard to binding of monoubiquitin to the S1 site, suggesting that the S1 site may not be coupled to the UBA domains in a manner similar to its interaction with the S1′ site.
Taken together, the UBA domain mutant data indicate that the UBA domains interact with the distal ubiquitin in the chains forming the S2 and S3 sites, and they do not modulate the binding of ubiquitin at the S1′-S1 sites.
DISCUSSION
Binding of Polyubiquitin Chain Isoforms to IsoT—Although it was previously known that IsoT can hydrolyze at least four different polyubiquitin isoforms, no detailed quantitative measure of the binding of polyubiquitin isoforms to IsoT had yet been described. The binding affinities reported here for two polyubiquitin isoforms reveal that IsoT binds K48-linked diubiquitin at least 50-fold tighter than linear diubiquitin. Removing Gly-76 from the C terminus of either form of polyubiquitin results in a decreased affinity for the chains, indicating that the proximal ubiquitin of both polyubiquitin isoforms contact the S1′ site. This requirement suggests that WT dimers bind across the S1′–S1 sites, indicating that both the ZnF UBP (S1′ site) and the UBP domain (S1 site), are responsible for the differential recognition between these two types of chains. In addition, the affinity for linear diubiquitin is weaker than for monoubiquitin, suggesting that a strain is induced upon binding of the linear chain. In contrast, the K48-linked diubiquitin binds with greater affinity than monoubiquitin suggesting that the K48-linked dimer adopts a more favorable conformation. The discrimination between the chains may be due to at least two different factors: the type of linkage, peptide versus isopeptide, between the ubiquitin subunits in linear and K48-linked chains and/or differences in conformation due to the spatial orientation of the ubiquitin in the different chains. Although linear and K63-linked diubiquitin adopt similar three-dimensional structures,4 IsoT interacts with them with different affinities. This suggests that IsoT may be discriminating between peptide and isopeptide-linked polyubiquitin. These data also suggest that the type of linkage connecting the ubiquitin subunits in polyubiquitin may also influence the discrimination between linear and K48-linked chains. K48- and K63-linked chains might display higher intrinsic flexibility required for optimal binding to IsoT, because of the greater rotational freedom and/or lower steric hindrance of the isopeptide bond versus the peptide bond.
Linear tri- and tetraubiquitin exhibit dramatically increased affinity to IsoT compared with linear diubiquitin, indicating that at least four ubiquitin binding sites contribute to the binding of linear polyubiquitin. These data show directly for the first time that IsoT has at least four ubiquitin binding sites for two common polyubiquitin isoforms.
Examination of the thermodynamic parameters calculated from the interaction between monoubiquitin and polyubiquitin with IsoT reveals that this interaction is characterized primarily by a large increase in enthalpy at the expense of a decrease in entropy (Fig. 6). Note that all the measured thermodynamic parameters fall on a straight line with a slope near the experimental temperature. This behavior has been called entropy-enthalpy compensation and is often attributed to the fact that, in order to make a new bond upon binding, conformational mobility is restricted and solvent molecules must be liberated causing a large decrease in entropy (44, 49). The binding affinities observed here are almost entirely enthalpy-driven indicating that a significant gain in enthalpy accompanies each binding event. Previous studies correlate this type of thermodynamic behavior with a decrease in the internal motion of both ligand and the receptor, in this case polyubiquitin and IsoT, respectively, which results in a decreased entropy (49).
FIGURE 6.
Correlation of the binding thermodynamic parameters that demonstrate enthalpy-entropy compensation in the binding of ubiquitin and polyubiquitin to IsoT. The square symbols correspond to the binding of monoubiquitin to IsoT. The circular symbols correspond to the binding of polyubiquitin to IsoT. The 0 in the square data symbols indicate the binding of monoubiquitin to either WT, C335A, or C335A/M643E/M711E IsoT. The letter A corresponds to the binding of monoubiquitin to WT IsoT-Ubal complex. The numbers 2, 3, and 4 in the square data symbols correspond to the binding of monoubiquitin to C335A IsoT complexed with either linear or K48-linked polyubiquitinΔG of 2, 3, or 4 ubiquitin subunits long, respectively. The numbers in the circular data symbols indicate the number of ubiquitin subunits (2, 3, and 4) in the polyubiquitin titrated into a solution containing IsoT. Error bars correspond to the S.D. of three measurements.
