Abstract
An OxlT homology model suggests R272 and K355 in transmembrane helices 8 and 11, respectively, are critical to OxlT-mediated transport. We offer positive evidence supporting this idea by studying OxlT function after cysteine residues were separately introduced at these positions. Without further treatment both mutant proteins had a null phenotype on reconstitution into proteoliposomes. By contrast, significant recovery of function occurred when proteoliposomes were treated with MTSEA (methanethiosulfonate ethylamine), a thiol-specific reagent that implants a positively charged amino group. In each case, there was a two-fold increase in the Michaelis constant (KM) for oxalate self-exchange (from 80 μM to 160 μM), along with a five-fold (K355C) or 100-fold (R272C) reduction in VMax compared to the cysteine-less parental protein. Analysis by MALDI-TOF confirmed that MTSEA introduced the desired modification. We also examined substrate selectivity for the treated derivatives. While oxalate remained the preferred substrate, there was a shift in preference among other substrates, so that the normal rank order (oxalate > malonate > formate) was altered to favor smaller substrates (oxalate > formate > malonate). This shift is consistent with the idea that the substrate-binding site is reduced in size by introduction of the -SCH2CH2NH3+ adduct, which generates a side chain about 1.85 Å longer than that of lysine or arginine. These findings lead us to conclude that R272 and K355 are essential components of the OxlT substrate-binding site.
In the anaerobic bacterium, Oxalobacter formigenes, the oxalate transporter, OxlT, allows the exchange of external divalent oxalate with the intracellular monovalent formate derived from oxalate decarboxylation (1, 2). The overall effect of these associated activities (exchange and decarboxylation) is generation of a proton-motive force to support membrane functions, including ATP synthesis, accumulation of growth substrates and extrusion of waste products (1–4).
The way that OxlT helps establish a proton-motive force clarifies the role of similar cycles in other bacteria (4, 5) and broadens the variety of mechanisms known to generate metabolic energy. Of equal interest, OxlT also serves as a model for understanding structure-function relationships in the Major Facilitator Superfamily (MFS)1, a collection of evolutionarily related transporters encompassing 30–40% of the so-called ‘secondary’ transport systems (6, 7). Thus, helix organization in the MFS was first described by electron crystallography of OxlT (8–10), confirming the presence of the 12 transmembrane α-helices predicted by less direct methods (11, 12). More detailed organizational features were revealed by later work, using x-ray crystallography of two other members of the MFS, LacY (13) and GlpT (14).
Analysis of OxlT hydropathy implicated K355 as a ligand-binding element, since this residue appeared to lie at the center of TM11 (11). Consistent with this idea, one finds that mutants lacking K355 are inactive (11, 15, 16) and that TM11 itself lines the transport pathway (15, 16). Since oxalate is a divalent anion at physiological pH (pH 7), it is of interest to ask if the OxlT hydrophobic sector has a second basic residue to facilitate substrate binding, and in this regard valuable insight comes from an OxlT homology model (17) based on the x-ray structure of GlpT (14). This model supports the idea that K355 is used in ligand binding and points to R272, in TM8, as a likely partner in this event; early work (17) shows that R272, too, is required for normal OxlT function.
The work presented here provides additional evidence bearing on the suggestion that K355 and R272 function in substrate binding and transport. In particular, we show these residues are essential for OxlT function, but not biogenesis or assembly, and we document that alterations of side-chain architecture at these sites lead to changes of substrate specificity.
EXPERIMENTAL PROCEDURES
Mutagenesis and Protein Expression
OxlT and its mutants were encoded within a 1.4 kb Xba1-HindIII fragment in pBluescript II SK+ (11, 18). To suppress uninduced protein expression, Ampr plasmids harboring OxlT were carried in Escherichia coli strain XL1 along with plasmid pMS421 (Specr, LacIq) (11). A fully functional cysteine-less variant of OxlT (C28G/C271A) with a C-terminal polyhistidine (His9) tail was used as parent to all single-cysteine mutations (15, 18). For protein expression, a colony from a fresh transformation was placed in LB broth with antibiotics and grown overnight at 37C. Cells were the diluted 100-fold into fresh medium and grown with shaking at 33–35C until A600 reached 0.12, at which point OxlT expression was induced by 1 mM IPTG. Cells were harvested after an additional 3 hr.
To detect OxlT in crude detergent extracts or after reconstitution into proteoliposomes, immunoblots were probed with antibody directed against polyhistidine; chemilumenescence was monitored using a Fuji LAS1000 gel documentation system (18).
