Abstract
The 33-kDa matrix protein SPARC (BM-40, osteonectin) binds several collagen types with moderate affinity. The collagen-binding site resides in helix αA of the extracellular calcium-binding domain of SPARC and is partially masked by helix αC. Previously, we found that the removal of helix αC caused a 10-fold increase in the affinity of SPARC for collagen, and we identified amino acids crucial for binding by site-directed mutagenesis. In this study, we used rotary shadowing, CNBr peptides, and synthetic peptides to map binding sites of SPARC onto collagens I, II, and III. Rotary shadowing and electron microscopy of SPARC-collagen complexes identified a major binding site ∼180 nm from the C terminus of collagen. SPARC binding was also detected with lower frequency near the matrix metalloproteinase cleavage site. These data fit well with our analysis of SPARC binding to CNBr peptides, denaturation of which abolished binding, indicating triple-helical conformation of collagen to be essential. SPARC binding was substantially decreased in two of seven α2(I) mutant procollagen I samples and after N-acetylation of Lys/Hyl side chains in wild-type collagen. Synthetic peptides of collagen III were used to locate the binding sites, and we found SPARC binding activity in a synthetic triple-helical peptide containing the sequence GPOGPSGPRGQOGVMGFOGPKGNDGAO (where O indicates 4-hydroxyproline), with affinity for SPARC comparable with that of procollagen III. This sequence is conserved among α chains of collagens I, II, III, and V. In vitro collagen fibrillogenesis was delayed in the presence of SPARC, suggesting that SPARC might modulate collagen fibril assembly in vivo.
Collagens are major components of the extracellular matrix, comprising ∼30% of the protein mass of vertebrates. At least 28 collagens have been identified so far (1–3) encoded by 46 separate genes. The defining feature of collagens is a triple-helical domain assembled from three polypeptide strands, called α-chains, which each consist of repeating Gly-X-Y sequences, where X is often proline and Y is often hydroxyproline. This primary structure facilitates left-handed poly-proline II helical conformation in each α-chain, and three α-chains form a right-handed triple helix stabilized by interchain hydrogen bonds (4). To form this structure, Gly is required as every third residue. Its lack of side chains allows contact between α-chains, whereas the side chains of residues X and Y are exposed on the surface of the triple helix where they can interact with various molecules. Collagens are major structural proteins, endowing mechanical strength upon the extracellular matrix and also regulating cell behavior through the binding of cell surface receptors and other proteins, such as discoidin domain receptors, a subset of the integrins α1β1, α2β1, α10β1, and α11β1, NG2 proteoglycan, and many matrix molecules (5–7). Nearly 50 different ligands have been shown to interact with collagen, and the binding sites for half of these have been mapped onto collagen I (8). The binding to integrins is particularly well studied using synthetic peptides (9–11), and the interaction between the I domain of integrin α2 and a collagen peptide has been elucidated at the atomic level (12).
SPARC (secreted protein acidic and rich in cysteine), also referred to as BM-40 or osteonectin, is an abundant 33-kDa extracellular calcium-binding protein expressed in various tissues. Particularly high expression is associated with morphogenesis, tissue repair, and remodeling. SPARC belongs to the matricellular protein family, which includes thrombospondins, osteopontin, CCN proteins (CRY61, CTGF, and NOV), and tenascin-C and -X. These proteins are thought to modulate cell-matrix interactions and cell function rather than to contribute to the structural integrity of the extracellular matrix (13). SPARC is a counter-adhesive and anti-proliferative protein that also regulates the activity of growth factors, such as platelet-derived growth factor, vascular endothelial growth factor, and fibroblast growth factor-2 (14, 15). It interacts with the fibrillar collagens (types I, II, III, and V), basement membrane collagen IV, vitronectin, thrombospondin 1, albumin, hydroxyapatite (16), and fibrinogen fragments (17). Barker et al. (18) reported that SPARC regulates fibronectin matrix assembly through its modulation of integrin-linked kinase activity. Interestingly, targeted disruption of the SPARC gene revealed an important role in lens transparency, because SPARC-null mice develop early onset cataracts (19, 20). Other phenotypes of SPARC-null mice are osteopenia (21), alteration of collagen fibrils in dermis (22), and accelerated dermal wound healing (23), suggesting that SPARC is involved in collagen fibrillogenesis. These data indicate that SPARC both modulates cellular phenotype and is involved in tissue organization.
SPARC consists of three domains, a flexible N-terminal domain I, a follistatin-like domain, and a C-terminal extracellular EF-hand calcium-binding (EC)3 domain (24). The structures of the follistatin-like and EC domains have been elucidated by x-ray crystallography (25, 26). The EC domain possesses a binding site of moderate affinity for several types of collagen located in helix αA, which is partially masked by helix αC (supplemental Fig. S1). The cleavage of a single peptide bond in helix αC either by unknown endogenous proteases (between Leu-197 and Leu-198) (27) or by MMPs (between Glu-196 and Leu-197) (28) increases affinity for collagen by a factor of 10. Based on these observations, we have expressed the helix αC deletion mutant (SPARC ΔI, αC, in which the residues Val-196 to Phe-203 are deleted) to mimic this effect of cleavage on affinity for collagen. X-ray crystallography of the deletion mutant demonstrated a rearrangement in the shortened link region between helix αA and the pair of EF hands, exposing the central part of helix αA, in which the collagen-binding site is located (29). It can be assumed that a similar situation exists in tissues when SPARC is nicked proteolytically at helix αC. Therefore, the deletion mutant, SPARC ΔI, αC can be a good model of the activated form of SPARC (29) (supplemental Fig. S1). This deletion mutant has allowed us to map the binding site and identify within it by site-directed mutagenesis those amino acids crucial for collagen binding (29). Meanwhile, we have developed two neoepitope-specific antibodies that distinguish between two adjacent cleavage sites and neither of which bind intact SPARC (30). Using these antibodies, a variable degree of cleavage was detected in SPARC obtained from several adult mouse tissues, suggesting this cleavage to be a physiological mechanism of modulating collagen binding. There is evidence that the N-glycan at Asn-99 also modulates the binding affinity for collagen (31–33), but the mechanism remains to be elucidated.
To date, most information on the SPARC-collagen interaction concerns SPARC alone, and much less is known about the binding site on collagens, apart from a single study on collagen I using atomic force microscopy (34). Further, the biological consequences of SPARC-collagen interaction in vivo are poorly understood. In the present study we mapped the SPARC-binding sites on collagens I, II, and III by using the intact collagens, CNBr peptides, and synthetic peptides. Rotary shadowing was used for the visualization of complexes. In this study the deletion mutant of SPARC (SPARC ΔI,αC) was used because recombinant wild-type SPARC has a lower affinity for collagen because of the absence of endogenous cleavage, whereas the native alternative, SPARC obtained from Engelbreth-Holm-Swarm tumor, is a mixture of intact and enzyme-nicked protein (27). We also showed that some mutations in the collagen α2(I) chain of patients with osteogenesis imperfecta (OI) caused decreased SPARC binding to collagen I. In vitro collagen fibrillogenesis assays demonstrated that SPARC may play a role for collagen fibril assembly.
EXPERIMENTAL PROCEDURES
Sources of Proteins—Acid-soluble collagen I from bovine skin, pepsin-soluble collagen II from bovine nasal septum, and their CNBr peptides were prepared and analyzed as described previously (35–37). Collagen III was purified from pepsin digests of fetal calf skin. Recombinant procollagen II (38) was kindly provided by Dr. Darwin Prockop (Tulane University). Recombinant human SPARC (39) and its deletion mutant ΔI, αC (SPARC ΔI, αC) (29) were prepared as described. Chemical modification of collagens and characterization of derivatives were described previously (40).