The binding of polyubiquitinΔG to the S1-S3 sites reveals a lack of discrimination between linear and K48-linked polyubiquitinΔG. Unlike WT chains, linear and K48-linked polyubiquitinΔG bind with the same affinity. This lack of discrimination is linked to the binding register occupied by polyubiquitinΔG. While WT chains bind across the S1′-S3 sites, linear and K48-linked polyubiquitinΔG predominantly bind across the S1-S3 sites. Interestingly, while monoubiquitin cannot compete for binding at the S1 site with linear polyubiquitinΔG, it can compete with K48-linked polyubiquitinΔG. Based on the thermodynamics of the binding of monoubiquitin to IsoT preloaded with excess K48-linked polyubiquitinΔG, it appears that this competition represents the displacement of only the proximal ubiquitin in the K48-linked polyubiquitinΔG while the remainder of the chain is bound to the S2-S3 sites.
The binding specificity of the four ubiquitin binding sites is in agreement with a predominant role for the S1′-S1 sites in discriminating between the polyubiquitin isoforms and suggests that the remaining ubiquitin binding sites do not contribute significantly to the differences in the binding of polyubiquitin.
Role of the UBA Domains in Polyubiquitin Recognition—Previous studies investigating the ubiquitin binding properties of UBA domains have focused predominantly on the binding characteristics of the isolated UBA domains. In one study, the two isolated UBA domains of IsoT were shown to have different binding properties (26). The isolated UBA1 failed to bind to either monoubiquitin or polyubiquitin, while UBA2 bound monoubiquitin and polyubiquitin with a preference for the later. However, the binding properties of UBA domains can differ when in the context of the host protein environment (26). Here we show that both UBA domains of IsoT are involved in polyubiquitin recognition, such that each one of them interacts with the distal ubiquitin subunits in the chain. Mutation of UBA1 results in defects in the binding to linear and K48-linked tri- and tetraubiquitinΔG, but not in the binding of both types of diubiquitinΔG. Because the binding of these two types of chains occurs predominantly across the S1-S3 sites, these data are consistent with UBA1 forming the S3 site of IsoT, and thus being responsible for the interaction of the penultimate ubiquitin in tetraubiquitinΔG. Mutation of UBA2 results in defects in the binding of di-, tri-, and tetraubiquitinΔG, indicating that this domain forms the S2 site of IsoT, and interacts with the second ubiquitin in tetraubiquitinΔG. Because similar binding defects are observed with each UBA domain mutant regardless of the chain linkage, we conclude that the UBA domains form the same sites for both polyubiquitin isoforms. However, the magnitudes of the defects are different depending on the type of ubiquitin linkage. The UBA1 mutant has a greater defect in binding linear polyubiquitinΔG than K48-linked polyubiquitinΔG, while the UBA2 mutant exhibits opposite behavior. These differences could arise from differences in the contacts made by the methionine in the MGF motif of the UBA domains of IsoT that were mutated relative to the ubiquitin subunits in the polyubiquitin isoforms. Structural studies would be needed to determine how these two methionines interact with the ubiquitins in the polyubiquitin isoforms.
UBA domains bind ubiquitin through the Leu-8, Ile-44, Val-70 hydrophobic patch on ubiquitin, and this interaction is also conserved in other UBA domains known to bind K48- and K63-linked polyubiquitin (24, 25). Given that Met-1 and Lys-63 are spatially adjacent to each other, linear diubiquitin adopts a three-dimensional structure similar to the one observed for K63-linked diubiquitin.4 K63-linked polyubiquitin adopts an “open conformation” where the hydrophobic patch residues are solvent-exposed and readily available to interact with the UBA domains (9). In contrast, K48-linked polyubiquitin can adopt two predominant conformations in solution: an “open” conformation in which the hydrophobic patch of ubiquitin is exposed or a “closed” conformation in which the hydrophobic patch is sequestered at the ubiquitin/ubiquitin interface (10, 40, 50, 51). The lack of discrimination between the two polyubiquitinΔG isoforms by IsoT could be explained by the stabilization of the open conformation in K48-linked polyubiquitin by the interaction of each UBA domain with the hydrophobic patch of the ubiquitin subunits in the chain. This would be analogous to the interaction of the ubiquitin-interacting motifs (UIM) of S5a with K48- and K63-linked diubiquitin, where each UIM domain contacts one ubiquitin at a time in the chain regardless of the type polyubiquitin linkage (52). This is in agreement with the larger decrease in entropy observed upon binding of C335A IsoT to linear polyubiquitinΔG in comparison with K48-linked polyubiquitinΔG (supplemental Table S3). The binding of linear polyubiquitinΔG by IsoT is less entropically favored than is K48-linked polyubiquitinΔG because the binding results in the displacement of solvent molecules from the hydrophobic patch of ubiquitin upon binding to the UBA domains. The binding of the K48-linked polyubiquitinΔGto IsoT is more entropically driven because fewer solvent molecules are displaced upon binding to the UBA domains.