Reconstitution and Assays of OxlT Function
Unless otherwise indicated, membranes prepared by osmotic lysis (1, 11) were solubilized in buffer containing 100 mM potassium oxalate, 50 mM potassium phosphate (pH 7), 20% (v/v) glycerol, 1.5% (w/v) octylglucoside and 0.5% (w/v) E. coli phospholipid (hydrated in distilled water). After clarification by centrifugation, solubilized protein (0.7–1 mg/ml) was reconstituted by detergent dilution (1, 19) to give proteoliposomes loaded with 100 mM potassium oxalate, 50 mM potassium phosphate (pH 7); after reconstitution, proteoliposomes were centrifuged, washed twice and suspended as a stock using the same buffer in which potassium oxalate was replaced by potassium sulfate. In a few cases, as noted, the suspension and assay buffer contained 50 mM MOPS/K plus 100 mM potassium gluconate instead of 50 mM potassium phosphate plus 100 mM potassium sulfate, with equivalent results. During studies of protease sensitivity, 4(2-aminoethyl)-benzylsulfonyl fluoride, normally present 0.25 mM, was omitted, and the solubilization buffer was altered so that the 100 mM potassium oxalate was replaced with either 50 mM potassium oxalate or 50 mM potassium sulfate (pH 7).
In routine experiments, [14C]oxalate transport was measured in duplicate at room temperature in 1 ml volumes containing 0.1 ml proteoliposome stock with 0.8 ml suspension buffer and additives. The reaction was initiated by addition of 0.1 ml of 0.1 mM labeled oxalate, after which 0.2 ml samples were filtered using Millipore GSTF filter (0.2 μm pore size) and washed twice with cold buffer. In some experiments, we evaluated apparent inhibition constants (Kiapp) for competing substrates. In these tests, 5 trace amounts of external unlabeled oxalate were removed by placing duplicate 0.2 ml aliquots of proteoliposomes onto the filter, followed by two washes using cold assay buffer. The vacuum was then disengaged, and samples were overlaid with 0.2 ml of a reaction mixture containing both [14C]oxalate and the competing compound (see 2). After a specified time (usually 30 s), the reaction was terminated by vacuum filtration, with washing as above. MTS-linked agents were normally added in excess (1 mM) from 1 M stocks prepared in dimethylsulfoxide; solvent-treated preparations served as controls.
MALDI-TOF Mass Spectrometry
Purified OxlT (0.3–1 mg/ml) was prepared by metal-chelate affinity chromatography (15, 18), with elution at pH 4.3 using 100 mM potassium oxalate, 50 mM potassium acetate, 20% glycerol and 0.25% diheptanolylphosphatidylcholine (8, 9). Purified protein was desalted and recovered in 1% glycerol (at times also with 0.1% diheptanolylphosphatidylcholine) using gel filtration (CentriSep, Princeton Separations). For mass determination using MALDI-TOF mass spectrometry (20), sample wells were layered alternately with 2 μl volumes of the experimental solution and the sinapinic acid matrix (10 mg/ml 3,5-dimethyoxy-4-hydroxycinnamic acid in 50% acetonitrile, 0.3% trifluoroacetic acid (w/v/v)). For calibration, bovine serum albumin (1 mg/ml) was used either as an internal standard, by mixing with the purified sample (1:10), or as an external control, by its placement in a well surrounded by samples; equivalent results were obtained with both methods. Spectra were acquired with a Voyager DE-STR MALDI-TOF mass spectrometer (Applied BioSystems).
RESULTS
R272 and K355 Facilitate OxlT Function
In the OxlT homology model (17), R272 and K355 are positioned so that their side chains extend into the permeation pathway, where they are presumed to interact with the divalent substrate, oxalate. This hypothesis is consistent with the findings that among derivatives at these sites, only the R272K and K355R variants retain measurable function (11, 17), although at relatively low levels (see below). Such work did not identify whether these null responses relate to OxlT biogenesis or OxlT function, and to address this issue we adopted a strategy resembling that used to identify a salt bridge in LacY (23, 24). Thus, we studied OxlT after reconstitution into proteoliposomes, where the already-assembled molecule is accessible to a variety of informative probes (see 15, 16, 21, 22). Accordingly, we generated the single-cysteine R272C and K355C derivatives and studied their responses to MTSEA, which is expected to generate a new side chain, resembling lysine, by addition of the -SCH2CH2NH3+ adduct. While the cysteine-substituted variants show no detectable function under normal conditions, in both cases treatment of proteoliposomes with excess (1 mM) MTSEA led to restoration of OxlT function (Fig. 1). This was especially striking for the K355C derivative, whose activity increased from effectively nil to nearly 20% of its parental protein (Fig. 1A). On the same time scale, functional recovery for the MTSEA-treated R272C variant was considerably less. Even so, extended assay showed evidence of functional recovery in this case as well (Fig. 1B).