Purification of Human Procollagen I—Human procollagen I was purified from serum-free conditioned medium of skin fibroblasts derived from human dermal punch biopsies, under Institutional Review Board-approved protocols. The cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum; serum-free conditioned medium was collected from confluent cells. Procollagen I was precipitated from the medium by adding solid ammonium sulfate to a final concentration of 176 mg/ml (30% saturation). The pellet was dissolved in 0.05 m Tris-HCl, pH 7.5, with 2 m urea, 2.5 mm EDTA in the presence of protease inhibitors (10 mm benzamidine, 2 mm N-ethylmaleimide, and 0.1 mm phenylmethylsulfonyl fluoride), dialyzed overnight against the same buffer, and loaded onto a DEAE-cellulose (DE52; Whatman) column (2.5 × 6.5 cm) equilibrated in the same buffer without inhibitors. Procollagen I was eluted with a linear gradient of 0–0.2 m NaCl. Fractions containing procollagen I were pooled, dialyzed against 0.05 m Tris-HCl, pH 7.5, 0.4 m NaCl, 0.01% NaN3, and concentrated by ultrafiltration. Purified procollagen I was stored at -80 °C.
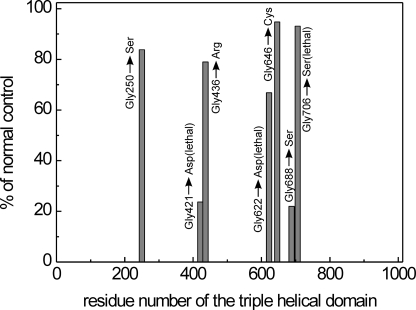
Seven differently mutated procollagen I samples were similarly purified from the culture media of skin fibroblasts originating from osteogenesis imperfecta patients. All of the mutations were single glycine substitutions in the triple-helical domain of the pro-α2(I) chain (Gly-250 → Ser, Gly-421 → Asp*, Gly-436 → Arg, Gly-622 → Asp*, Gly-646 → Cys, Gly-688 → Ser, and Gly-706 → Ser*, where the asterisks indicate lethal mutations; amino acids are numbered from the first glycine of the triple-helical domain). Data describing the phenotype and the characteristics of these mutations are given in Ref. 41. In principle, all of the mutated procollagen samples are a ∼1:1 mixture of normal and overmodified trimers, containing a normal or a mutated α2(I) chain, respectively, as determined by SDS-PAGE analysis with and without pepsin treatment. Under these conditions, a band with normal mobility and a band with delayed mobility for both the α1(I) and α2(I) chains are detected (data not shown).
Recombinant Production and Purification of Human Procollagen III—Human embryonic kidney 293 cells, which were stably co-transfected with human α1(III) procollagen chain and with murine HSP47 (42), were used to collect serum-free conditioned medium in the presence of l-ascorbic acid phosphate at 77 μg/ml (Wako Pure Chemical Industries, Osaka, Japan). The medium (400–500 ml) was dialyzed against 0.05 m Tris-HCl, pH 8.6, with 2 m urea, and passed over a DEAE-cellulose (DE52; Whatman) column (2.5 × 15 cm) equilibrated in the same buffer as used for dialysis. Elution was carried out with a linear gradient of 0–0.5 m NaCl (400 ml) and procollagen III eluted at ∼0.15 m NaCl. Procollagen III was further purified on a Superose 6 HR16/50 column (GE Healthcare) equilibrated in 0.05 m Tris-HCl, pH 8.6, 0.5 m NaCl. The purity and CD spectrum of procollagen III are shown in supplemental Fig. S2.
Proteolytic Digestions—Pepsin digestion was performed in 0.1 m acetic acid at 20 °C for 2 h at an enzyme-substrate ratio of 1:100 and stopped by adding 1 m NaOH to inactivate pepsin. Cleavage was examined by SDS-PAGE under reducing conditions.
To obtain fragments generated by MMP13, collagen was dissolved in 0.05 m Tris-HCl, pH 7.4, containing 0.5 m NaCl, 10 mm CaCl2, and digested with recombinant human proMMP13 (kindly provided by Dr Gillian Murphy, University of Cambridge) (43) at an enzyme-substrate ratio of 1:100 at 25 °C for 24 h. Prior to the digestion, proMMP13 was activated by 2 mm p-aminophenylmercuric acetate at room temperature for 1 h. The cleavage products were separated on Superose 6 HR10/30 equilibrated in 0.05 m Tris-HCl, pH 7.4, containing 0.5 m NaCl. Purified fragments were analyzed by SDS-PAGE (supplemental Fig. S2).
To test whether SPARC interferes with MMP cleavage of collagens I, II, and III, 2 μg of collagen samples (collagen I, procollagen II, collagen II, procollagen III, and collagen III) were incubated with 0.02 μg of MMP1 (kindly provided by Dr Hideaki Nagase, Imperial College, London) or MMP13 in the presence of SPARC ΔI, αC at different ratios at 25 °C and different time intervals and then analyzed by SDS-PAGE.
Analytical Methods—Protein concentrations were determined after hydrolysis with 6 m HCl (16 h at 110 °C) on a LC 3000 amino acid analyzer (Biotronik). SDS-polyacrylamide electrophoresis followed standard protocols. The runs were calibrated with protein standards. Edman degradation was performed on 473 and Procise sequencers following the manufacturer's instructions (Applied Biosystems). Samples for CD analysis were dissolved in 0.1 m acetic acid, and protein concentrations were determined by amino acid analysis. CD spectra in the far UV region were recorded at 10 °C on a JASCO J-715 CD spectropolarimeter in a thermostatted quartz cell with an optical pathlength of 1 mm. Scans were performed at 20 nm/min, collecting data points every 0.1 nm and averaging the data over at least five scans, with background subtraction. Molar ellipticities [Θ] were calculated assuming a mean residue molecular mass and were expressed in degrees cm2 dmol-1.
Rotary Shadowing—Electron microscopy followed standard protocols for rotary shadowing of proteins (44). In experiments to visualize collagen/SPARC complexes, procollagen and SPARC ΔI, αC were incubated in 1:1–1:3 ratios at 30–50 μg/ml for 2 h at room temperature in 0.05 m Tris-HCl, pH 7.4, containing 0.15 m NaCl, 2 mm CaCl2, 0.05% Tween 20. The solutions were dialyzed against 0.2 m ammonium bicarbonate before proceeding to rotary shadowing. The lengths of collagen molecules and the locations of binding sites were measured using NIH Image J 1.55 software.
Binding Assays—The collagens were immobilized by adsorption on plastic wells, and SPARC ΔI, αC was used as a soluble ligand as established previously (45). Binding tests were performed in the presence of 2 mm CaCl2 at room temperature unless mentioned. For denaturing conditions, collagenous samples were heated at 50 °C for 5 min prior to coating.
Synthesis of Collagen Peptides—The Collagen III Toolkit was used to map the SPARC-binding site onto collagen III. Peptide synthesis, using an AB Systems Pioneer automated synthesizer and N-(9-fluorenyl)methoxycarbonyl (Fmoc) chemistry, and the sequences have been described in detail (11); briefly, the peptides were purified after synthesis by high pressure liquid chromatography, identity, and purity proven by matrix-assisted laser desorption ionization time-of-flight mass spectrometry, and triple-helical conformation in aqueous solution demonstrated by polarimetry. The host-guest strategy was applied (46) to prepare a set of overlapping homotrimeric peptides from the entire triple-helical domain of collagen III. The guest sequence of 27 amino acids (primary collagen sequence) are placed between (GPP)5 hosts, and the C-terminal 9 amino acids of each peptide overlap with the first 9 amino acids of the next peptide.