All Four Ubiquitin Binding Domains of IsoT Coordinate the Binding of K48-linked and Linear Polyubiquitin—Our data support a model in which all of the four ubiquitin binding domains coordinate the binding and hydrolysis of chains. As shown in Fig. 7, each domain has a specific role in polyubiquitin recognition; in both linear and K48-linked polyubiquitin, the ZnF UBP domain interacts with the proximal ubiquitin in the chain, the UBP domain interacts with the second ubiquitin from the proximal end, UBA2 interacts with the third ubiquitin, and UBA1 with the fourth ubiquitin. Although there is no structure of the UBP domain of IsoT, the structures of six different UBP domains unliganded and/or bound to ubiquitin or the active site inhibitor Ubal have been described (41, 47, 48, 53, 54). In five of the structures, the UBP domain adopts a fold that is highly conserved and consists of three domains named Finger, Palm, and Thumb, which together resemble a right hand. The structure of the CYLD UBP domain diverges in that it lacks a Finger domain (41). The surface that contacts ubiquitin is highly conserved; the C terminus of ubiquitin sits in a groove formed by the junction between the Palm and the Thumb domains, and the globular body of ubiquitin contacts the Finger, and Palm-Thumb scaffold. K48 of ubiquitin, which participates in polyubiquitin formation, is oriented toward α-helix 5 of the Thumb domain with its side chain partially solvent-exposed (47, 48, 54). Met-1, which is linked to Gly-76 in distal ubiquitins in linear polyubiquitin, is oriented toward the Finger domain and is fully solvent-exposed (47, 48, 54). The results of the current study identified two additional ubiquitin binding sites formed by the UBA domains of IsoT and that are distally located from the ubiquitin binding site at the UBP domain (S1 site). Sequence alignments predict that the UBA domains of IsoT would be inserted in the loop connecting β9 with β10 in the Palm domain of USP7 (Fig. S2). This region varies among UBP domains. Although the UBP domains of USP2 and USP8 do not have insertions in this region, USP14 and its yeast homolog, UBP6, contain an insertion of α-helices between β9 and β10, while CYLD has an insertion of a zinc binding domain that resembles a B-box (41, 47, 53, 54). For the third ubiquitin in polyubiquitin to bind UBA2 of IsoT (S2 site), the UBA domain must be spatially adjacent to the junction between the Finger and Thumb domains. Interestingly, this region varies among UBP domains. Three different UBP domain structures have an insertion between α5 and α6 helices of the Thumb domain (Fig. S2) (47, 53, 54). Two of them have an α-helix, while one UBP domain contains a disordered loop (47, 53, 54). These insertions could interfere with an ubiquitin molecule located distal to the ubiquitin bound to the UBP domain in K48-linked polyubiquitin. Sequence alignments suggest that IsoT also has an insertion in this region, and this insertion could help position the UBA domains in the right orientation to bind the third ubiquitin in tetraubiquitin (supplemental Fig. S2). Because the UBA1 of IsoT forms the S3 site, it should be positioned spatially adjacent to UBA2 to be able to contact the fourth ubiquitin in the chain. The presence of linker regions between UBA1 and 2 and between the UBA domains and the UBP domain should allow enough flexibility for the interaction with linear and K48-linked polyubiquitin to occur with similar affinities. In addition, polyubiquitin chains are known to adopt multiple conformations in solution due to the flexibility of the ubiquitin C terminus (9, 10, 50, 51). The combination of these two properties may allow IsoT to bind and hydrolyze multiple polyubiquitin isoforms.
FIGURE 7.
Model for the role of each ubiquitin binding domain in polyubiquitin binding by IsoT. The S1′, S1, S2, and S3 sites are formed that the ZnF UBP, UBP, UBA2, UBA1, respectively. The solid and dashed lines correspond to a schematic representation of IsoT. The solid lines correspond to domains of IsoT for which currently there are structural models. The dashed lines indicate regions of IsoT for which currently there are no structural models. These regions are the N terminus of IsoT, residues that link the ZnF UBP domain to the UBP domain, residues that link the UBA domains to the UBP domain, and, finally, residues that link the UBA domains to each other. N and C correspond to the N and C terminus of IsoT, respectively.
