Abstract
Caspase-8 is the initiator caspase of the extrinsic apoptosis pathway and also has a role in non-apoptotic physiologies. Identifying endogenous substrates for caspase-8 by using integrated bioinformatics and biological approaches is required to delineate the diverse roles of this caspase. We describe a number of novel putative caspase-8 substrates using the Prediction of Protease Specificity (PoPS) program, one of which is histone deacetylase 7 (HDAC7). HDAC7 is cleaved faster than any other caspase-8 substrate described to date. It is also cleaved in primary CD4+CD8+ thymocytes undergoing extrinsic apoptosis. By using naturally occurring caspase inhibitors that have evolved exquisite specificity at concentrations found within the cell, we could unequivocally assign the cleavage activity to caspase-8. Importantly, cleavage of HDAC7 alters its subcellular localization and abrogates its Nur77 repressor function. Thus we demonstrate a direct role for initiator caspase-mediated proteolysis in promoting gene transcription.
During apoptosis cells are systematically dismantled and packaged into small membrane-bound particles ready for removal by professional phagocytes, by a process that is driven by members of the caspase family of proteases. Members of the caspase family have generally been separated into two groups as follows: those involved in apoptosis (caspase-2, -3, and -6–10) and those involved in non-apoptotic processes such as inflammation and differentiation (caspase-1, -4, -5, and -14) (1). However, this simple demarcation is complicated by evidence suggesting that some apoptotic caspases may have functions in non-apoptotic physiologies, including, but not limited to, cell differentiation, migration, proliferation, T and B cell activation, and nuclear factor-κB (NF-κB)3 activation (2).
Of the initiator apoptotic caspases, the strongest evidence for alternative non-apoptotic roles is for caspase-8. The proteolytic activity of the caspase may be dispensable for some of these processes. For instance, tumor necrosis factor (TNF)-mediated NF-κB activation in T cells, fibroblasts, and epithelial cells is dependent on caspase-8 protein but not its proteolytic activity. In contrast, NF-κB activation in response to T cell receptor ligation in T cells does require caspase-8 activity (3). Regarding differentiation, caspase-8 is required for maturation of monocytes into macrophages, and pan-caspase-inhibitors block this process (4, 5). In addition, a requirement for caspase-8 activity has been demonstrated during differentiation of placental villous trophoblasts (6). Targeted deletion reveals that caspase-8 protein is also required for T cell activation, formation of blood vessels, and maintenance of hemopoietic progenitor cells in mice (4, 7, 8). More significantly, caspase-8 null mice and humans manifest a complex condition, including immunodeficiency early in their life and autoimmunity as the individuals age (9, 10).
If the cell utilizes lethal pro-apoptotic proteases such as caspase-8 to perform other cellular functions, an immediate challenge it must overcome is how to survive while harboring active caspase-8. One possibility is to sequester the “apoptotic” substrates (e.g. procaspase-3, procaspase-7, and Bid) from active caspase-8, while leaving non-apoptotic substrates available for proteolysis. To test this hypothesis, we need to identify the elusive non-apoptotic caspase-8 substrates. To this end we employed a bioinformatic approach. Searching the human proteome with a matrix model based on the well defined substrate specificity of caspase-8 revealed a number of potential substrates. We formally tested three of these and confirmed that one, HDAC7, is very efficiently cleaved by caspase-8 both in vitro and in vivo. HDAC7 is a class II histone deacetylase, and like other members of the family, it represses transcription of MEF2-dependent genes in a cell type-specific manner (11). We provide evidence that cleavage of HDAC7 is a loss of function event, abrogating its transcription repressor activity. Finally we discuss the implications of HDAC7 cleavage in both apoptotic and non-apoptotic scenarios.
EXPERIMENTAL PROCEDURES
PoPS Search Parameters—The matrix model of caspase-8 specificity (C14.009 > Boyd > 1.2) was used to search the human proteome using the Web-based PoPS program. The model was designed based on studies using positional scanning libraries and fluorescence-quenched substrates, and each amino acid was given a score of -5.0 to +5.0 (12, 13). Unprofiled amino acids were given a value of 0. Each subsite was given equal weighting. The threshold was set to 24 (maximum score is 25 for five independent subsites).
Phylogeny—Phylogenic tree and multiple sequence alignment were generated using ClustalW at the San Diego Super-computer Center Biology Workbench.
Materials and Antibodies—Benzyloxycarbonyl-Val-Ala-(O-methyl)-Asp-fluoromethyl ketone (Z-VAD-fmk) and acetyl-Asp-Glu-Val-Asp-amino-4-trifluoromethylcoumarin (Ac-DEVD-afc) were from Enzyme System Products. SuperSignal was from Pierce. DC protein assay was from Bio-Rad. Killer-TRAIL, FasL, and TNFα were from Alexis Biochemicals. Cycloheximide and MG132 were from Calbiochem. Etoposide was from Biomol. Annexin V-PE was from Caltag. Luciferase reporter assay was from Promega. Monoclonal antibodies were specific for XIAP/hILP, mouse caspase-3, and HSP90 (BD Transduction Laboratories), FLAG (M2), mouse CD8a-FITC and mouse CD4-FITC (Sigma), caspase-8 (C15, kind gift from Dr. Markus Peter, University of Chicago), and poly(ADP-ribose) polymerase and GST (Pharmingen). Polyclonal rabbit antibodies were specific for caspase-3 and hemagglutinin epitope tag (HA) (Santa Cruz Biotechnology), cleaved caspase-3 (Cell Signaling Technology), p35 (kind gift from Dr. Stan Krajewski, Burnham Institute for Medical Research, La Jolla, CA), cleaved poly(ADP-ribose) polymerase (New England Biolabs), and CrmA (kind gift from Dr. David Pickup, Duke University, Durham, NC). Polyclonal goat antibody was against HDAC7 (N-18, Santa Cruz Biotechnology). Secondary antibodies were horseradish peroxidase-conjugated donkey anti-rabbit IgG, donkey anti-mouse IgG (Amersham Biosciences), and donkey anti-goat IgG (Santa Cruz Biotechnology). All other chemicals were from Sigma.
Plasmids—RAB9A with a C-terminal His6 tag in pET-3a was a kind gift from Dr. Suzanne Pfeiffer (Stanford University, San Francisco). Rat TRIM3/BERP with an N-terminal FLAG tag in pcDNA3 was a kind gift from Dr. Steve Vincent (University of British Columbia, Vancouver, Canada). Histone deacetylase 7 (HDAC7) splice variant 3 with a C-terminal FLAG tag in pcDNA3.1 was a kind gift from Dr. Eric Verdin (The Gladstone Institute, San Francisco, CA). Myc-XIAP plasmid was described previously (14). Bax plasmid was already described (15). FLAG-Bid, Bcl-xL-HA (in pcDNA3), and pCMV-β-galactosidase were the kind gifts from Dr. John Reed (Burnham Institute for Medical Research, La Jolla, CA). MEF2D plasmid was a kind gift from Dr. Xiao-kun Zhang (Burnham Institute for Medical Research, La Jolla, CA). The pcDNA3.1/caspase-8 wild type and dominant-negative catalytic mutant C285A (caspase-1 numbering system) was described previously (16). The pcDNA3/p35-FLAG and pcDNA3/CrmA plasmid was described previously (17). Luciferase reporter construct under control of the Nur77 promoter (Nur77-luc) was described previously (18). Human Bid was PCR-amplified with primers containing flanking EcoRI sites and cloned into the EcoRI site of pGEX-4T-1. HDAC7 D375A mutant was generated by site-directed mutagenesis using QuickChange (Stratagene).