Collagen Fibrillogenesis Assay—Acid soluble calf collagen I in 50 mm acetic acid (1.22 mg/ml) was diluted into 150 mm sodium phosphate buffer, pH 7.8, with 150 mm NaCl, at a final concentration of 122 μg/ml. Fibril formation in the absence and presence of SPARC or SPARC ΔI, αC at different molar ratios was measured by monitoring absorbance at 313 nm in a Cary4 spectrophotometer at 34 °C.
RESULTS
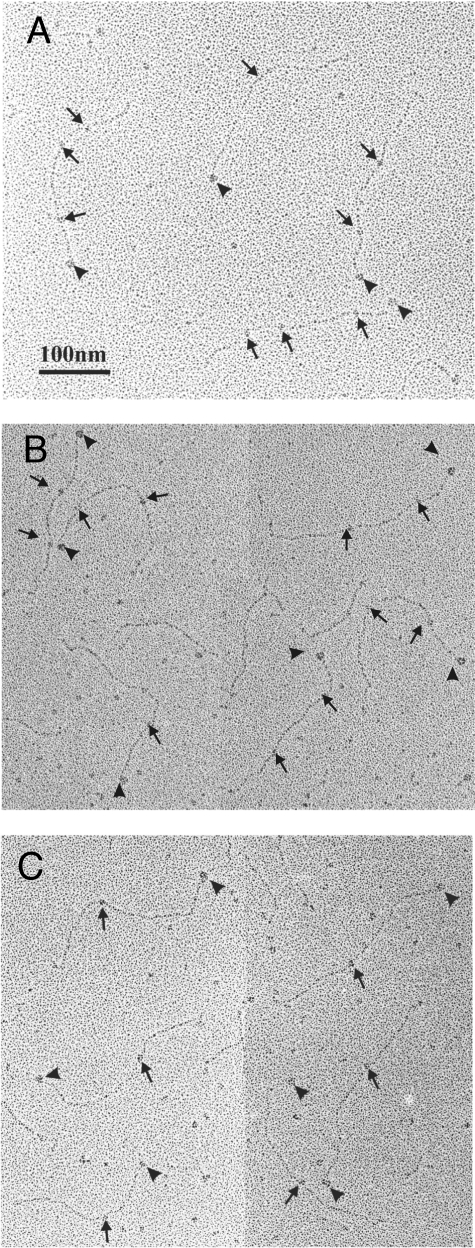
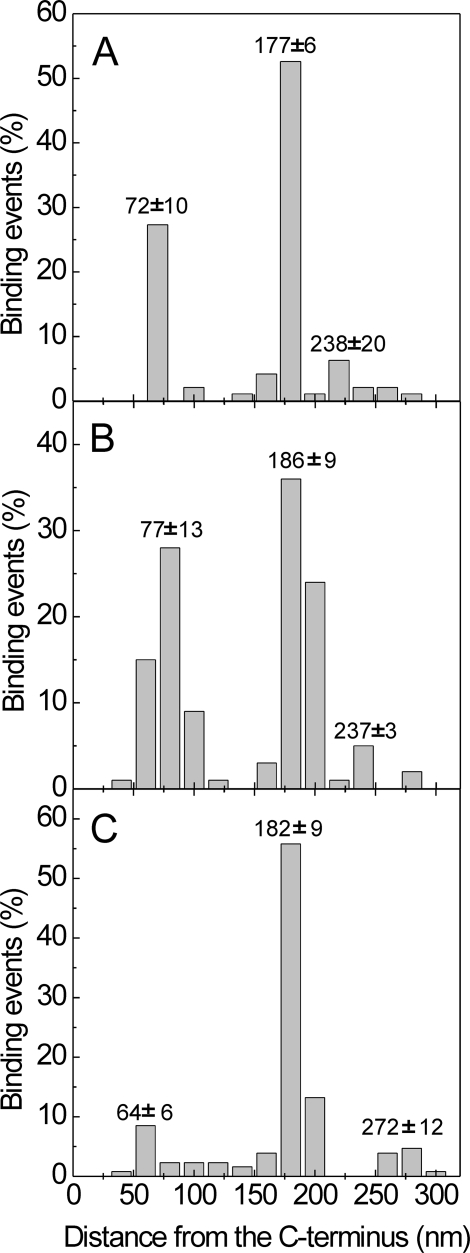
Localization of SPARC-binding Regions in Collages I, II, and III—To locate the SPARC-binding regions on collagens, we examined mixtures of SPARC ΔI, αC and collagen by rotary shadowing followed by electron microscopy (Fig. 1). The use of procollagens allowed the C terminus of collagen molecules to be identified, because the C-propeptides appeared as globular structures. The total length of collagen molecules was found to be ∼300 nm, indicating that the majority of collagen molecules are intact (304 ± 12 nm for procollagen I, 321 ± 24 nm for procollagen II, and 320 ± 18 nm for procollagen III). SPARC molecules were found at two or three different sites on procollagen I (Fig. 1A) and procollagen II (Fig. 1B). Usually only a single SPARC molecule was observed on procollagen III, with infrequent binding of one or two additional molecules (Fig. 1C). The position of SPARC was calculated as the distance from the procollagen C terminus, and histograms are shown in Fig. 2. For procollagen I, frequent binding was found at 72 ± 10 and 177 ± 6 nm from the C terminus. Almost equal frequencies of occupation of collagen II were observed at 77 ± 13 and 186 ± 9nm from the C terminus. These data suggested the presence of two binding sites on collagen I and II and one binding site on collagen III, at 182 ± 9 nm from its C terminus. Sites observed to be occupied by SPARC with ∼10% of the frequency of the most prominent sites may represent in all three collagens either low affinity sites or artifacts of identification.
FIGURE 1.
Rotary shadowing electron micrographs of procollagens in complex with SPARC ΔI, αC. A, human procollagen I; B, human procollagen II; C, human procollagen III. The C-terminal propeptide was seen as a globular structure at one end of procollagen molecule as indicated by arrowhead, and bound SPARC ΔI, αC molecules are indicated by arrows.
FIGURE 2.
Histograms of the binding events of SPARC ΔI, αC along collagen molecules. The position of bound SPARC ΔI, αC was measured from the C terminus. A, human procollagen I (n = 63); B, human procollagen II (n = 70); C, human procollagen III (n = 90). The averages of total length of collagen molecules were 304 nm for procollagen I, 321 nm for procollagen II, and 320 nm for procollagen III. For type I, a total of 95 binding events, for type II, a total of 125 events, and for type III, a total of 126 events were counted. The mean distance from the C terminus ± S.D. is shown in the figure.