Humans have three IsoT isoforms; however, it remains to be tested whether the polyubiquitin binding properties described for IsoT also apply to IsoT-L and IsoT-3. Because IsoT differs from IsoT-L by the substitution of Ala-629 in IsoT with 23 amino acids, Gly-629 to Ser-652 in IsoT-L, it is possible that these differences may affect the binding to polyubiquitin isoforms. This insertion is located at the junction between the UBP domain and UBA1 and it could affect the binding of the fourth ubiquitin in the chain because the additional residues may make IsoT-L more flexible than IsoT. IsoT-3 is the product of a different gene that shares 54.8% identity with IsoT. Although IsoT-3 contains all four putative ubiquitin binding domains (ZnF UBP, a UBP, and two UBA domains) present in IsoT, it has not been shown to hydrolyze polyubiquitin (31). It remains to be tested whether the four putative ubiquitin binding domains of IsoT-3 bind ubiquitin and can interact with polyubiquitin similar to IsoT.
The recognition of polyubiquitin of four or more subunits by the four ubiquitin binding domains of IsoT may allow this specialized deubiquitinating enzyme to efficiently process polyubiquitin isoforms that are long enough to inhibit the proteasome or that could compete for binding to other cellular polyubiquitin receptors (55). In the case of proteasomal inhibition, this would be very important physiologically, because protein degradation by the proteasome is an essential cellular function. Similar inhibition of other ubiquitin receptors is likely, and it is probably important to keep the levels of all unanchored chains low. The presence of four ubiquitin binding domains in IsoT may have optimized this enzyme to accomplish this function.
Supplementary Material
Acknowledgments
We thank Dale E. Edmondson and James E. Mullally for critically reading the manuscript and Xiaodong Cheng for helpful discussions. We would also like to thank Maxim Balakirev for generously providing the E1 enzyme used to make the chains.
This work was supported, in whole or in part, by National Institutes of Health Ruth Kirschstein Predoctoral Award GM075426 (to F. E. R.-T.) and Grant GM30308 (to K. D. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S8.
Footnotes
The abbreviations used are: UBA, ubiquitin-associated; IsoT, Isopeptidase T; DUB, deubiquitinating enzymes; WT, wild type; ITC, isothermal titration calorimetry; PDB, Protein Data Bank; UBP, ubiquitin-specific processing protease; ZnF, zinc finger.
D. Komander, F. Reyes-Turcu, K. D. Wilkinson, and D. Barford, manuscript in preparation.
References
- 1.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425-479 [DOI] [PubMed] [Google Scholar]
- 2.Pickart, C. M. (2001) Mol. Cell 8 499-504 [DOI] [PubMed] [Google Scholar]
- 3.Daniel, J. A., Torok, M. S., Sun, Z. W., Schieltz, D., Allis, C. D., Yates, J. R., 3rd, and Grant, P. A. (2004) J. Biol. Chem. 279 1867-1871 [DOI] [PubMed] [Google Scholar]
- 4.Hicke, L. (2001) Nat. Rev. Mol. Cell Biol. 2 195-201 [DOI] [PubMed] [Google Scholar]
- 5.Haglund, K., Di Fiore, P. P., and Dikic, I. (2003) Trends Biochem. Sci. 28 598-603 [DOI] [PubMed] [Google Scholar]
- 6.Pickart, C. M. (2000) Trends Biochem. Sci. 25 544-548 [DOI] [PubMed] [Google Scholar]
- 7.Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D., and Gygi, S. P. (2003) Nat. Biotechnol. 21 921-926 [DOI] [PubMed] [Google Scholar]