Fig. 1.
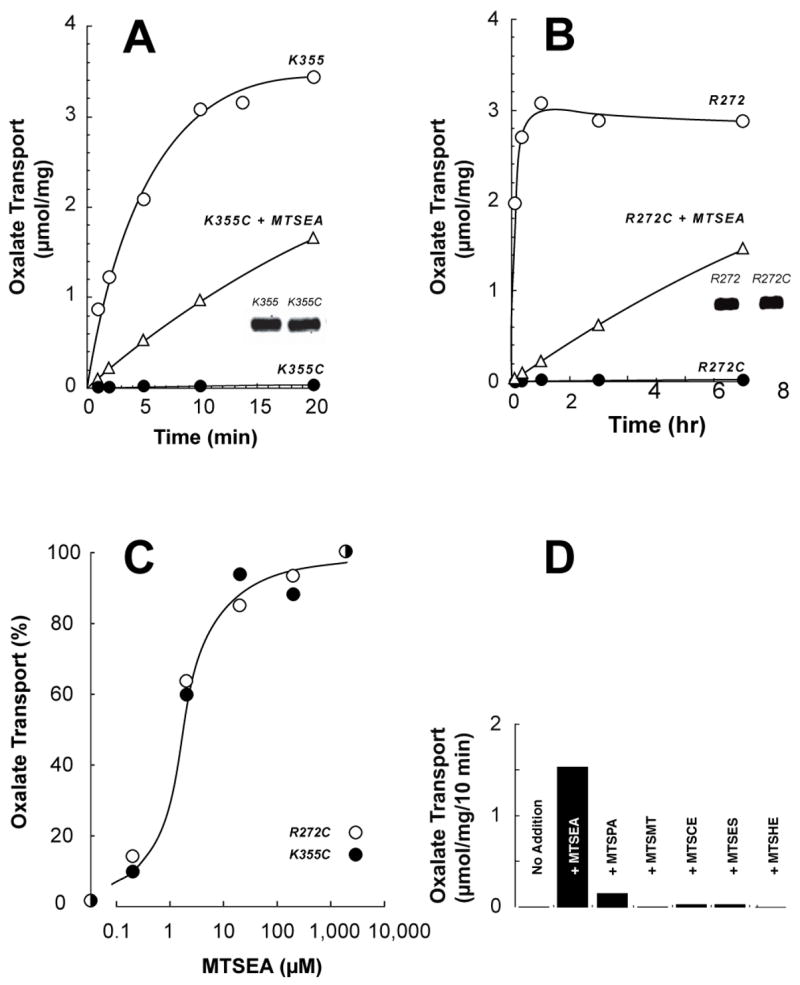
Functional rescue of OxlT by modification of K355C and R272C with MTSEA. (A, B) Oxalate-loaded proteoliposomes were prepared using membranes from cells expressing the cysteine-less parent and its K355C (panel A) or R272C (panel B) derivatives. Ten min before assay, proteoliposomes were given either 1 mM MTSEA (triangles) or an equivalent volume of the dimethylsulfoxide solvent (circles). After addition of 0.1 mM [14C]oxalate, samples were taken for filtration and washing as indicated. Insets: Immunoblot analysis shows reconstitution of comparable levels of cysteine-less OxlT and the K355C (panel A) or R272C (panel B) derivative. (C) Proteoliposomes were exposed to solvent or MTSEA. Initial rates of [14C]oxalate transport are given as a percentage of that found with excess (2 mM) MTSEA. (D) As in Part A or B, using proteoliposomes with the K355C variant exposed to solvent (no addition) or the indicated agents at 1 mM: MTSEA; MTSPA, aminopropyl methanethiosulfonate; MTSMT, (trimmethylammonium)methyl methanethiosulfonate; MTSCE, carboxyethyl methanethiosulfonate methanethiosulfonate; MTSES, sulfonatoethyl methanethiosulfonate methanethiosulfonate; and MTSHE, hydroxyethyl methanthiosulfonate.
Further study revealed, for both variants, that half-maximal responses were elicited by relatively low (ca. 1 μM) levels of MTSEA (Fig. 1C). Moreover, we also noted that similar functional recovery could be achieved by treatment before reconstitution. Thus, when normalized for protein recovered during reconstitution, activity found after MTSEA treatment of membranes (before solubilization) or detergent extract (before reconstitution) ranged from 84% to 117% (97 ± 8%, mean ± SE) of that observed when proteoliposomes were treated (as in Fig. 1). These data indicate that MTSEA modification may be studied in a variety of settings, and that the reaction proceeds to the same extent when using membranes, detergent extracts or proteoliposomes. Such data also suggest that MTSEA modification is not influenced by the presence of substrate, since 100 mM potassium oxalate was present in the detergent extracts, but not in the buffers used to suspend membranes or proteoliposomes (see Experimental Procedures).