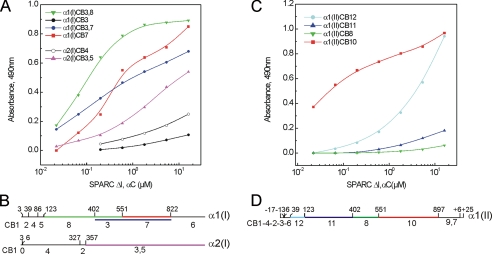
Characterization of the Binding of SPARC to CNBr Peptides Derived from Collagens I and II and Modified Collagens—CNBr peptides from bovine collagens I and II were purified and characterized previously (35–37). CB8, CB3, CB7, CB6, CB3,7, and CB3,8 of the α1(I) chain, CB4 and CB3,5 of the α2(I) chain (Fig. 3B), and CB12, CB11, CB8, and CB10 of the α1(II) chain (Fig. 3D) were immobilized for solid phase binding assays. The activated form of SPARC (SPARC ΔI, αC) was used as soluble ligand. The results are shown in Fig. 3. Prominent binding to α1(I) CB3,8, which consists of uncleaved CB3 and CB8, was observed, and the concentration of SPARC ΔI, αC required for half-maximal binding was 85 nm. The individual peptides CB3 (Fig. 3A) and CB8 (not shown) did not interact with SPARC, suggesting that its binding site is located near the junction of these two peptides, at residue 402 (Fig. 3B). Other peptides, CB7, CB3,7 of α1(I), and CB3,5 of α2(I) showed weaker binding that did not reach a plateau (Fig. 3A). CB7 and CB3,7 of α1(I) contain the second binding site (72 ± 10 nm from the C terminus) located by rotary shadowing. This site was occupied with lower frequency compared with the site in α1(I) CB3,8. CB3,5 of α2(I) contains two binding sites demonstrated by rotary shadowing but has much lower affinity than homotrimeric peptides (CB3,8, CB7, and CB3,7) derived from α1(I) chain. CNBr peptides of bovine collagen II, CB10 and CB12 but not CB11 and CB8, interacted with SPARC, with CB10 displaying higher affinity (Fig. 3C). Although we observed the same binding profile for CB12 reproducibly, we cannot interpret this binding because it does not correlate with the rotary shadowing data. All of the binding shown in Fig. 3 (A and C) was abolished after denaturation of peptides at 50 °C for 5 min, indicating that triple-helical conformation is required for the interaction (data not shown).
FIGURE 3.
Binding of SPARC ΔI, αC to CNBr peptides derived from bovine collagen I and II. A, solid phase binding assay with CNBr peptides derived from bovine collagen I. Immobilized ligands used were CB3,8 (green inverted triangles), CB3 (black filled circles), CB3,7 (dark blue circles), CB7 (red squares) from α1(I) chain, and CB4 (black open circles), and CB3,5 (violet triangles) fromα2(I) chain. The binding to CB8 and CB6 were negative (data not shown). B, the schema shows the location of CNBr peptides in collagen α1(I) and α2(I) chains. C, solid phase binding assay with immobilized CNBr peptides from collagen II, CB12 (light blue circles), CB11 (dark blue triangles), CB8 (green inverted triangles), and CB10 (red squares). D, the schema shows the location of CNBr peptides in collagen α1(II). CNBr peptides (10 μg/ml in phosphate-buffered saline) were coated onto 96-well plate at 4 °C overnight and then coated wells were blocked with 1% bovine serum albumin/phosphate-buffered saline at room temperature for 1 h. The uncoated wells blocked with bovine serum albumin were used as negative control. The incubation with SPARC ΔI, αC was performed at room temperature for 2 h in the presence of 2 mm CaCl2, and the bound SPARC ΔI, αC were detected using a specific antibody. Binding assays were done several times, and representative data are shown here. Heat denaturation of CNBr peptides before coating and the presence of EDTA abolished the bindings. Shown is a representative of five experiments, each performed in duplicate.
We also found that chemical modification of the primary amino groups of collagens I and II reduced their affinity for SPARC ΔI, αC, with N-methylation having a lesser effect than N-acetylation (supplemental Fig. S3).
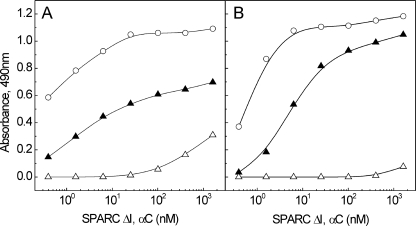
Binding of SPARC to the Fragments Obtained by MMP13 Cleavage—Mammalian collagenases are known to cleave collagens and yield two fragments consisting of ¾ and ¼ of collagen molecules. In this study, MMP13 was used to obtain the fragments of collagen I and procollagen III, and the resulting fragments were separated by size exclusion chromatography and analyzed by SDS-PAGE (electrophoretograms of the fragments obtained from procollagen III are shown in supplemental Fig. S2). These fragments were used in a solid phase assay to locate the SPARC-binding site on the collagen I (Fig. 4A) and III molecule (Fig. 4B). SPARC ΔI, αC interacted well with the larger fragment corresponding to the N-terminal three-quarters of collagen molecule; however, ∼10 times higher concentration of SPARC ΔI, αC was required to achieve the same half-maximal binding than to full-length collagens. The short fragment (one-quarter) of collagen I showed marginal binding (Fig. 4A), whereas the same fragment of collagen III had no binding (Fig. 4B). However, the binding is insignificant, and the data did not give any conclusion as to where the second binding site is located on collagen I.
FIGURE 4.
Binding of SPARC ΔI, αC to the fragments obtained by MMP13. Solid phase binding assay with collagen I (A) and procollagen III fragments (B) is shown. Immobilized ligands were collagen I or procollagen III (○), the N-terminal three-quarters fragment (▴), and the C-terminal one-quarter fragment (▵), and SPARC ΔI, αC was used as a soluble ligand. Shown is a representative of three experiments, each performed in duplicate.
Rotary shadowing showed a prominent binding to the region ∼80 nm from the C terminus of procollagens I and II but was much less obvious in procollagen III. Because this is in the area of the MMP cleavage site, we wondered whether SPARC may interfere with the binding of MMPs to collagens. Therefore, different molar ratios of SPARC ΔI, αC were incubated with collagens at room temperature for 30 min prior to addition of MMP1 or MMP13. The reaction mixtures were analyzed by SDS-PAGE to evaluate cleavage of collagens by MMPs. Only marginal inhibition was detected when SPARC ΔI, αC was present at more than 10-fold molar excess, and no difference was observed among collagen I, II and III (data not shown), suggesting that SPARC did not bind to the same site as MMPs.
Binding of Synthetic Peptides of Human Collagen III—Collagen III is a homotrimer composed of three α1(III) chains, and a set of synthetic peptides of human collagen III was used to map the SPARC-binding sites. The design of these peptides (Collagen III Toolkit) is described under “Experimental Procedures” and was characterized previously (11). We have tested that part of the Toolkit that covers residues 361–1029 of the triple-helical domain, with (GPP)10 as control. Only peptide 23 (GPC(GPP)5 GPOGPSGPRGQOGVMGFOGPKGNDGAO (GPP)5 GPC-NH2; residues 397–423, where O is 4-hydroxyproline) showed high affinity binding comparable with the full-length procollagen III. Truncation by four triplets, retaining the sequences GPSGPRGQOGVMGFO and GPRGQOGVMGFOGPK, reduced binding activity by 2 orders of magnitude. Further truncation to the sequences GPSGPRGQOGVM, GPSGPRGQO, and GFOGPK reduced binding yet further (Fig. 5).
FIGURE 5.
Solid phase assay using synthetic peptides of collagen III. Immobilized ligands used are procollagen III (○), peptide 23 containing the amino acids residues 397–423 (GPOGPSGPRGQOGVMGFOGPKGNDGAO) of the triple-helical domain of human collagen III (•), peptide 23GPS-GFO (GPSGPRGQOGVMGFO) (▵), peptide 23 GPS-GVM (GPSGPRGQOGVM) (▴), peptide 23GPS-GQO (GPSGPRGQO) (▿), peptide 23GFOGPK (▾), and peptide 23 GPRGPK (GPRGQOGVMGFOGPK) (⋄). Procollagen III and peptides were coated at 10 μg/ml at 4 °C overnight, and the binding assays were performed at room temperature. The peptide 23 showed high affinity binding, whereas the truncation to GPSGPRGQOGVM (▴) and GPSGPRGQO (▿) almost abolished the binding (these two lines overlap). Shown is a representative of five experiments, each performed in duplicate.