FLAG-tagged HDAC7 Constructs—HDAC7-FLAG/pcDNA3.1 was cut with EcoRI to remove full-length HDAC7 and produce linearized pcDNA3.1 containing a C-terminal FLAG tag. The N-terminal (1–375) fragment (600 primer, GAGAACCCACTGCTTACTGGC, and 1700 primer, CCCGCGGAATTCTGTCTCCAGGTCTTCAGCCG) and C-terminal (376–915) fragment (1687 primer, GGCGAATTCGCCATGGGCGGGGGACCGGGCCAG, and 755 primer, TAGAAGGCACAGTCGAGG) of HDAC7 were generated by PCR, digested with EcoRI, and cloned into pcDNA3.1 with a C-terminal FLAG tag. The N-terminally FLAG-tagged HDAC7 construct, and double-tagged FLAG-HDAC7-FLAG, were PCR-amplified from HDAC7-FLAG/pcDNA3.1 and cloned into pFLAG-CMV4.
GFP-tagged HDAC7 Constructs—Full-length HDAC7 was subcloned into the EcoRI site of pEGFP-N2 (Clontech). The N-terminal (1–375) fragment (600 primer and 1688 primer, CGCGCGGTACCCTGTCTCCAGGTCTTCAGCCG) was amplified by PCR and cloned into the EcoRI and KpnI sites of pEGFP-N2. The C-terminal fragment of HDAC7 (376–915) was amplified as described above and cloned into the EcoRI site of pEGFP-N2. All plasmids were verified by sequencing.
Recombinant Proteins—His6-tagged wild type caspases, caspase-3 catalytic mutant (C285A, caspase-1 numbering system), p35 C2A, and RAB9A were expressed in BL21(DE3) Escherichia coli and purified by nickel-affinity chromatography as described previously (19). Caspases were titrated with Z-VAD-fmk to determine the concentration of catalytic sites as described previously (20). GST-Bid was expressed and purified as described previously (14). Recombinant HDAC7 and TRIM3 were purified from transfected HEK293. Cells were lysed with modified radioimmunoprecipitation buffer containing 200 μm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 2 μg/ml leupeptin, 1 μg/ml pepstatin, and 2 μm E-64 as described previously (21). Lysates were clarified by centrifugation and combined with 50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 0.05% (v/v) Nonidet P-40, 0.25% (w/v) gelatin at a 1:1 ratio. For every 1 ml of lysate, 20 μl of anti-FLAG M2 affinity gel beads (Sigma) were added and immunoprecipitated for up to 4 h at 4 °C, and the beads were washed three times in PBS. Beads were either used directly in caspase cleavage assays or resuspended in 50 mm Tris-Cl, 150 mm NaCl, pH 7.4, and eluted with 150 μg/ml 3xFLAG peptide (Sigma) for 30 min at 4 °C.
Protein Cleavage Assay—Putative protein substrates in caspase buffer (20 mm PIPES, 100 mm NaCl, 10% (w/v) sucrose, 0.1% (w/v) CHAPS, 10 mm dithiothreitol, 1 mm EDTA, pH 7.2) were incubated for 30 min at 37 °C with the indicated concentration of active site titrated caspase. In some instances caspase buffer was supplemented with 10% (w/v) PEG 6000. Cleavage assays were terminated by the addition of reducing Laemmli sample buffer and boiled for 5 min. Substrate cleavage was monitored by SDS-PAGE and immunoblotting.
Cell Lines and Transfections—The DPK cell line is a CD4+CD8+ thymocyte precursor cell line (a kind gift from Dr. John Kaye, The Scripps Research Institute, La Jolla, CA) and was maintained in Click's medium with 10% heat-inactivated fetal bovine serum, 50 μm β-mercaptoethanol, nonessential amino acids, 20 mm Hepes, penicillin/streptomycin, and 2 mm l-glutamine. HEK293 and COS7 cells were transfected with FuGENE 6 transfection reagent (Roche Applied Science). Immature primary thymocytes were purified from the thymus of a 7-week-old FVB/N mouse. Greater than 75% of cells were CD4+CD8+. Thymocytes were cultured for up to 2 days in RPMI with 10% heat-inactivated fetal bovine serum, 50 μm β-mercaptoethanol, penicillin/streptomycin, and 2 mm l-glutamine.
Induction and Monitoring of Apoptosis—HEK293 cells were treated with KillerTRAIL, TNFα with cycloheximide, etoposide, staurosporine, MG132, or transfected with pcDNA3/Bax for the indicated times and concentrations. Primary mouse thymocytes were treated with 100 ng/ml FasL with 1 μg/ml Enhancer (Alexis Biochemicals). Activation-induced cell death (AICD) was induced in primary thymocytes with 5 μg/ml biotinylated anti-mouse CD3ε chain (145-2C11; Pharmingen) with or without 5 μg/ml biotinylated anti-mouse CD28 (Pharmingen) and 25 μg/ml streptavidin (Southern Biotech) for 24 or 48 h. All cells were harvested, washed in ice-cold PBS, and cell pellets were stored at -20 °C. Frozen pellets were lysed in ice-cold modified radioimmunoprecipitation buffer (containing protease inhibitors, as described above), normalized for protein concentration, and analyzed by SDS-PAGE and immunoblotting. A portion of the lysate was added to caspase buffer containing 100 μm Ac-DEVD-afc, and executioner caspase activity was monitored as described previously (21). In some experiments a portion of the cells was reserved prior to storage, stained with annexin V-PE or annexin V-FITC, and analyzed by FACS (FACSort, BD Biosciences). In other experiments, the sub-G1 population was identified by harvesting cells in PBS containing 0.1% (v/v) Triton X-100, 0.1% (w/v) sodium citrate, 50 μg/ml propidium iodide, and analyzed by FACS on the FL3 channel.
Electrophoresis and Immunoblotting—Samples were analyzed by 8–18% linear gradient acrylamide SDS-PAGE under reducing conditions, and immunoblotting was as described (22).
Fluorescent Microscopy—COS7 cells grown on glass coverslips in 24-well plates were transfected with FuGENE 6. Cells transfected with C-terminal GFP-tagged HDAC7 constructs were washed in PBS and fixed with 4% paraformaldehyde. Cells co-transfected with 200 ng of C-terminal FLAG-tagged HDAC7, 200 ng of caspase-8, and 1.6 μg of XIAP-myc were washed twice with PBS, fixed with ice-cold methanol for 2 min, blocked with 5% nonfat powdered milk (w/v) in PBS for 1 h at 37 °C, and incubated with mouse anti-FLAG monoclonal antibody (M2, 10 μg/ml) and a FITC-conjugated anti-mouse IgG (Molecular Probes, Eugene, OR). Samples were stained with 250 nm 4′,6-diamidino-2-phenylindole and mounted with VectaShield (Burlingame, CA). Images were captured with a Color CCD SPOT RT Camera (Diagnostic Instruments Inc.) attached to an Inverted TE300 Nikon microscope.