In other work, we modified K355C with alternative probes (Fig. 1D). With one exception, we found no positive response for agents that introduce either a positive charge ((trimethylammonium)methyl methanethiosulfonate, (trimethylammonium)ethyl methanethiosulfonate (not shown) and 2-guanidinoethyl 2-guanidinoethanethiosulfanate (not shown)), a negative charge (carboxyethyl methanethiosulfonate and sulfonatoethyl methanethiosulfonate) or a neutral substituent (hydroxyethyl methanethiosulfonate). In the exceptional case, when aminopropyl methanethiosulfonate was used, the positive response was about 10% that found with MTSEA. In these trials, we did not confirm that each probe introduced the expected modification. Instead, we presumed considerations of size and/or charge did not interfere with probe accessibility or reactivity, because (i) we could document the expected modification after treatment with MTSEA itself (see below), and (ii) each probe can function as an inhibitor of variants with cysteine substitutions in pathway-lining residues in TM2, TM5 or TM11 (15, 16, 22; data not given).
The low level of functional recovery for the R272C variant (Fig. 1B) was of some concern. On the one hand, since the modified target resembles lysine more closely than arginine, one might expect reduced activity, as for the R272K derivative (17). It was also feasible, however, that MTSEA modified only a small fraction of the target population. To address this issue, we relied on early work showing that in crude detergent extracts substrate binding protects OxlT from denaturation and proteolysis (18, 25; unpublished). We reasoned that if MTSEA failed to modify the R272C variant, addition of substrate would fail to stabilize the population. In preliminary work, we confirmed that the cysteine-less parent showed the expected behavior (not shown). We then used immunoblot analysis to examine protease sensitivity for samples treated or untreated with MTSEA, in the absence and presence of oxalate. For example, when the K355C derivative was examined, incubation at room temperature led proteolysis, and this did not appear markedly altered by MTSEA modification (Fig. 2A). And as expected, the MTSEA treated sample was stabilized by oxalate, while proteolysis of the unmodified protein was unaffected by substrate (Fig. 2B). A somewhat different pattern emerged on study of the R272 variant. Thus, without MTSEA treatment, this mutant did not appear susceptible to proteolysis, yet after MTSEA modification, marked protease sensitivity was evident (Fig. 2C). It is also clear that addition of substrate (oxalate) promotes stability of MTSEA-modified material, as it did for the K355C variant (Fig. 2D). We draw two conclusions from these findings. First, it appears that part of the marked instability of OxlT may be attributed to the influence of R272, since in its absence (i.e., the R272C variant) OxlT has an extended lifetime in crude extracts (Fig. 2C). Importantly, such stability was lost on modification by MTSEA, unless oxalate (Fig. 2C) or formate (not shown) is present. Such observations focus directly on the entire population of probe-treated materials, without requiring assays of transport to report MTSEA modification. For this reason, we conclude that for the conditions of our experiments (Figs. 1) use of MTSEA leads to modification of a most, if not all, of the target population.
Fig. 2.
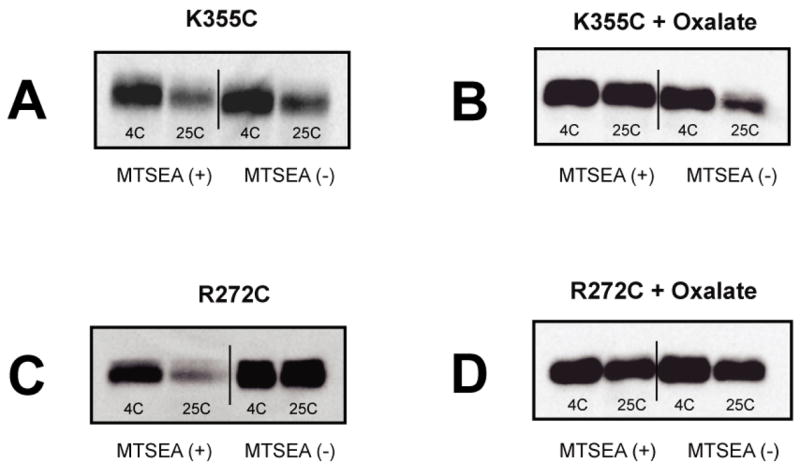
MTSEA modification influences OxlT stability. Membranes containing the indicated OxlT variants were treated with either solvent (−) or MTSEA (+) as indicated, before preparation of detergent extracts (without protease inhibitor) with potassium oxalate omitted. After solubilization, equal portions of each extract were supplemented with 50 mM potassium sulfate (A and C) or 50 mM potassium oxalate (B and D) and placed at 4C or 25C for either 1 hr (R272C) or 2 hr (K355C) before removing samples for SDS-PAGE and immunoblot analysis.