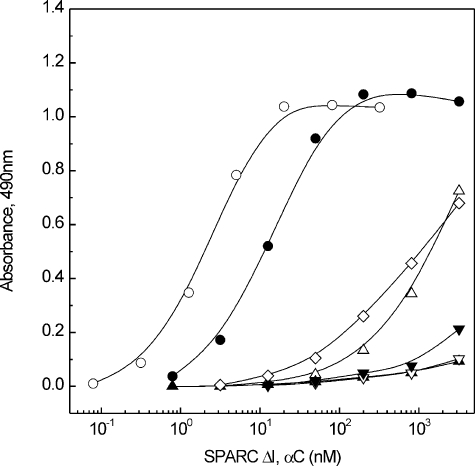
Effect of Collagen Mutations on SPARC Binding—We examined the effect of seven different mutations in the triple-helical domain of the pro-α2(I) chain of procollagen I samples obtained from patients affected with OI. The mutations are listed under “Experimental Procedures.” The data clearly showed a selective effect: two mutations (lethal Gly-421 → Asp and nonlethal Gly-688 → Ser) caused a large decrease in the affinity for SPARC ΔI, αC, whereas the other mutations had a smaller or no effect (Fig. 6). Saturation curves (supplemental Fig. S4) and dissociation constants for all OI samples (supplemental Table S1) are reported in the supplemental material.
FIGURE 6.
The effect of mutations in the α2 (I) chain of collagen I on the binding to SPARC ΔI, αC. Binding assays with mutated procollagen I samples compared with a normal procollagen I, and procollagen samples were used as immobilized ligands. The data were obtained at 50 μg/ml (1.7 μm) of SPARC ΔI, αC, and the binding to normal procollagen I was set to 100%. Mutations are disposed along the triple-helical domain according to the mutation site. The experiments were performed three times, and the S.D. of each datum was within 5–10%.
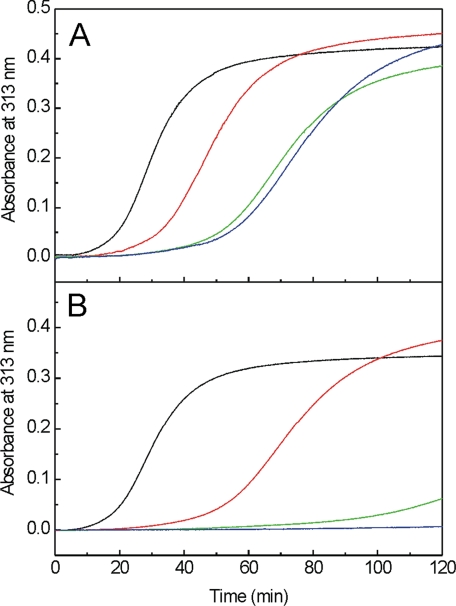
Effect of SPARC on Collagen I Fibrillogenesis—We examined collagen I fibril formation in vitro in the presence of SPARC or SPARC ΔI, αC. When SPARC was added, lag phases were lengthened by ∼40 min at 10- or 17-fold molar excess of SPARC over collagen I and by 20 min using a 1.7-fold excess (Fig. 7A). In the presence of SPARC ΔI, αC at equimolar or 2.6-fold excess, fibrillogenesis was nearly or completely inhibited over 2 h (Fig. 7B). With collagen I in 3.8-fold excess over SPARC ΔI, αC, fibrillogenesis was delayed by 40 min. No effect was observed when either SPARC or SPARC ΔI, αC were added in the middle of the fiber growth phase. These data suggest that SPARC may control the nucleation of collagen fiber assembly.
FIGURE 7.
The effect of SPARC and SPARC ΔI, αC on collagen I fibrillogenesis assay. Collagen fibrillogenesis were monitored at 313 nm. In A, in the absence of SPARC (black line), SPARC was added at the ratio of collagen/SPARC, 1:1.7 (red line), 1:10 (green line), and 1:17 (blue line). In B, in the absence of SPARC ΔI, αC(black line), SPARC ΔI, αC was added at the ratio of collagen/SPARC ΔI, αC, 3.8:1 (red line), 1:1 (green line), and 1:2.6 (blue line). Shown is a representative of three experiments.
DISCUSSION
In this study we have used several different strategies to identify the binding sites for SPARC on fibrillar collagens type I, II, and III. Rotary shadowing demonstrated two binding sites on procollagens I and II, and one prominent binding site on the procollagen III molecule (Fig. 8A). We found a major site located at ∼180 nm from the C terminus (circa residue 400 in the triple-helical domain). A less preferred binding site is located 60–100 nm from the C terminus. These data are in partial agreement with the binding regions mapped by atomic force microscopy (34), in which the major binding of SPARC to procollagen I was identified 87.5–125 nm from the C terminus. No binding was reported 180 nm from the C terminus, but a second putative binding site was described at 237.5–262.5 nm from the C terminus (34), which might correspond to our low frequency binding region on procollagen III (260–284 nm from C terminus, residues 160–190) and to the low affinity binding to α1(II) CB12. The discrepancy could be due to the different experimental methods or the source of SPARC. Wang et al. (34) used recombinant SPARC expressed in insect cells, and such proteins tend to undergo mannose-rich glycosylation (47). SPARC with high mannose N-glycans, purified from bone or expressed in an osteosarcoma cell line, were shown to have a higher affinity for collagen I (33). On the other hand, prevention of N-glycosylation by introducing a mutation caused an insignificant change in the binding of SPARC to collagens I and IV (28). These data suggest that mannose-rich N-glycans might interact with collagen I directly, whereas our HEK293-expressed SPARC may interact differently with collagen.
FIGURE 8.
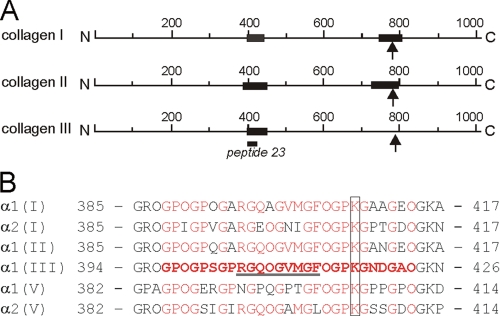
Schematic representation of the SPARC-binding sites (A) and sequence comparison of putative SPARC binding regions on human collagens (B). A, the black bars show the binding regions based on the data obtained from this study. The arrows indicate the MMP cleavage site and the position of peptide 23 is shown. B, amino acid sequences of human collagens were obtained from the Swiss-Prot data base. The accession numbers are P02452 for α1(I) chain, P08123 for α2(I) chain, P02458 for α1(II) chain, P02461 for α1(III) chain, P20908 for α1(V), and P05997 for α2(V) chain. The amino acid residues are numbered from the first glycine of the triple-helical domain. The peptide sequence that showed a binding is shown in red bold letters. Identical residues in other collagens are indicated in red. The conserved lysine residues are marked. The binding site for VWF (52) is underlined. O represents 4-hydroxyproline.