Nur77 Promoter Activity Assays—HEK293 cells in 6-well trays were transfected with the indicated plasmids for 48 h. The amount of DNA was kept constant by balancing with empty pcDNA3.1 plasmid. Cells were lysed in 100 μl of Passive Lysis Buffer (Promega), and luciferase activity was assessed using the luciferase assay system (Promega) according to the manufacturer's instruction. β-Galactosidase activity was assayed to monitor transfection efficiency and viability of cells (hydrolysis of 800 μg/ml o-nitrophenyl β-d-galactopyranoside in PBS with 1 mm dithiothreitol, 1 μm MgCl2, emission at 405 nm). Luciferase activity was normalized to β-galactosidase activity. For experiments involving transfection of caspase-8, if the β-galactosidase activity for a given experiment had a standard deviation of more than 10%, it was assumed that expression of caspase-8 had killed too many cells and that data set was excluded.
Quantitative RT-PCR—Total RNA was purified from primary thymocytes with TRIzol (Invitrogen) according to the manufacturer's instructions. Residual genomic DNA was removed with DNA-free (Ambion), according to manufacturer's instructions. 1–5 μg of RNA was reverse-transcribed with Superscript First Strand Synthesis System for RT-PCR (Invitrogen). Nur77 mRNA was quantified with the SYBR green fluorogenic detection system on a Stratagene Mx3000p. PCRs were performed in duplicate with the following primers: Nur77 sense (5′-CTTGAGTTCGGCAAGCCTAC-3′) and Nur77 antisense (5′-CGAGGATGAGGAAGAAGACG-3′). Standard curves were plotted, and Nur77 expression was normalized to cyclophilin A.
RESULTS
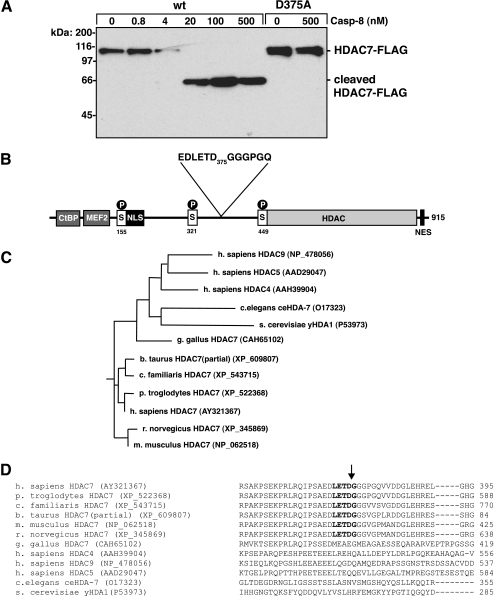
Identification of HDAC7 as a Potential Caspase-8 Substrate in Silico—The distinct specificity of caspases has been established using positional scanning substrate approaches (12, 13). Empirical data from these studies were used to generate a matrix model of caspase-8 specificity for the P4-P3-P2-P1-P1′ sub-sites. This model was used to search the human proteome for potential caspase-8 substrates using PoPS, a web-based program for prediction of protease substrates. Using this five subsite model (number C14.009 > Boyd > 1.2 in the model data base at the PoPS website), the maximum score for a potential caspase-8 substrate is 25. Extracellular proteins were eliminated from further consideration, as were candidate substrates whose predicted cleavage sites were within extracellular domains (supplemental Table 1). Three high scoring substrates, whose cleavage may have interesting biological consequences, were selected for in vitro analysis with recombinant caspase-8: HDAC7, RAB9A, and tripartite motif-containing 3 (TRIM3). Recombinant proteins were purified and incubated with recombinant caspase-8. Neither RAB9A nor TRIM3 was cleaved by caspase-8, despite the high scoring prediction (data not shown). For RAB9A it is clear why it was not cleaved; crystal structures show the predicted cleavage site is in a rigid tight turn, with P4–P2 within a β-strand, making it inaccessible to the protease active site (23). There is no structural information about TRIM3 to explain why it was not cleaved by caspase-8. HDAC7 was cleaved by caspase-8, generating a C-terminal FLAG-tagged fragment of ∼63 kDa (Fig. 1A). This fragment is in accordance with that predicted by PoPS.
FIGURE 1.
HDAC7 is cleaved by caspase-8 in vitro. A, HDAC7 was incubated with caspase-8 for 30 min at 37 °C. Reactions were resolved by SDS-PAGE and immunoblotted with anti-FLAG antibody. B, schematic representation of HDAC7 (splice variant 3). Like all class IIa HDACs, the N terminus interacts with the transcriptional co-repressor CtBP and transcription factors, including MEF2 family members. The conserved phosphorylation sites at Ser155, Ser321, and Ser449 mediate nuclear/cytoplasmic shuttling through interactions with 14-3-3 proteins. The NLS, NES, and the conserved histone deacetylase domain are indicated. The PoPS predicted caspase-8 cleavage site at Asp375 is shown. Cleavage at this position would separate the N-terminal transcription factor binding domain and NLS from the C-terminal deacetylase domain and NES. C, phylogenic tree analysis of HDAC7 proteins with human class IIa HDACs and yeast HDA1. D, multiple sequence alignment of class II HDACs across the PoPS-predicted cleavage site showing conservation in mammalian HDAC7 orthologues. Caspase-8 recognition sequence is in boldface type. wt, wild type.
Although we initially set our threshold at 24 to focus on proteins most likely to be caspase-8 substrates, using this matrix model for caspase-8 specificity, which is based on data using small synthetic substrates, PoPS attributes much lower scores for well characterized cleavage sites within in vivo caspase-8 substrates (supplemental Table 1). According to the PoPS prediction, HDAC7 may also harbor additional cleavage sites with lower PoPS score (Asp228, PoPS 15.31; Asp589, PoPS 14.57; Asp385, PoPS 14.33). To confirm the cleavage site allocated ant the highest PoPS score was indeed the one utilized by caspase-8 in vitro, we generated recombinant HDAC7 with Asp375 mutated to alanine. This mutant was not cleaved by caspase-8 (Fig. 1A).
Nucleocytoplasmic shuttling regulates the transcription repressor function of HDAC7. Phosphorylation by protein kinase D or Ca2+/CaM-dependent protein kinase I promote 14-3-3 binding and cytoplasmic accumulation of HDAC7, such that it no longer functions as a transcription repressor (24, 25). Dephosphorylation by myosin phosphatase promotes nuclear import and transcription repression (26). Cleavage of HDAC7 at Asp375 would yield fragments containing distinct functional domains; the N-terminal transcription factor binding elements and nuclear localization signal (NLS) would be separated from the histone deacetylase (HDAC) domain and nuclear export signal (NES; Fig. 1B). Cleavage may also disrupt the 14-3-3-binding site. Phylogenic and primary sequence analysis demonstrate that the caspase-8 cleavage site is conserved in other mammalian HDAC7 proteins, although it is not found in the more distantly related HDAC7 from chickens (Fig. 1, C and D). Indeed the chicken HDAC7 protein appears to be more closely related to the other class IIa histone deacetylase family members and the ancestral yeast class II histone deacetylase gene product (yHDA1), which do not contain the predicted cleavage site.