In more detailed work, we next characterized the kinetic behavior of both the R272C and K355C derivatives after modification by MTSEA. In both instances, it appeared that the major kinetic effect was to alter the maximal velocity for oxalate self-exchange, with modest effect (2-fold increase) on the Michaelis constants for transport (Table 1). By contrast, the individual substitutions (R272K and K355R) gave substantial changes in both parameters (Table 1).
Table 1.
Kinetic Parameters for Mutants of R272 and K355
| Samplea | KM (μM) | VMax (μmol/min/mg) |
|---|---|---|
| Cysteine-less OxlT | 82 ± 7 | 1720 ± 210 |
| R272Kb | 690 ± 72 | 50 ± 5 |
| K355Rb | 5400 | 66 |
| R272K/K355R | ndc | |
| R272C | ndc | |
| K355C | ndc | |
| R272C + MTSEA | 165 ± 9 | 16 ± 1 |
| K355C + MTSEA | 160 ± 4 | 370 ± 110 |
The C28G/C271A cysteine-less OxlT was parent to all variants except the K355R mutant, which was in wild-type background. Data are mean values ± SE from 4–7 separate trials. Where indicated, MTSEA was used at 1 mM (as in Fig. 1AB).
Summary data for the R272K mutant were noted earlier (17); analysis of the K355R mutant has been reported (16).
Not detectable on extended assay (as in Fig. 1B) (< 0.01% parental specific activity)
MALDI-TOF Mass Spectrometry
In the work summarized above we presumed that cysteine modification restored the positive character of the target side chain by addition of the -SCH2CH2NH3+ group, yielding a mass increment of 76 Da. Preliminary work suggested MALDI-TOF could monitor such changes in OxlT (45 kDa) without requiring analysis of peptide fragments, since in a series of eight comparisons involving multiple individual trials, the mean difference between observed and expected masses was 11 ± 12 Da (mean ± SD) (see Table 2).
Table 2.
MALDI-TOF Mass Determinations of OxlT and Its Derivatives
| Samplea | Expected Mass (Da) b | Observed Mass (Da) | ||
|---|---|---|---|---|
| No Addition | + MTSEA | Differencec | ||
| Wild Type | 45,620 | 45,635 ± 16 | 45,629 ± 14 | − 6 |
| Cysteine-less OxlT | 45,542 | 45,537 ± 15 | 45,553 ± 20 | 16 |
| Single-cysteine R272C | 45,489 | 45,494 ± 17 | 45,565 ± 14 | 71 |
| Single-cysteine K355C | 45,517 | 45,541 ± 13 | 45,620 ±3 | 79 |
Mean values ± SD for mass comparisons in 3–6 independent trials, except in a single case that reports the average (± range) of duplicate measurements.
Mass expected for OxlT lacking the N-terminal methionine, but with a C-terminal polyhistidine extension (see text).
Difference between “+ MTSEA” and “No Addition”
MALDI-TOF mass spectrometry of wild type and cysteine-less OxlT (Fig. 3) was instructive. In both instances, mass determinations were lower (by 115 Da) than expected for the full OxlT sequence with its C-terminal His9 extension. Given this result, and the reproducibility of the measurement (above), we concluded that affinity-purified material lacked the N-terminal methionine (131 Da), and all subsequent calculations were based on this assumption (Table 2). This post-translational change was not unexpected, since N-terminal microsequencing revealed the same modification after OxlT purification from its natural host, O. formigenes, using conventional techniques (2).
Fig. 3.