We have prepared and used a set of CNBr peptides of bovine collagen I and II for SPARC binding analyses. CB3,8 of α1(I) exhibited significant binding, whereas individual CB3 and CB8 showed no binding, suggesting that the binding site is located close to methionine 402. Similarly, homologous peptides CB8 and CB11 in collagen II did not bind SPARC. For the α2(I) chain, we could test only CB3,5, a peptide that embraces both binding sites visualized by rotary shadowing of procollagen I. However, its binding to SPARC was much weaker than CB3,8, CB3,7, and CB7 of α1(I), suggesting that the contribution of the α2(I) chain to binding is low or that fully competent sites require a contribution from the α1(I) chain as well as α2(I), as might be the case in native heterotrimers. Such a scenario has been proposed for the binding of von Willebrand factor (VWF) to collagen I (48).
Using the Collagen III Toolkit (11), we identified GPOGPSGPRGQOGVMGFOGPKGNDGAO as the minimal peptide sequence for SPARC binding. We searched for this sequence among the collagens shown to interact with SPARC and found highly conserved sequences in the α chains of collagen I, II, and V (Fig. 8B). SPARC also binds to basement membrane collagen type IV. Although the electron microscopy after rotary shadowing demonstrated one SPARC ΔI, αC molecule on collagen IV ∼185 nm from the C terminus (data not shown), we failed to find homologous sequences in the α1(IV) and α2(IV) chains. The localization of the SPARC-binding sequence in collagen I and II fitted well with the data obtained by rotary shadowing and CNBr peptides (Figs. 2 and 3). The reduced binding after N-acetylation of Lys/Hyl residues may reflect loss of a positive charge involved in binding or steric hindrance of the interaction by the more bulky, N-acetylated side chains (supplemental Fig. S3). The presence of a conserved lysine residue in the binding sequence of these collagen chains suggests that this lysine residue may be involved in SPARC binding (Fig. 8B).
Several extracellular matrix proteins possess EC domains related to SPARC (49). A sequence comparison of those regions of SPARC critical for collagen binding shows that quail retina protein QR1 (50) and SC1/Hevin (51, 52) are remarkably conserved, including all five essential residues (Arg-149, Asn-156, Leu-242, Met-245, and Glu-246), and we predict that they may share binding sites on collagens with SPARC. The crystal structure of the EC domain of SPARC ΔI, αC revealed that these five residues are arranged in a ring of ∼15-Å diameter, surrounded by residues that made little contribution to collagen binding, and also suggested that two additional residues, Trp-153 and Pro-241, may be part of the collagen-binding site. This model suggested that the complementary SPARC-binding site on collagens does not extend beyond two Gly-X-Y triplets (29). Recently the binding site for VWF in collagens II and III was identified using the Collagen III Toolkit (48). The minimal sequence for VWF binding is RGQOGVMGF, which overlaps with the SPARC-binding sequence identified here. Interestingly, the collagen-binding receptor tyrosine kinase, discoidin domain receptor 2 also binds to the corresponding site on collagen II and to peptide 23 of collagen III tool kit (53). Those truncated peptides of peptide 23 that still bound VWF failed to bind fully to SPARC, indicating that the binding sites for the two proteins are close but not identical. The smallest peptide found to bind fully to SPARC spans nine Gly-X-Y triplets, and because truncation to five triplets reduced the binding of SPARC (Fig. 5), the full SPARC-binding site must embrace a longer stretch of collagen. In contrast, the collagen-binding sites of integrins (12), glycoprotein VI (54), and discoidin domain receptor 2 (55) as well as of VWF accommodate no more than four triplets. However, the EC domain of SPARC is no larger than the collagen-binding domains of any of these proteins. The data obtained in this study may suggest substantial extra contributions from SPARC regions other than its mapped binding site. Currently we have no explanation of this discrepancy. Structural analysis of the SPARC-collagen complex at the atomic level is required to elucidate the precise molecular interaction.
One of us4 proposed a regional model of OI mutations in the α2(I) chain, which occur in clusters of alternating lethal and nonlethal domains; the function of such domains is unknown but may reflect the pattern of binding sites for collagen ligands (56). We have used seven differently mutated type I procollagen samples from OI patients, all of the mutations being single glycine substitutions in the α2(I) chain. The results obtained here demonstrate a selective effect among the mutations on the SPARC ΔI, αC binding capacity of procollagen I; two mutations, one lethal and one nonlethal, cause a large decrease in affinity, whereas the other mutations (two lethal and three nonlethal) are largely without effect (Fig. 6). These results provide a possible correlation for one of the α2 (I) lethal clusters but do not appear to be related to the remaining clusters.
Collagen pathology is a complex field where mutations may have a local structural effects (e.g. the formation of a kink, described for some mutations) and/or long range influence (overmodifications in most cases, lower susceptibility to N-proteinase in some cases) (57); disruption of collagen epitopes and register alterations of the three chains cannot be excluded for single glycine substitutions. For a subset of Gly to Cys or Asp substitutions that also cause kinking, there is apparently a register shift that is propagated as far as all the way down the N-terminal end of the helix and disrupts N-proteinase processing, for example, Gly-718 (58) and Gly-748 (59) of α1 (I) chain. A local effect may be sufficient to explain the effect of the Gly-421 → Asp mutation, very close to the main binding site for SPARC (Fig. 8B); the very low binding capacity to SPARC may be attributed to the presence of a structural abnormality at the mutation site (the formation of a kink) (60). Furthermore, overhydroxylation of the conserved Lys-408 in the SPARC-binding site, present in both α1(I) and α2(I) chains, and/or of Lys-420 present in α2(I) only, and overglycosylation of the resultant Hyl might contribute to reduced binding. On the other hand, only a long range effect can be attributed to the Gly-688 → Ser substitution, because it is far from the SPARC-binding sites (Fig. 8A). We speculate that it may decrease binding through a long range effect, such as kinking or shifting the register between the α1 and/or α2 chains. This speculation of a discrete long range effect is strengthened by the fact that the nearby Gly-706 mutation does not disrupt SPARC binding.
One biological implication of SPARC-collagen interaction is the regulation of collagen fibrillogenesis as described in a recent publication by Rentz et al. (61). It was found that fibril aggregation in collagens in the dermis of SPARC-null mice was inefficient, and they assembled with diameters of 60–70 nm, a proposed intermediate range in collagen fibril growth. In vitro analysis also demonstrated that SPARC regulates processing of procollagen I and collagen fibrillogenesis. In this study, we analyzed collagen I fibrillogenesis in vitro in the presence of SPARC or SPARC ΔI, αC. Although collagen fibrillogenesis was delayed in the presence of SPARC, the effect of SPARC ΔI, αC was more pronounced, and the fibrillogenesis was inhibited almost completely even at levels equimolar with collagen. On the other hand, Hevin, which shares the critical amino acids for the collagen binding in the homologous EC domain with SPARC, was shown to enhance the collagen fibril formation (62), and these proteins have opposite effects on the collagen assembly. The data obtained in this study suggest that SPARC may regulate the initiation of collagen fibril formation. This process might be modulated by proteolytic activation of SPARC. However, the mechanism for collagen fibrillogenesis in vivo is quite complex, and further analyses in vitro and in vivo are necessary to elucidate the function of SPARC.
Peptides derived from the C-telopeptides of the α1(I) and α2(I) chain were shown to inhibit collagen I self-assembly, and these peptides bound to a region between residues 776 and 822 of the α1(I) chain, narrowed to residues 781–794 by molecular modeling (63). This region coincides with the second SPARC-binding site on collagen I and II demonstrated by both rotary shadowing and binding analysis using CNBr peptides. It is possible that SPARC binds to this self-assembly initiation site, but we cannot exclude the possibility that its binding to a region between 391 and 412 of α1(I) and α2(I) chains caused delay/inhibition of collagen fibrillogenesis.