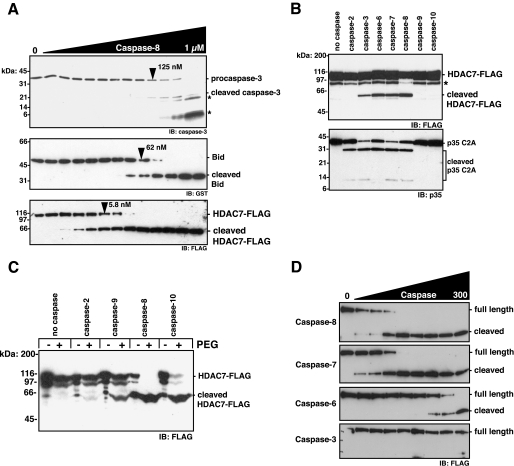
Caspase-8 Cleaves HDAC7 with Physiological Efficiency—Procaspase-3, procaspase-7, and Bid are physiological caspase-8 substrates, whose cleaved counterparts have integral roles as pro-apoptotic cell death mediators. We therefore determined the catalytic efficiency for caspase-8 against HDAC7 compared with these physiological substrates. HDAC7 was cleaved more efficiently than either Bid or procaspase-3. Under these experimental conditions, the EC50 value for HDAC7 cleavage was achieved with 10- and 20-fold less caspase-8 than for Bid and procaspase-3, respectively (Fig. 2A). Accordingly, HDAC7 is the best caspase-8 substrate described to date.
FIGURE 2.
HDAC7 is cleaved as efficiently as physiological caspase-8 substrates. A, caspase-3 C285A (5 nm, top), GST-Bid (25 nm, middle), or HDAC7-FLAG (bottom) was incubated with 0–1 μm caspase-8 in a ½ dilution series for 30 min at 37 °C. Asterisks indicate nonspecific recognition of caspase-8 large and small subunits by the caspase-3 antibody. Arrowheads indicate caspase-8 concentration at which half the substrate was cleaved (EC50). B, HDAC7-FLAG or 25 nm p35 C2A was incubated with 50 nm of the indicated active site-titrated caspase for 30 min at 37 °C. Asterisk represents HDAC7-FLAG that may be translated from a minor internal initiator methionine. C, conditions are as in B, with (+) or without (-) 10% PEG 6000. D, HDAC7-FLAG was incubated with 0–150 nm caspase in a ½ dilution series for 30 min at 37 °C. Reactions were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. IB, immunoblot.
Considering all caspases can cleave substrates after aspartic acid residues, it was important to determine whether caspases other than caspase-8 can also cleave HDAC7. In standard caspase buffer, HDAC7 was also cleaved by the executioner caspases-3, -6, and -7 (Fig. 2B). An inactive mutant of p35, a baculovirus protein with broad reactivity with caspases, served as a positive control for caspase activity. Recombinant initiator caspases-2 and -8–10 are in a dynamic equilibrium between an inactive monomer and an active dimer. In a buffer that promotes full initiator caspase activity (caspase buffer plus 10% (w/v) PEG 6000), caspase-9 could also cleave HDAC7 to some degree, with caspases-8 and -10 being most efficient (Fig. 2C). When comparing the efficiency with which other caspases cleave HDAC7, the ranking is caspase-8 = caspase-7 > caspase-6 > caspase-3 (Fig. 2D).
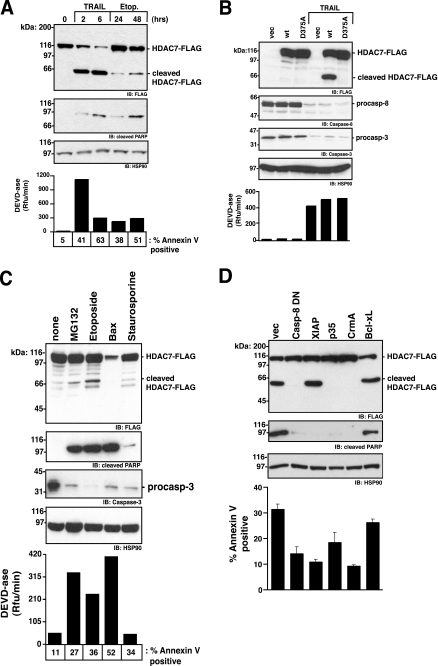
Exogenous HDAC7 Is Cleaved in Cells Undergoing Caspase-8-mediated Apoptosis—To determine whether endogenously activated caspase-8 can cleave HDAC7, we transfected HEK293 cells with HDAC7-FLAG and induced apoptosis with TRAIL. Greater than 50% of HDAC7-FLAG was cleaved within 2 h of TRAIL treatment (Fig. 3A). Indeed HDAC7-FLAG cleavage was evident before significant PARP cleavage, indicating it is an early event in TRAIL-mediated apoptosis. Only one C-terminal FLAG-tagged fragment was detected at ∼63 kDa, consistent with a single cleavage at Asp375. Transfection of HDAC7-FLAG D375A confirmed that cleavage was because of proteolysis at Asp375 (Fig. 3B). Disappearance of procaspase-3 and -8, increase in executioner caspase activity (Ac-DEVD-afc hydrolysis), and annexin V staining are controls for apoptosis. In contrast with TRAIL-induced apoptosis, HDAC7-FLAG was not significantly processed in cells undergoing apoptosis induced by etoposide (Fig. 3, A and C) or other intrinsic death pathway activators, including MG132, staurosporine, and ectopic Bax expression (Fig. 3C). The reduced HDAC7 expression in conjunction with Bax expression is because cells undergo apoptosis before HDAC7 expression is maximal.
FIGURE 3.
HDAC7 is cleaved following engagement of the caspase-8-dependent extrinsic apoptosis pathway. HEK293 cells were transfected with HDAC7-FLAG and treated with 200 ng/ml TRAIL or 100 μm etoposide (Etop) for the indicated times (A); wild type (wt) or D375A HDAC7-FLAG and treated with 200 ng/ml TRAIL for 3 h (B); HDAC7-FLAG and treated with 5 μm MG132, 200 μm etoposide, 200 nm staurosporine for 18 h, or co-transfected with 50 ng Bax cDNA during the initial transfection (C); 0.2 μg of HDAC7-FLAG and 0.8 μg of the indicated cDNA and were treated with 10 ng/ml TNFα and 1 μm cycloheximide for 20 h (D). Average and the standard deviation are shown (n = 3). A–D, cell lysates were normalized for protein concentration, resolved by SDS-PAGE, and immunoblotted (IB) with the indicated antibodies. A, B, and C, executioner caspase activity was assessed by hydrolysis of 100 μm Ac-DEVD-afc. A, C, and D, apoptosis was assessed by annexin V binding and flow cytometry. Unless indicated, the data represent one of three independent experiments.
Finally, to confirm HDAC7-FLAG was directly processed by caspase-8 as opposed to caspase-7, which is activated downstream from caspase-8, and can efficiently cleave HDAC7 in a purified system (Fig. 2D), we employed various apoptosis inhibitors. We used natural proteinaceous inhibitors because they are more specific than small synthetic peptidyl inhibitors (27). The catalytic mutant, caspase-8 C285A (caspase-1 numbering system), acts as a dominant-negative, preventing activation of caspase-8 at the death-inducing signaling complex, whereas Cowpox response modifier A (CrmA) directly and irreversibly inhibits the catalytic activity of caspase-8 (28). XIAP specifically inhibits the activity of caspases-3, -7, and -9, leaving caspase-8 activity intact, whereas p35 inhibits all human caspases (15, 29). Bcl-xL blocks the intrinsic apoptosis pathway, preventing cytochrome c release from the mitochondria, apoptosome formation, and caspase-9 activation and serves as a negative control. Expression of the inhibitors was confirmed by immunoblot (data not shown). CrmA, caspase-8 C285A, and p35 blocked cleavage of HDAC7-FLAG (Fig. 3D). XIAP, which inhibited the morphological changes associated with the executioner caspase activity, did not prevent HDAC7-FLAG cleavage. This result provides strong evidence that caspase-8 directly cleaves HDAC7-FLAG in cells undergoing caspase-8-initiated apoptosis. It indicates that caspase-7, which is activated downstream from caspase-8 and can efficiently cleave HDAC7 in a purified system, was not responsible for HDAC7 processing in this cellular context.