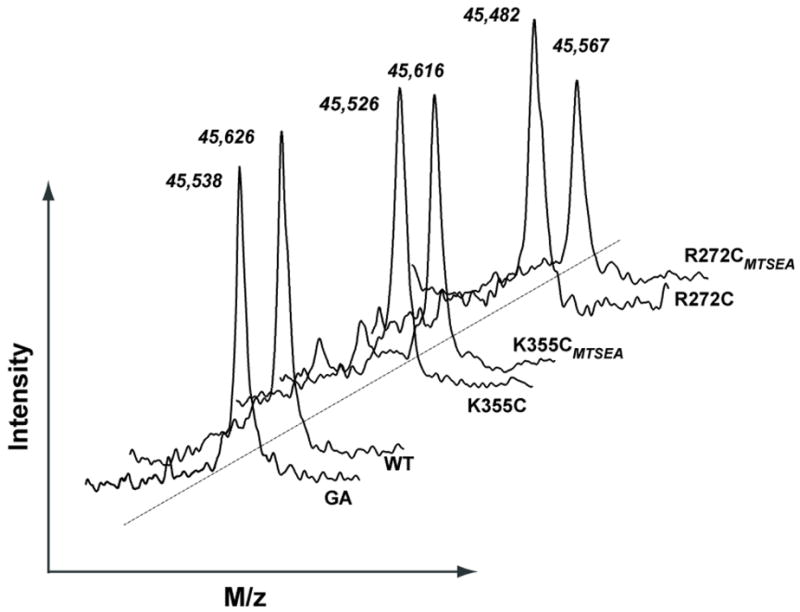
Mass determination of OxlT and its derivatives after exposure to MTSEA. (Lower left) Tracings show MALDI-TOF mass spectrometry for wild type OxlT (WT) and its C28G/C271A derivative (GA), both treated with excess MTSEA (1 mM) prior to solubilization and purification. (Center) Tracings show mass determinations for the K355C variant exposed to excess MTSEA (K355CMTSEA) and then treated with excess DTT (K355C); (Upper right) Mass determinations for the R272C derivative exposed to MTSEA (R272CMTSEA) and then to excess DTT (R272C).
Mass spectrometry of the R272C and K355C variants also proved informative (Fig. 3). For this analysis, we exposed membranes to excess (1 mM) MTSEA, so that the presence of oxalate during solubilization and purification could enhance protein stability (see Fig. 2). We then incubated purified material with or without 200 mM DTT and used the mass difference between control and reduced samples as an index of the modification introduced by MTSEA. As anticipated (see 17), there was no significant modification by MTSEA of either the wild type protein (mean difference of −6 Da) or of cysteine-less OxlT (+16 Da) (Table 2). But for the R272C and K355C variants, exposure to MTSEA led to mass gains of 71 and 79 Da, respectively, values well beyond those expected of normal variation (11 ± 12 Da) but comparable to the increments expected after MTSEA modification (76 Da). We conclude that MTSEA modification of R272C and K355C yields the desired product, with restoration of the electropositive character of the permeation pathway.
Active-Site Residues Determine Substrate Specificity
The experiments described above suggest that R272 and K355 function in the binding of oxalate, the normal substrate of OxlT. We note that functional recovery after MTSEA modification was achieved by generating a new side-chain (-CH2SSCH2CH2NH3+) whose full extension is greater than that of the normal partners, R272 or K355. If this novel side-chain contributes to substrate binding, as implied by the results summarized earlier (Fig 1), one might expect MTSEA modification to be associated with altered substrate specificity due to changes in binding site architecture. To test this idea, we screened a number of potential substrates as competitors of [14C]oxalate transport by parental and derivative proteins (Table 3). In such work, proteoliposomes were treated with 1 mM MTSEA, and initial rates of the oxalate self-exchange (0.1 mM substrate) were measured in the absence and presence of potential competitors present at a 100-fold excess (10 mM). As expected, oxalate itself (−OOCCOO−) exhibited the highest degree of inhibition, since among known OxlT substrates oxalate has the highest apparent affinity (11, 27). Malonate (−OOCCH2COO−) and formate (HCOO−) also gave marked inhibition, consistent with earlier studies that identified them as OxlT substrates (11, 27). Less effective inhibitors included glyoxylate (HCOCOO−), acetate (H3CCOO−), lactate (CH3HCOHCOO−), bicarbonate (HCO3−) and glycolate (H2COHCOO−), none of which appeared to be effective substrates by this test. A similar rank order was observed for the R272K variant (the low activity of the K355R mutant (Table 1) precluded its use in this work), but significant differences emerged in tests with the MTSEA-treated cysteine-substitution variants. For example, in both the R272C and K355C proteins, there was a blunted response to malonate, suggesting these variants have a significantly reduced affinity for this substrate. By contrast, the response to formate was strengthened, especially for the K355C protein, suggesting an enhanced affinity for this anion.
Table 3.