The biological consequences of SPARC binding to collagens in vivo may include the regulation of collagen assembly, but sequestration or complexing with collagen may also modulate the many biological activities reported for SPARC. These include anti-adhesive and anti-proliferative activities and the regulation of growth factor function (14, 15). Furthermore, VWF and discoidin domain receptor 2 were shown to bind to the same region as SPARC (48, 53), suggesting that SPARC might modulate their activities by competing with their binding to collagens. The data obtained in this study will provide defined ligands with which to manipulate and further study the function of SPARC.
Supplementary Material
Acknowledgments
We are grateful for the excellent technical assistance of Vera van Delden, Christa Wendt, Mischa Reiter, and Dorothea Jahn. We thank Dr. Darwin Prockop, Dr. Gillian Murphy, and Dr. Hideaki Nagase for providing valuable reagents, Dr. Karlheinz Mann for protein sequencing, Dr. Kazunori Mizuno for preparing supplemental Fig. S1, and Dr. Jürgen Engel and Dr. Erhard Hohenester for critically reading the manuscript.
This work was authored, in whole or in part, by National Institutes of Health staff. This work was supported by Deutsche Forschungsgemeinschaft Grants Sa 1003/1 and 2 (to T. S.), European Community Project QLK3-CT2000-00084 (to R. T. and T. S.), and grants from the Shriners Hospital for Children (to H. P. B.), the Wellcome Trust (to R. W. F.), and by University of Pavia (to R. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
Footnotes
The abbreviations used are: EC, extracellular calcium-binding; MMP, matrix metalloproteinase; OI, osteogenesis imperfecta; VWF, von Willebrand factor.
J. C. Marini, unpublished observations.
References
- 1.Myllyharju, J., and Kivirikko, K. I. (2004) Trends Genet. 20 33-43 [DOI] [PubMed] [Google Scholar]
- 2.Veit, G., Kobbe, B., Keene, D. R., Paulsson, M., Koch, M., and Wagner, R. (2006) J. Biol. Chem. 281 3494-3504 [DOI] [PubMed] [Google Scholar]
- 3.Gara, S. K., Grumati, P., Urciuolo, A., Bonaldo, P., Kobbe, B., Koch, M., Paulsson, M., and Wagner, R. (2008) J. Biol. Chem. 283 10658-10670 [DOI] [PubMed] [Google Scholar]
- 4.Brodsky, B., and Shah, N. K. (1995) FASEB J. 9 1537-1546 [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. C., and Timpl, R. (1995) Int. Arch. Allergy Immunol. 107 484-490 [DOI] [PubMed] [Google Scholar]
- 6.Olsen, B. R., and Ninomiya, Y. (1999) Guidebook to the Extracellular Matrix, Anchor, and Adhesion Proteins, 2nd Ed., pp. 380-398, Oxford University Press, Oxford
- 7.Leitinger, B., and Hohenester, E. (2007) Matrix Biol. 26 146-155 [DOI] [PubMed] [Google Scholar]
- 8.Di Lullo, G. A., Sweeney, S. M., Korkko, J., Ala-Kokko, L., and San Antonio, J. D. (2002) J. Biol. Chem. 277 4223-4231 [DOI] [PubMed] [Google Scholar]
- 9.Knight, C. G., Morton, L. F., Peachey, A. R., Tuckwell, D. S., Farndale, R. W., and Barnes, M. J. (2000) J. Biol. Chem. 275 35-40 [DOI] [PubMed] [Google Scholar]
- 10.Siljander, P. R. M., Hamaia, S., Peachey, A. R., Slatter, D. A., Smethurst, P. A., Ouwehand, W. H., Knight, C. G., and Farndale, R. W. (2004) J. Biol. Chem. 279 47763-47772 [DOI] [PubMed] [Google Scholar]
- 11.Raynal, N., Hamaia, S. W., Siljander, P. R. M., Maddox, B., Peachey, A. R., Fernandez, R., Foley, L. J., Slatter, D. A., Jarvis, G. E., and Farndale, R. W. (2006) J. Biol. Chem. 281 3821-3831 [DOI] [PubMed] [Google Scholar]
- 12.Emsley, J., Knight, C. G., Farndale, R. W., Barnes, M. J., and Liddington, R. C. (2000) Cell 101 47-56 [DOI] [PubMed] [Google Scholar]
- 13.Bornstein, P., and Sage, E. H. (2002) Curr. Opin. Cell Biol., 14 608-616 [DOI] [PubMed] [Google Scholar]
- 14.Brekken, R. A., and Sage, E. H. (2001) Matrix Biol. 19 816-827 [DOI] [PubMed] [Google Scholar]
- 15.Bradshaw, A. D., and Sage, E. H. (2001) J. Clin. Investig. 107 1049-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan, Q., and Sage, E. H. (1999) J. Histochem. Cytochem. 47 1495-1506 [DOI] [PubMed] [Google Scholar]
- 17.Wang, H., Workman, G., Chen, S., Barker, T. H., Ratner, B. D., Sage, E. H., and Jiang, S. (2006) Matrix Biol. 25 20-26 [DOI] [PubMed] [Google Scholar]
- 18.Barker, T. H., Baneyx, G., Cardo-Vila, M., Workman, G. A., Weaver, M., Menon, P. M., Dedhar, S., Rempel, S. A., Arap, W., Pasqualini, R., Vogel, V., and Sage, E. H. (2005) J. Biol. Chem. 280 36483-36493 [DOI] [PubMed] [Google Scholar]
- 19.Gilmour, D. T., Lyon, G. J., Carlton, M. B. L., Sanes, J. R., Cunningham, J. M., Anderson, J. R., Hogan, B. L. M., Evans, M. J., and Colledge, W. H. (1998) EMBO J. 17 1860-1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norose, K., Clark, J. I., Syed, N. A., Basu, A., Heber-Katz, E., Sage, E. H., and Howe, C. C. (1998) Investig. Ophthalmol. Vis. Sci. 39 2674-2680 [PubMed] [Google Scholar]
- 21.Delany, A. M., Kalajzic, I., Bradshaw, A. D., Sage, E. H., and Canalis, E. (2003) Endocrinology 144 2588-2596 [DOI] [PubMed] [Google Scholar]
- 22.Bradshaw, A. D., Puolakkainen, P., Dasgupta, J., Davidson, J. M., Wight, T. N., and Sage, E. H. (2003) J. Investig. Dermatol. 120 949-955 [DOI] [PubMed] [Google Scholar]
- 23.Bradshaw, A. D., Reed, M. J., and Sage, E. H. (2002) J. Histochem. Cytochem. 50 1-10 [DOI] [PubMed] [Google Scholar]
- 24.Maurer, P., Hohenadl, C., Hohenester, E., Göhring, W., Timpl, R., and Engel, J. (1995) J. Mol. Biol. 253 347-357 [DOI] [PubMed] [Google Scholar]
- 25.Hohenester, E., Maurer, P., Hohenadl, C., Timpl, R., Jansonius, J. N., and Engel, J. (1996) Nat. Struct. Biol. 3 67-73 [DOI] [PubMed] [Google Scholar]