- 8.Pickart, C. M., and Fushman, D. (2004) Curr. Opin. Chem. Biol. 8 610-616 [DOI] [PubMed] [Google Scholar]
- 9.Varadan, R., Assfalg, M., Haririnia, A., Raasi, S., Pickart, C., and Fushman, D. (2004) J. Biol. Chem. 279 7055-7063 [DOI] [PubMed] [Google Scholar]
- 10.Varadan, R., Walker, O., Pickart, C., and Fushman, D. (2002) J. Mol. Biol. 324 637-647 [DOI] [PubMed] [Google Scholar]
- 11.Chau, V., Tobias, J. W., Bachmair, A., Marriott, D., Ecker, D. J., Gonda, D. K., and Varshavsky, A. (1989) Science 243 1576-1583 [DOI] [PubMed] [Google Scholar]
- 12.Ozkaynak, E., Finley, D., Solomon, M. J., and Varshavsky, A. (1987) EMBO J. 6 1429-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirisako, T., Kamei, K., Murata, S., Kato, M., Fukumoto, H., Kanie, M., Sano, S., Tokunaga, F., Tanaka, K., and Iwai, K. (2006) EMBO J. 25 4877-4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker, R. T., and Board, P. G. (1987) Nucleic Acids Res. 15 443-463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiborg, O., Pedersen, M. S., Wind, A., Berglund, L. E., Marcker, K. A., and Vuust, J. (1985) EMBO J. 4 755-759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicke, L., Schubert, H. L., and Hill, C. P. (2005) Nat. Rev. Mol. Cell Biol. 6 610-621 [DOI] [PubMed] [Google Scholar]
- 17.Hurley, J. H., Lee, S., and Prag, G. (2006) Biochem. J. 399 361-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, K., and Bucher, P. (1996) Trends Biochem. Sci. 21 172-173 [PubMed] [Google Scholar]
- 19.Ohno, A., Jee, J., Fujiwara, K., Tenno, T., Goda, N., Tochio, H., Kobayashi, H., Hiroaki, H., and Shirakawa, M. (2005) Structure 13 521-532 [DOI] [PubMed] [Google Scholar]
- 20.Mueller, T. D., and Feigon, J. (2002) J. Mol. Biol. 319 1243-1255 [DOI] [PubMed] [Google Scholar]
- 21.Kozlov, G., Nguyen, L., Lin, T., De Crescenzo, G., Park, M., and Gehring, K. (2007) J. Biol. Chem. 282 35787-35795 [DOI] [PubMed] [Google Scholar]
- 22.Swanson, K. A., Hicke, L., and Radhakrishnan, I. (2006) J. Mol. Biol. 358 713-724 [DOI] [PubMed] [Google Scholar]
- 23.Lowe, E. D., Hasan, N., Trempe, J. F., Fonso, L., Noble, M. E., Endicott, J. A., Johnson, L. N., and Brown, N. R. (2006) Acta Crystallogr. D Biol. Crystallogr. 62 177-188 [DOI] [PubMed] [Google Scholar]
- 24.Varadan, R., Assfalg, M., Raasi, S., Pickart, C., and Fushman, D. (2005) Mol. Cell 18 687-698 [DOI] [PubMed] [Google Scholar]
- 25.Trempe, J. F., Brown, N. R., Lowe, E. D., Gordon, C., Campbell, I. D., Noble, M. E., and Endicott, J. A. (2005) EMBO J. 24 3178-3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raasi, S., Varadan, R., Fushman, D., and Pickart, C. M. (2005) Nat. Struct. Mol. Biol. 12 708-714 [DOI] [PubMed] [Google Scholar]
- 27.Amerik, A., Swaminathan, S., Krantz, B. A., Wilkinson, K. D., and Hochstrasser, M. (1997) EMBO J. 16 4826-4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkinson, K. D., Tashayev, V. L., O'Connor, L. B., Larsen, C. N., Kasperek, E., and Pickart, C. M. (1995) Biochemistry 34 14535-14546 [DOI] [PubMed] [Google Scholar]
- 29.Lindsey, D. F., Amerik, A., Deery, W. J., Bishop, J. D., Hochstrasser, M., and Gomer, R. H. (1998) J. Biol. Chem. 273 29178-29187 [DOI] [PubMed] [Google Scholar]
- 30.Doelling, J. H., Yan, N., Kurepa, J., Walker, J., and Vierstra, R. D. (2001) Plant J. 27 393-405 [DOI] [PubMed] [Google Scholar]
- 31.Timms, K. M., Ansari-Lari, M. A., Morris, W., Brown, S. N., and Gibbs, R. A. (1998) Gene 217 101-106 [DOI] [PubMed] [Google Scholar]