Substrate Specificity of OxlT and Its Derivatives
| Sample a | Parent | R272K | R272C + MTSEA | K355C + MTSEA |
|---|---|---|---|---|
|
Residual [14C]Oxalate Transport (%)
|
||||
| No Addition | 100 | 100 | 100 | 100 |
| + Oxalate | 1 ± 0.1 | 9 ± 0.8 | 5 ± 1.4 | 3 ± 0.5 |
| + Malonate | 34 ± 3.1 | 39 ± 5.9 | 80 ± 3.1 | 70 ± 8 |
| + Nitrate b | 51 ± 10 | 36 ± 4.5 | 28 ± 4 | 11 ± 3 |
| + Formate | 59 ± 4.4 | 55 ± 3.6 | 40 ± 1.6 | 7 ± 0.4 |
| + Glyoxylate | 69 ± 6.1 | 69 ± 1.7 | 52 ± 1.3 | 64 ± 1.0 |
| + Bicarbonate b | 80 ± 8.8 | 95 ± 6.6 | 72 ± 3.7 | 84 ± 4.2 |
| + Acetate | 81 ± 6.5 | 72 ± 1.2 | 93 ± 2.0 | 45 ± 1.7 |
| + Lactate | 82 ± 2.8 | 103 ± 2.7 | 119 ± 3.7 | 86 ± 4.3 |
| + Glycolate | 88 ± 8.9 | 86 ± 1.9 | 95 ± 3.4 | 62 ± 3.4 |
Mean values ± SD from 3 separate experiments reporting relative accumulation of labeled oxalate by proteoliposomes for initial rate determinations, using 0.1 mM [14C]oxalate with potassium sulfate (no addition) or the potassium salt of the indicated competing substrates present at 10 mM.
Using assay buffer in which 50 mM MOPS/K plus 100 mM potassium gluconate replaced 50 mM potassium phosphate plus 100 mM potassium sulfate. Control experiments showed this substitution had no significant effect on OxlT function.
Recent study of the MFS nitrate transporter, NtrA, suggested that paired arginine residues, one each in TM2 and TM8, might serve as substrate (anion) binding sites (26) as in GlpT and UhpT (14, 25), and this raised the possibility that OxlT and its variants might accept inorganic as well as organic anions. In 10 added tests, therefore, we examined a panel of inorganic anions as possible OxlT substrates, including bicarbonate, chloride, dithionite, fluoride, nitrate, nitrite, perchlorate, perchlorite, phosphate, sulfate, sulfite, and thiocyanate. Among these, only nitrate and nitrite gave significant inhibition of the oxalate self-exchange (Table 3 and data not shown), suggesting a limited capacity of OxlT to accept inorganic anions.
The crude screen used to identify potential substrate specificity alterations (Table 3) prompted more explicit tests of MTSEA-modified variants in regard to their responses to malonate, formate and nitrate (Fig. 4). In each case we confirmed the discrepancies noted in the preliminary screen. Thus, for malonate we obtained Kiapp values (mean ± SE) of 6.1 ± 0.4 mM for the parental protein, comparable to that found earlier (25), but significantly elevated values of 115 ± 3.6 mM and 51 ± 13 mM for the modified R272C and K355C proteins, respectively (Fig. 4A). By contrast, enhanced inhibition of K355C by formate and nitrate (Table 3) correlated with 3 to 4-fold lowered Kiapp values for this derivative relative to its unmodified parent (Fig. 4B & C). (The modified R272C variant was not examined.) For formate, Kiapp for these conditions was lowered from 1.6 ± 0.3 mM to 0.5 ± 0.1 mM; for nitrate, Kiapp fell from 4.1 ± 0.8 mM to 1.1 ± 0.3 mM. Such modifications of substrate specificity provide further evidence that R272 and K355 play direct roles in substrate binding.
Fig. 4.
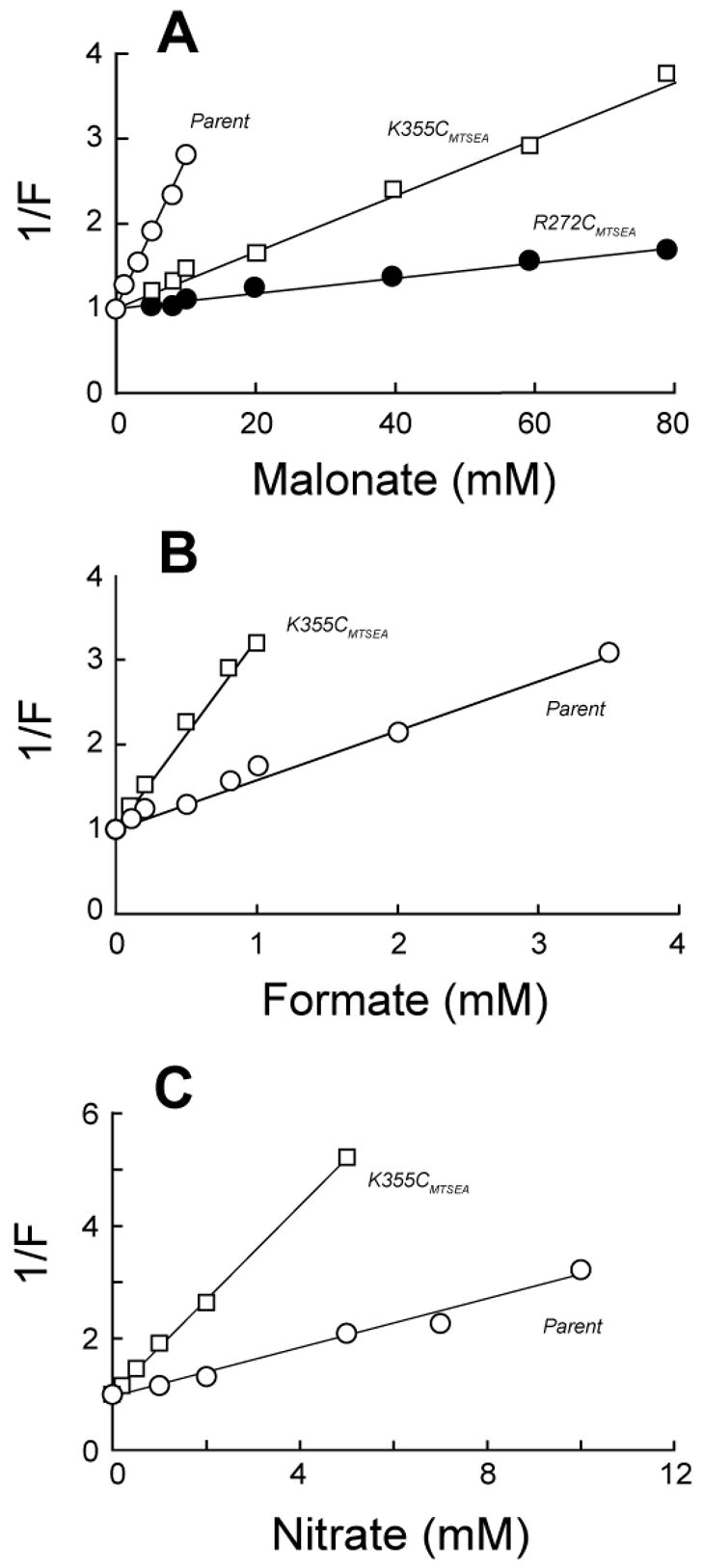