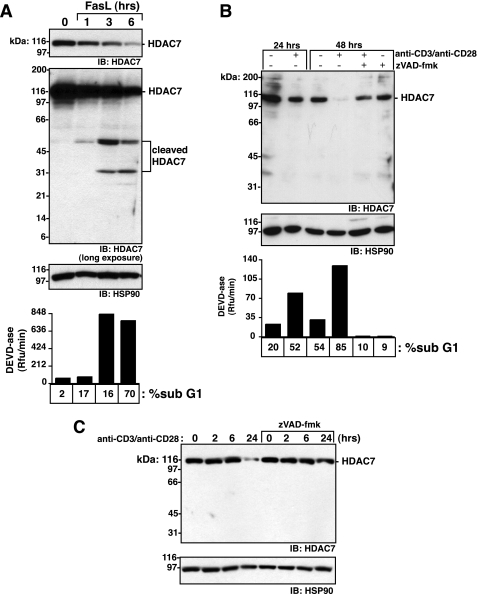
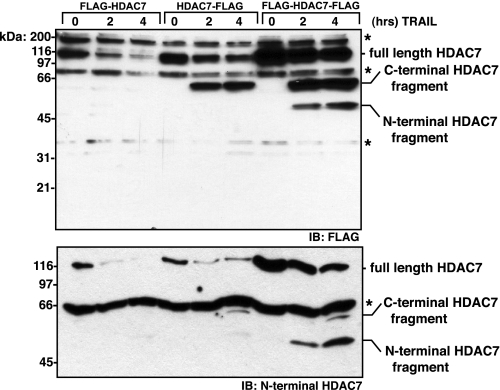
Endogenous HDAC7 Is Cleaved in CD4+CD8+ Thymocytes Undergoing Caspase-8-mediated Apoptosis—HDAC7 is primarily expressed in CD4+CD8+ immature thymocytes (18) and was recently reported in the endothelial cells of the heart and lungs (30). To determine whether endogenous HDAC7 is cleaved in cells upon caspase-8 activation, we purified primary immature CD4+CD8+ thymocytes and treated them with FasL. Full-length HDAC7 disappeared over time, coincident with executioner caspase activation and DNA fragmentation, as evidenced by an increase in cells with sub-G1 DNA content (Fig. 4A). Cleavage products of ∼50 and 31 kDa were only detected upon extended exposure of the film (Fig. 4A). The 50-kDa species probably corresponds to the N-terminal fragment generated by cleavage at Asp405 of mouse HDAC7, which is equivalent to Asp375 of human HDAC7. As apoptosis progressed, the HDAC7 N-terminal antibody detected another species of ∼31 kDa. This may result from a secondary cleavage at Asp251 or cleavage by an unrelated protease. Together this indicates that the N-terminal cleavage product produced during FasL-induced apoptosis is unstable. This was confirmed by expressing N-terminally FLAG-tagged HDAC7 and assessing its stability during extrinsic apoptosis. Compared with the caspase-8-generated C-terminal fragment of HDAC7, the N-terminal fragment, as detected by anti-FLAG antibody or anti-N-terminal HDAC7 antibody, is significantly less stable than the C-terminal fragment (Fig. 5). The N-terminal fragment was only detected in the form of FLAG-HDAC7-FLAG, which was expressed at significantly higher levels than the singularly tagged versions.
FIGURE 4.
Endogenous HDAC7 is cleaved in CD4+CD8+ thymocytes following engagement of the extrinsic apoptosis pathway. Primary CD4+CD8+ thymocytes were treated with 100 ng/ml FasL (A) or 5 μg/ml anti-CD3 and 5 μg/ml anti-CD28 antibody, ±1 h preincubation with 100 μm Z-VAD-fmk (B). C, DPK cells were treated with 5 μg/ml anti-CD3 and 5 μg/ml anti-CD28 antibody for 0, 2, 6, and 24 h, ±100 μm Z-VAD-fmk. Cell lysates were normalized for protein concentration, resolved by SDS-PAGE, and immunoblotted (IB) with the indicated antibodies. Executioner caspase activity was assessed by hydrolysis of 100 μm Ac-DEVD-afc. Apoptosis as measured by DNA content was assessed by propidium iodide staining of permeabilized cells. Data represent one of three independent experiments.
FIGURE 5.
The apoptosis-generated N-terminal fragment of HDAC7 is unstable. HEK293 cells transfected with FLAG-HDAC7, HDAC7-FLAG, or FLAG-HDAC7-FLAG were treated with 200 ng/ml TRAIL for the indicated times. Cell lysates were normalized for protein concentration, resolved by SDS-PAGE, and immunoblotted (IB) with the indicated antibody. Asterisks represent nonspecific bands and serve as loading controls. Although the polyclonal HDAC7 antibody is specific and recognizes the N terminus (Santa Cruz Biotechnology, N-18) the position of the epitope(s) is unknown.
FasL treatment of a CD4+CD8+ thymocyte cell line, DPK, also down-regulated HDAC7 protein, and this was caspase-dependent as Z-VAD-fmk blocked this process. However, with the DPK cells we could not detect any cleavage products, even in the presence of a proteasome inhibitor (data not shown).
Recent reports suggest that HDAC7 may play a role in negative selection of double-positive thymocytes by preventing expression of Nur77, a pro-apoptotic orphan receptor (18). An in vitro model of negative selection is AICD and can be engaged by cross-linking CD3 and CD28 on the surface of immature thymocytes with antibody. In vitro AICD induced DEVDase activity, DNA cleavage, and annexin V binding (Fig. 4B and data not shown). HDAC7 was removed from the cells in a caspase-dependent manner (Fig. 4B). However, no cleavage products were detected, even upon long exposure of the film (data not shown). These results were confirmed in the immature thymocyte DPK cell line (Fig. 4C).
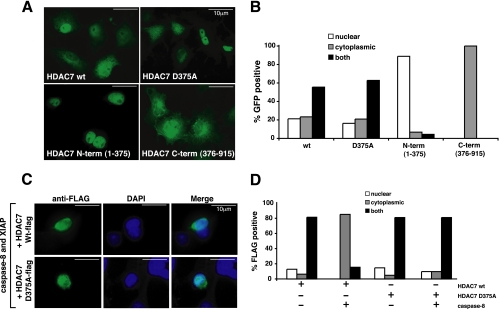
HDAC7 Cleavage Alters Its Subcellular Localization—In COS7 cells, both wild type and D375A HDAC7 are predominantly expressed in both the nucleus and cytoplasm (Fig. 6, A and B). Cleavage of HDAC7 at Asp375 would separate the N-terminal NLS from the C-terminal NES (Fig. 1B). We predict this would dramatically influence the localization of each cleavage fragment. Indeed the N terminus (amino acids 1–375) fused to GFP resided almost completely in the nucleus, whereas the C terminus (amino acids 376–915) fused to GFP was excluded from the nucleus (Fig. 6, A and B).