- 26.Hohenester, E., Maurer, P., and Timpl, R. (1997) EMBO J. 16 3778-3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurer, P., Göhring, W., Sasaki, T., Mann, K., Timpl, R., and Nischt, R. (1997) Cell Mol. Life Sci. 53 478-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki, T., Göhring, W., Mann, K., Maurer, P., Hohenester, E., Knäuper, V., Murphy, G., and Timpl, R. (1997) J. Biol. Chem. 272 9237-9243 [DOI] [PubMed] [Google Scholar]
- 29.Sasaki, T., Hohenester, E., Göhring, W., and Timpl, R. (1998) EMBO J. 17 1625-1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki, T., Miosge, N., and Timpl, R. (1999) Matrix Biol. 18 499-508 [DOI] [PubMed] [Google Scholar]
- 31.Xie, R. L., and Long, G. L. (1995) J. Biol. Chem. 270 23212-23217 [DOI] [PubMed] [Google Scholar]
- 32.Kelm, R. J., and Mann, K. G. (1991) J. Biol. Chem. 266 9632-9639 [PubMed] [Google Scholar]
- 33.Kaufmann, B., Müller, S., Hanisch, F. G., Hartmann, U., Paulsson, M., Maurer, P., and Zaucke, F. (2004) Glycobiology 14 609-619 [DOI] [PubMed] [Google Scholar]
- 34.Wang, H., Fertala, A., Ratner, B. D., Sage, E. H., and Jiang, S. (2005) Anal. Chem. 77 6765-6771 [DOI] [PubMed] [Google Scholar]
- 35.Rossi, A., Zuccarello, L. V., Zanaboni, G., Monzani, E., Dyne, K. M., Cetta, G., and Tenni, R. (1996) Biochemistry 35 6048-6057 [DOI] [PubMed] [Google Scholar]
- 36.Rossi, A., Zanaboni, G., Cetta, G., and Tenni, R. (1997) J. Mol. Biol. 269 488-493 [DOI] [PubMed] [Google Scholar]
- 37.Tenni, R., Viola, M., Welser, F., Sini, P., Giudici, C., Rossi, A., and Tira, M. E. (2002) Eur. J. Biochem. 269 1428-1437 [DOI] [PubMed] [Google Scholar]
- 38.Fertala, A., Sieron, A. L., Ganguly, A., Li, S. W., Ala-Kokko, L., Anumula, K. R., and Prockop, D. J. (1994) Biochem. J. 298 31-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nischt, R., Pottgiesser, J., Krieg, T., Mayer, U., Aumailley, M., and Timpl, R. (1991) Eur. J. Biochem. 200 529-536 [DOI] [PubMed] [Google Scholar]
- 40.Giudici, C., Viola, M., Tira, M. E., Forlino, A., and Tenni, R. (2003) FEBS Lett. 547 170-176 [DOI] [PubMed] [Google Scholar]
- 41.Marini, J. C., Forlino, A., Cabral, W. A., Barnes, A. M., San Antonio, J. D., Milgrom, S., Hyland, J. C., Korkko, J., Prockop, D. J., De Paepe, A., Couche, P., Symoens, S., Glorieux, F. H., Roughley, P. J., Lund, A. M., Kuurilla-Svahn, K., Hartikka, H., Cohn, D. H., Krakow, D., Mottes, M., Schwarze, U., Chen, D., Yang, K., Kuslich, C., Troendle, J., Dalgleish, R., and Byers, P. H. (2007) Hum. Mutat. 28 209-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosokawa, N., Hohenadl, C., Satoh, M., Kühn, K., and Nagata, K. (1998) J. Biochem. (Tokyo) 124 654-662 [DOI] [PubMed] [Google Scholar]
- 43.Knäuper, V., Lopez-Otin, C., Smith, B., Knight, G., and Murphy, G. (1996) J. Biol. Chem. 271 1544-1550 [DOI] [PubMed] [Google Scholar]
- 44.Engel, J. (1994) Methods Enzymol. 245 469-488 [DOI] [PubMed] [Google Scholar]
- 45.Aumailley, M., Wiedemann, H., Mann, K., and Timpl, R. (1989) Eur. J. Biochem. 184 241-248 [DOI] [PubMed] [Google Scholar]
- 46.Shah, N. K., Ramshaw, J. A., Kirkpatrick, A., Shah, C., and Brodsky, B. (1996) Biochemistry 35 10262-10268 [DOI] [PubMed] [Google Scholar]
- 47.Altmann, F., Staudacher, E., Wilson, I. B. H., and März, L. (1999) Glycoconj. J. 16 109-123 [DOI] [PubMed] [Google Scholar]
- 48.Lisman, T., Raynal, N., Groeneveld, D., Maddox, B., Peachey, A. R., Huizinga, E. G., de Groot, P. G., and Farndale, R. W. (2006) Blood 108 3753-3756 [DOI] [PubMed] [Google Scholar]
- 49.Hohenester, E., and Timpl, R. (2004) Handbook of Metalloproteins, Vol. 3, pp. 509-515, John Wiley & Sons, Ltd, Chichester, UK [Google Scholar]
- 50.Guermah, M., Crisanti, P., Laugier, D., Dezelee, P., Bidou, L., Pessac, B., and Calothy, G. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 4503-4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnston, I. G., Paladino, T., Gurd, J. W., and Brown, I. R. (1990) Neuron 4 165-176 [DOI] [PubMed] [Google Scholar]
- 52.Girard, J. P., and Springer, T. A. (1995) Immunity 2 113-123 [DOI] [PubMed] [Google Scholar]
- 53.Konitsiotis, A. D., Raynal, N., Bihan, D., Hohenester, E., Farndale, R. W., and Leitinger, B. (2008) J. Biol. Chem., 283 6861-6868 [DOI] [PubMed] [Google Scholar]
- 54.Horii, K., Kahn, M. L., and Herr, A. B. (2006) Blood 108 936-942 [DOI] [PubMed] [Google Scholar]
- 55.Ichikawa, O., Osawa, M., Nishida, N., Goshima, N., Nomura, N., and Shimada, I. (2007) EMBO J. 26 4168-4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, Q., Orrison, B. M., and Marini, J. C. (1993) J. Biol. Chem. 268 25162-25167 [PubMed] [Google Scholar]
- 57.Kuivaniemi, H., Tromp, G., and Prockop, D. J. (1991) FASEB J. 5 2052-2060 [DOI] [PubMed] [Google Scholar]
- 58.Lightfoot, S. J., Holmes, D. F., Brass, A., Grant, M. E., Byers, P. H., and Kadler, K. E. (1992) J. Biol. Chem. 267 25521-25528 [PubMed] [Google Scholar]
- 59.Vogel, B. E., Doelz, R., Kadler, K. E., Hojima, Y., Engel, J., and Prockop, D. J. (1988) J. Biol. Chem. 263 19249-19255 [PubMed] [Google Scholar]
- 60.Forlino, A., Keene, D. R., Schmidt, K., and Marini, J. C. (1998) Matrix Biol. 17 575-584 [DOI] [PubMed] [Google Scholar]
- 61.Rentz, T. J., Poobalarahi, F., Bornstein, P., Sage, E. H., and Bradshaw, A. D. (2007) J. Biol. Chem. 282 22062-22071 [DOI] [PubMed] [Google Scholar]
- 62.Sullivan, M. M., Barker, T. H., Funk, S. E., Karchin, A., Seo, N. S., HööK, M., Sanders, J., Starcher, B., Wight, T. N., Puolakkkainen, P., and Sage, E. H. (2006) J. Biol. Chem. 281 27621-27632 [DOI] [PubMed] [Google Scholar]
- 63.Prockop, D., and Fertala, A. (1998) J. Biol. Chem. 273 15598-15604 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.