- 32.Ansari-Lari, M. A., Muzny, D. M., Lu, J., Lu, F., Lilley, C. E., Spanos, S., Malley, T., and Gibbs, R. A. (1996) Genome Res. 6 314-326 [DOI] [PubMed] [Google Scholar]
- 33.Melandri, F., Grenier, L., Plamondon, L., Huskey, W. P., and Stein, R. L. (1996) Biochemistry 35 12893-12900 [DOI] [PubMed] [Google Scholar]
- 34.Stein, R. L., Chen, Z., and Melandri, F. (1995) Biochemistry 34 12616-12623 [DOI] [PubMed] [Google Scholar]
- 35.Dang, L. C., Melandri, F. D., and Stein, R. L. (1998) Biochemistry 37 1868-1879 [DOI] [PubMed] [Google Scholar]
- 36.Gabriel, J. M., Lacombe, T., Carobbio, S., Paquet, N., Bisig, R., Cox, J. A., and Jaton, J. C. (2002) Biochemistry 41 13755-13766 [DOI] [PubMed] [Google Scholar]
- 37.Reyes-Turcu, F. E., Horton, J. R., Mullally, J. E., Heroux, A., Cheng, X., and Wilkinson, K. D. (2006) Cell 124 1197-1208 [DOI] [PubMed] [Google Scholar]
- 38.Russell, N. S., and Wilkinson, K. D. (2005) Methods Mol. Biol. 301 207-219 [DOI] [PubMed] [Google Scholar]
- 39.Larsen, C. N., Krantz, B. A., and Wilkinson, K. D. (1998) Biochemistry 37 3358-3368 [DOI] [PubMed] [Google Scholar]
- 40.Cook, W. J., Jeffrey, L. C., Kasperek, E., and Pickart, C. M. (1994) J. Mol. Biol. 236 601-609 [DOI] [PubMed] [Google Scholar]
- 41.Komander, D., Lord, C. J., Scheel, H., Swift, S., Hofmann, K., Ashworth, A., and Barford, D. (2008) Mol. Cell 29 451-464 [DOI] [PubMed] [Google Scholar]
- 42.Raasi, S., and Pickart, C. M. (2005) Methods Mol. Biol. 301 47-55 [DOI] [PubMed] [Google Scholar]
- 43.Gill, S. C., and von Hipple, P. H. (1989) Anal. Biochem. 182 319-326 [DOI] [PubMed] [Google Scholar]
- 44.Hunter, C. A., and Tomas, S. (2003) Chem. Biol. 10 1023-1032 [DOI] [PubMed] [Google Scholar]
- 45.Lacombe, T., and Gabriel, J. M. (2002) FEBS Lett. 531 469-474 [DOI] [PubMed] [Google Scholar]
- 46.Mueller, T. D., Kamionka, M., and Feigon, J. (2004) J. Biol. Chem. 279 11926-11936 [DOI] [PubMed] [Google Scholar]
- 47.Hu, M., Li, P., Song, L., Jeffrey, P. D., Chenova, T. A., Wilkinson, K. D., Cohen, R. E., and Shi, Y. (2005) EMBO J. 24 3747-3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu, M., Li, P., Li, M., Li, W., Yao, T., Wu, J. W., Gu, W., Cohen, R. E., and Shi, Y. (2002) Cell 111 1041-1054 [DOI] [PubMed] [Google Scholar]
- 49.Williams, D. H., O'Brien, D. P., Sandercock, A. M., and Stephens, E. (2004) J. Mol. Biol. 340 373-383 [DOI] [PubMed] [Google Scholar]
- 50.Phillips, C. L., Thrower, J., Pickart, C. M., and Hill, C. P. (2001) Acta Crystallogr. D Biol. Crystallogr. 57 341-344 [DOI] [PubMed] [Google Scholar]
- 51.Eddins, M. J., Varadan, R., Fushman, D., Pickart, C. M., and Wolberger, C. (2007) J. Mol. Biol. 367 204-211 [DOI] [PubMed] [Google Scholar]
- 52.Haririnia, A., D'Onofrio, M., and Fushman, D. (2007) J. Mol. Biol. 368 753-766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avvakumov, G. V., Walker, J. R., Xue, S., Finerty, P. J., Jr., Mackenzie, F., Newman, E. M., and Dhe-Paganon, S. (2006) J. Biol. Chem. 281 38061-38070 [DOI] [PubMed] [Google Scholar]
- 54.Renatus, M., Parrado, S. G., D'Arcy, A., Eidhoff, U., Gerhartz, B., Hassiepen, U., Pierrat, B., Riedl, R., Vinzenz, D., Worpenberg, S., and Kroemer, M. (2006) Structure 14 1293-1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thrower, J. S., Hoffman, L., Rechsteiner, M., and Pickart, C. M. (2000) EMBO J. 19 94-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.