FIGURE 6.
Cleavage of HDAC7 alters intracellular localization. A, COS7 cells were transfected with HDAC7 mutants containing C-terminal (C-term) tagged GFP. B, cells were counted and scored for subcellular GFP localization, n >100 cells. C, COS7 cells were co-transfected with HDAC7-FLAG (wild type (wt) or D375A) together with caspase-8 and XIAP. Cells were fixed and stained with mouse anti-FLAG antibody, FITC-conjugated anti-mouse IgG, and 4′,6-diamidino-2-phenylindole (DAPI). D, cells were scored blind for subcellular FLAG staining, n > 100 cells. A representative of two independent experiments is shown. Caspase-8 processing of HDAC7 causes relocalization of C-terminal fragment to the cytoplasm. Scale bar represents 10 μm.
To determine whether in vivo cleavage of HDAC7 changes its localization, HDAC7-FLAG was co-expressed with caspase-8 and XIAP, the latter to maintain cellular integrity without affecting the proteolytic activity of caspase-8. In the presence of caspase-8, anti-FLAG immunostaining was excluded from the nucleus, in agreement with cleavage-dependent generation of a C-terminal FLAG-tagged fragment lacking an NLS (Fig. 6, C and D). However, HDAC7 D375A-FLAG maintained a nuclear and cytoplasmic distribution in the presence of caspase-8, consistent with a lack of cleavage.
HDAC7 Cleavage Deregulates Its Nurr77 Repressor Activity—In seeking an assay for the functional significance of HDAC7 cleavage, we settled on its role in repressing MEF2-dependent gene transcription (11). Nur77 is a pro-apoptotic orphan receptor that is induced by MEF2D and plays a role in AICD in immature thymocytes during negative selection.
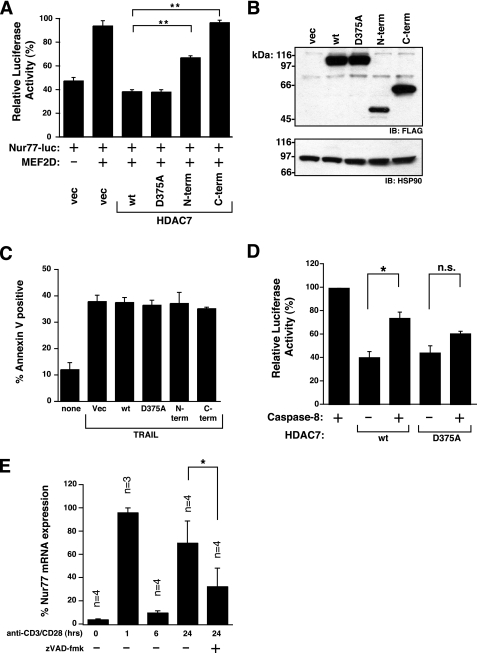
Our data show that cleavage of HDAC7 disrupts the re-entry of the whole molecule back into the nucleus. We therefore predict that cleavage of HDAC7 would disrupt its transcription repressor activity. To test this hypothesis, we transfected HEK293 cells with MEF2D and a luciferase reporter construct under control of the Nur77 promoter. Expression of only the C-terminal portion of HDAC7 has absolutely no Nur77 transcription repressor activity, whereas the N-terminal portion has reduced activity (Fig. 7, A and B). In addition, neither HDAC7 nor the cleavage fragments influence TRAIL-induced apoptosis (Fig. 7C). This is expected as ligation of the TRAIL receptor directly activates caspase-8 to rapidly kill the cell, in a transcription-independent manner.
FIGURE 7.
Cleavage of HDAC7 abolishes its transcription repressor function. A, HEK293 cells were co-transfected with 50 ng of β-galactosidase, 200 ng of Nur77-luciferase reporter construct, 200 ng of MEF2D, and 20 ng of the indicated HDAC7-FLAG variant (n = 4). B, to control for expression levels, HEK293 cells were transfected 1 μg of the indicated HDAC7-FLAG variant. C, HEK293 cells were transfected with HDAC7 variants for 24 h and then treated with 100 ng/ml TRAIL for 4 h (n = 3). D, HEK293 cells were co-transfected with 50 ng of β-galactosidase, 2.5 μg of XIAP, 200 ng of Nur77-luciferase reporter construct, and 200 ng of MEF2D with 20 ng of HDAC7-FLAG wild type (wt) or D375A, ±8 ng of caspase-8 plasmid (n = 3). E, primary thymocytes were treated with 5 μg/ml anti-CD3 and 5 μg/ml anti-CD28 antibody ±100 μm Z-VAD-fmk for 0, 1, 6, and 24 h. Nur77 mRNA expression was assessed by quantitative RT-PCR (n = 3 or 4). Luciferase activity was normalized to β-galactosidase activity. Apoptosis was assessed by annexin V binding and flow cytometry. Average and mean ± S.E. is shown. Paired Student's t test: **, p < 0.01; *, p < 0.05; n.s., p > 0.05. IB, immunoblot.
Significantly, co-expression of trace amounts of caspase-8, in the presence of XIAP (an inhibitor of downstream pro-apoptotic caspases), significantly reduced the Nur77-repressor activity of wild type HDAC7 (Fig. 7D). The ratio of HDAC7-FLAG plasmid to caspase-8 in this experiment was 2.5:1. In other experiments, we have determined HDAC7 cleavage to be greater that 50% at ratios as high as 16.7:1 (data not shown). Therefore, we are confident that a substantial portion of HDAC7 is cleaved in the assay, even though the HDAC7 expression level is beyond the detection limits by Western blot with anti-FLAG antibody (data not shown). Importantly, the repressor activity of HDAC7 D375A was not significantly altered in the presence of caspase-8. Therefore, in cells that contain nonlethal amounts of active caspase-8, HDAC7 can be cleaved, abolishing its transcription repressor activity and promoting transcription from downstream genes. Importantly, no apoptosis was detected under these conditions.
Finally, we assessed whether caspase inhibition could influence Nur77 expression in primary thymocytes undergoing AICD induced by anti-CD3 and anti-CD28 antibody. In agreement with previous reports, Nur77 mRNA is induced as an early response gene and then rapidly decreases. A second wave of Nur77 expression at 24 h of AICD induction is substantially blocked by caspase inhibition (Fig. 7E).
DISCUSSION
In recent years a number of bioinformatics tools have been developed for the identification of caspase and other protease substrates. We used the PoPS program to predict potential caspase-8 substrates because it takes into account the structural nature of the predicted cleavage site during the proteome search. HDAC7 was confirmed as a caspase-8 substrate both in vitro and in cells undergoing caspase-8-dependent apoptosis. By taking advantage of naturally occurring caspase inhibitors that have evolved exquisite specificity at concentrations found within the cell, we could confidently assign the HDAC7 cleavage activity to caspase-8. Only a handful of caspase-8 substrates have been proposed to date, and even fewer have been confirmed in cells using natural inhibitors. Caspase-8 cleavage of the executioner caspases-3 and -7 and Bid has well documented pro-apoptotic consequences (31, 32). Our study shows that HDAC7 is cleaved ∼10 and 20 times more efficiently than Bid and procaspase-3, respectively (Fig. 2), indicating that HDAC7 is the best caspase-8 substrate described to date. In addition, this is the first protein involved in transcription regulation that can be assigned as a caspase-8 substrate. We predict that generation of small amounts of active caspase-8 may be sufficient to cleave HDAC7 without activating caspase-3 and engaging the apoptotic pathway.
Histone acetylation and deacetylation are important epigenetic mechanisms that control gene expression. Histone acetylases catalyze the transfer of an acetyl group to specific lysine residues of histones, promoting gene expression. HDACs remove these acetyl groups, condensing the chromatin and repressing transcription. Class I HDACs (HDAC1–3, -8, and -11) are primarily localized in the nucleus, are ubiquitously expressed, and most data suggest that class I HDACs account for the majority of the histone deacetylase activity in mammalian cells. Class II HDACs (HDAC4–7, -9, and -10) display a restricted cell- and tissue-specific expression pattern.
HDACs are regulated by phosphorylation, ubiquitination, and sumolyation (reviewed in Refs. 11, 33, 34). More recently, caspase-mediated proteolysis of HDAC3 and HDAC4 has been reported (35, 36). However, our study is the first to show caspase-8-specific cleavage of HDAC7. The rapid and specific cleavage of HDAC7 by caspase-8 in cells suggests a role of this initiator caspase in regulating the transcription of HDAC7-regulated genes. This would be quite different to the conventional blanket shutdown of transcription observed in the execution phase of the apoptotic program.
The most thoroughly described function of HDAC7 is its ability to bind and repress members of the MEF2 transcription factor family, which participate in numerous developmental events, including heart development, muscle differentiation, T cell apoptosis, and growth factor signaling. In immature CD4+CD8+ thymocytes HDAC7 represses transcription of Nur77, a pro-apoptotic protein involved in the death of T lymphocytes during negative selection (37, 38). T cell receptor stimulation of immature thymocytes triggers nuclear export of HDAC7. This leads to a loss of transcription repression and an MEF2D-dependent induction of Nur77 expression and apoptosis (18, 39).
We can envisage two possible outcomes of caspase-8-mediated HDAC7 cleavage in thymocytes. First, cleavage could promote expression of Nur77 and feed forward to enhance apoptosis. This may be important for promoting apoptosis in cells that have reduced levels of executioner caspases or high levels of apoptosis inhibitors such as XIAP. Alternatively, if the N-terminal cleavage fragment is transported back into the nucleus, a dominant-negative effect may result. The caspase-generated N-terminal fragment of HDAC7 may bind MEF2D, preventing transcription of Nur77 in a way that cannot be switched off because it cannot be exported from the nucleus, promoting thymocyte cell survival. Currently, our data do not support this latter possibility, as the caspase-generated HDAC7 N-terminal fragment is unstable. In addition, a previous study reporting ubiquitination of HDAC7 and the presence of two predicted PEST sites supports the notion that cleaved HDAC7 is unstable (40).
It is noteworthy that the proposed role of HDAC7 in negative selection could not be confirmed in HDAC7 knock-out mice because they die in utero at E11 because of a failure of the developing cardiovascular system. This is postulated to be due to up-regulation of matrix metalloproteinase 10, a secreted protease that cleaves extracellular matrix components (30). Interestingly, mice with a targeted deletion of caspase-8 in the whole animal, or specifically in endothelial cells, also die at E11–12 because of a failure of the developing circulatory system (4, 8). Although there was no gross defect in the endothelial cells themselves, there is an elevation in extravascular cell apoptosis suggesting a disruption of cell-cell contacts (4). It will be interesting to see if endothelial cells deficient for caspase-8, or expressing a caspase-8 inhibitor such as CrmA, have altered expression of matrix metalloproteinase 10.
Finally, we note that caspase cleavage of HDAC7 results in cytosolic accumulation of the deacetylase-containing C-terminal domain. In addition to histones, other targets for histone acetylases and HDACs have been identified, including a number of cytosolic proteins (41–43). Indeed, it has been proposed that the modification of lysine residues by acetylation and deacetylation regulates ubiquitination and sumoylation, influencing protein stability, subcellular localization, and activity, depending on the target protein (34). Currently, specific cytosolic targets of HDAC7-mediated deacetylation have not been identified and await further investigation. The C-terminal domain of HDAC7 corresponding to that generated by caspase-8 may be a useful tool to look for such targets.
Supplementary Material
Acknowledgments
We thank Scott Snipas and Annamarie Price for expert technical assistance, Dr. Imre Kovacs for assistance with fluorescence microscopy, and Dr. Christina Pop for insightful discussion. We greatly appreciate the kind gift of plasmids from Dr. Suzanne Pfeiffer, Dr. Steve Vincent, Dr. Eric Verdin, Dr. John Reed, and Dr. Xiao-kun Zhang.
This work was supported, in whole or in part, by National Institutes of Health Grants RR20843 and CA69381. This work was also supported by Australian Research Council Grant DP0771183. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and additional references.
Footnotes
The abbreviations used are: NF-κB, nuclear factor-κB; Ac, acetyl; afc, amino-4-trifluoromethylcoumarin; AICD, activation-induced cell death; CrmA, cowpox response modifier A; HDAC, histone deacetylase; NES, nuclear export signal; NLS, nuclear localization signal; PoPS, prediction of protease specificity; RT, reverse transcriptase; TNF, tumor necrosis factor; XIAP, X-linked inhibitor of apoptosis; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; PBS, phosphate-buffered saline; FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; GST, glutathione S-transferase; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; PIPES, 1,4-piperazinediethanesulfonic acid; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand.
References
- 1.Fuentes-Prior, P., and Salvesen, G. S. (2004) Biochem. J. 384 201-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamkanfi, M., Festjens, N., Declercq, W., Vanden Berghe, T., and Vandenabeele, P. (2007) Cell Death Differ. 14 44-55 [DOI] [PubMed] [Google Scholar]
- 3.Su, H., Bidere, N., Zheng, L., Cubre, A., Sakai, K., Dale, J., Salmena, L., Hakem, R., Straus, S., and Lenardo, M. (2005) Science 307 1465-1468 [DOI] [PubMed] [Google Scholar]
- 4.Kang, T. B., Ben-Moshe, T., Varfolomeev, E. E., Pewzner-Jung, Y., Yogev, N., Jurewicz, A., Waisman, A., Brenner, O., Haffner, R., Gustafsson, E., Ramakrishnan, P., Lapidot, T., and Wallach, D. (2004) J. Immunol. 173 2976-2984 [DOI] [PubMed] [Google Scholar]
- 5.Sordet, O., Rebe, C., Plenchette, S., Zermati, Y., Hermine, O., Vainchenker, W., Garrido, C., Solary, E., and Dubrez-Daloz, L. (2002) Blood 100 4446-4453 [DOI] [PubMed] [Google Scholar]
- 6.Black, S., Kadyrov, M., Kaufmann, P., Ugele, B., Emans, N., and Huppertz, B. (2004) Cell Death Differ. 11 90-98 [DOI] [PubMed] [Google Scholar]
- 7.Salmena, L., Lemmers, B., Hakem, A., Matysiak-Zablocki, E., Murakami, K., Au, P. Y., Berry, D. M., Tamblyn, L., Shehabeldin, A., Migon, E., Wakeham, A., Bouchard, D., Yeh, W. C., McGlade, J. C., Ohashi, P. S., and Hakem, R. (2003) Genes Dev. 17 883-895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varfolomeev, E. E., Schuchmann, M., Luria, V., Chiannilkulchai, N., Beckmann, J. S., Mett, I. L., Rebrikov, D., Brodianski, V. M., Kemper, O. C., Kollet, O., Lapidot, T., Soffer, D., Sobe, T., Avraham, K. B., Goncharov, T., Holtmann, H., Lonai, P., and Wallach, D. (1998) Immunity 9 267-276 [DOI] [PubMed] [Google Scholar]
- 9.Chun, H. J., Zheng, L., Ahmad, M., Wang, J., Speirs, C. K., Siegel, R. M., Dale, J. K., Puck, J., Davis, J., Hall, C. G., Skoda-Smith, S., Atkinson, T. P., Straus, S. E., and Lenardo, M. J. (2002) Nature 419 395-399 [DOI] [PubMed] [Google Scholar]
- 10.Salmena, L., and Hakem, R. (2005) J. Exp. Med. 202 727-732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdin, E., Dequiedt, F., and Kasler, H. G. (2003) Trends Genet. 19 286-293 [DOI] [PubMed] [Google Scholar]
- 12.Thornberry, N. A., Rano, T. A., Peterson, E. P., Rasper, D. M., Timkey, T., Garcia-Calvo, M., Houtzager, V. M., Nordstrom, P. A., Roy, S., Vaillancourt, J. P., Chapman, K. T., and Nicholson, D. W. (1997) J. Biol. Chem. 272 17907-17911 [DOI] [PubMed] [Google Scholar]
- 13.Stennicke, H. R., Renatus, M., Meldal, M., and Salvesen, G. S. (2000) Biochem. J. 350 563-568 [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi, R., Deveraux, Q., Tamm, I., Welsh, K., Assa-Munt, N., Salvesen, G. S., and Reed, J. C. (1998) J. Biol. Chem. 273 7787-7790 [DOI] [PubMed] [Google Scholar]
- 15.Deveraux, Q. L., Leo, E., Stennicke, H. R., Welsh, K., Salvesen, G. S., and Reed, J. C. (1999) EMBO J. 18 5242-5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boatright, K. M., Renatus, M., Scott, F. L., Sperandio, S., Shin, H., Pedersen, I., Ricci, J.-E., Edris, W. A., Sutherlin, D. P., Green, D. R., and Salvesen, G. S. (2003) Mol. Cell 11 529-541 [DOI] [PubMed] [Google Scholar]
- 17.Ryan, C. A., Stennicke, H. R., Nava, V. E., Lewis, J., Hardwick, J. M., and Salvesen, G. S. (2002) Biochem. J. 366 595-601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dequiedt, F., Kasler, H., Fischle, W., Kiermer, V., Weinstein, M., Herndier, B. G., and Verdin, E. (2003) Immunity 18 687-698 [DOI] [PubMed] [Google Scholar]
- 19.Stennicke, H. R., and Salvesen, G. S. (1999) Methods (San Diego) 17 313-319 [DOI] [PubMed] [Google Scholar]
- 20.Stennicke, H. R., and Salvesen, G. S. (2000) Methods Enzymol. 322 91-100 [DOI] [PubMed] [Google Scholar]
- 21.Scott, F. L., Denault, J. B., Riedl, S. J., Shin, H., Renatus, M., and Salvesen, G. S. (2005) EMBO J. 24 645-655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denault, J. B., and Salvesen, G. S. (2003) J. Biol. Chem. 278 34042-34050 [DOI] [PubMed] [Google Scholar]
- 23.Wittmann, J. G., and Rudolph, M. G. (2004) FEBS Lett. 568 23-29 [DOI] [PubMed] [Google Scholar]
- 24.Dequiedt, F., Van Lint, J., Lecomte, E., Van Duppen, V., Seufferlein, T., Vandenheede, J. R., Wattiez, R., and Kettmann, R. (2005) J. Exp. Med. 201 793-804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao, H. Y., Verdel, A., Tsai, C. C., Simon, C., Juguilon, H., and Khochbin, S. (2001) J. Biol. Chem. 276 47496-47507 [DOI] [PubMed] [Google Scholar]
- 26.Parra, M., Mahmoudi, T., and Verdin, E. (2007) Genes Dev. 21 638-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McStay, G. P., Salvesen, G. S., and Green, D. R. (2007) Cell Death Differ. 15 322-331 [DOI] [PubMed] [Google Scholar]
- 28.Zhou, Q., Snipas, S., Orth, K., Dixit, V. M., and Salvesen, G. S. (1997) J. Biol. Chem. 273 7797-7800 [DOI] [PubMed] [Google Scholar]
- 29.Zhou, Q., Krebs, J. F., Snipas, S. J., Price, A., Alnemri, E. S., Tomaselli, K. J., and Salvesen, G. S. (1998) Biochemistry 37 10757-10765 [DOI] [PubMed] [Google Scholar]
- 30.Chang, S., Young, B. D., Li, S., Qi, X., Richardson, J. A., and Olson, E. N. (2006) Cell 126 321-334 [DOI] [PubMed] [Google Scholar]
- 31.Stennicke, H. R., Jurgensmeier, J. M., Shin, H., Deveraux, Q., Wolf, B. B., Yang, X., Zhou, Q., Ellerby, H. M., Ellerby, L. M., Bredesen, D., Green, D. R., Reed, J. C., Froelich, C. J., and Salvesen, G. S. (1998) J. Biol. Chem. 273 27084-27090 [DOI] [PubMed] [Google Scholar]
- 32.Li, H., Zhu, H., Xu, C. J., and Yuan, J. (1998) Cell 94 491-501 [DOI] [PubMed] [Google Scholar]
- 33.Gill, G. (2005) Curr. Opin. Genet. Dev. 15 536-541 [DOI] [PubMed] [Google Scholar]
- 34.Caron, C., Boyault, C., and Khochbin, S. (2005) Bioessays 27 408-415 [DOI] [PubMed] [Google Scholar]
- 35.Escaffit, F., Vaute, O., Chevillard-Briet, M., Segui, B., Takami, Y., Nakayama, T., and Trouche, D. (2007) Mol. Cell. Biol. 27 554-567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paroni, G., Mizzau, M., Henderson, C., Del Sal, G., Schneider, C., and Brancolini, C. (2004) Mol. Biol. Cell 15 2804-2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He, Y. W. (2002) J. Leukocyte Biol. 72 440-446 [PubMed] [Google Scholar]
- 38.Siggs, O. M., Makaroff, L. E., and Liston, A. (2006) Curr. Opin. Immunol. 18 175-183 [DOI] [PubMed] [Google Scholar]
- 39.Parra, M., Kasler, H., McKinsey, T. A., Olson, E. N., and Verdin, E. (2005) J. Biol. Chem. 280 13762-13770 [DOI] [PubMed] [Google Scholar]
- 40.Li, X., Song, S., Liu, Y., Ko, S. H., and Kao, H. Y. (2004) J. Biol. Chem. 279 34201-34208 [DOI] [PubMed] [Google Scholar]
- 41.Hubbert, C., Guardiola, A., Shao, R., Kawaguchi, Y., Ito, A., Nixon, A., Yoshida, M., Wang, X. F., and Yao, T. P. (2002) Nature 417 455-458 [DOI] [PubMed] [Google Scholar]
- 42.Luo, J., Su, F., Chen, D., Shiloh, A., and Gu, W. (2000) Nature 408 377-381 [DOI] [PubMed] [Google Scholar]
- 43.Glozak, M. A., Sengupta, N., Zhang, X., and Seto, E. (2005) Gene (Amst.) 363 15-23 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.