Altered substrate specificity for OxlT variants exposed to MTSEA. To determine apparent inhibition constants, the initial rates of 0.1 mM [14C]oxalate transport were determined in the presence of malonate (panel A), formate (panel B) or nitrate (panel C). Proteoliposomes treated with 1 mM MTSEA contained either cysteine-less OxlT (open circles), or its K355C (open squares) or R272C (closed circles) derivative. Observed rates were normalized by expressing them as a fraction (F) of the rate found in the absence of inhibitor, and apparent Ki values (Kiapp) were determined as the inhibitor concentrations yielding 50% inhibition of [14C]oxalate transport. Kiapp values cited in the text summarize three independent experiments as shown here. Assuming simple competitive inhibition and the kinetic parameters in Table I, the observed Kiapp values for malonate would reflect true Ki values of 72 mM, 53 mM, and 2.7 mM for R272CMTSEA, K355CMTSEA and cysteine-less OxlT, respectively; for formate, the respective Ki values would be 0.31 mM (K355CMTSEA) and 0.71 mM (cysteine-less OxlT); for nitrate, these Ki values would correspond to 0.7 mM (K355CMTSEA) and 1.8 mM (cysteine-less OxlT).
DISCUSSION
Examination of the OxlT homology model (17), as well as earlier biochemical work (11, 15, 16) suggests that two positively charged residues (R272 and K355) serve as ligand-binding elements for the anionic OxlT substrates. The work reported here supports this hypothesis, in part by showing that a requirement for positive charge at these positions reflects requirements for OxlT function, rather than biogenesis. More important, we document that side-chain modifications at these sites can have an impact on substrate specificity.
Early work showed that substitution at positions 272 or 355 abolished OxlT function unless the basic character of the original residue was preserved (15–17). But even like substitutions (e.g., R272K or K355R) yield substantially diminished catalytic activity (Table 1), suggesting that side-chain architecture in this region is critical. Those findings did not clarify whether such effects reflect roles crucial to substrate transport alone, or whether long-range effects in protein folding and/or assembly might also be relevant to the observed phenotype. To address such issues, we analyzed the R272C and K355C null mutants after reconstitution, using MTSEA to restore the positive character of these positions. That function was restored in both instances (Fig. 1) argues against the idea that the normal side chains contribute importantly to protein assembly, and instead points to their participation in OxlT function. This conclusion is reinforced by findings related to OxlT stability, which show that MTSEA modification restores the capacity for substrate (oxalate) stabilization (Fig. 2), a phenomenon that requires ligand binding (18, 25).
Several well-characterized anion transporters in the MFS, such as GlpT, UhpT, NtrA and OxlT, share a common feature in that substrate binding appears to be require interactions with two positively charged residues on the permeation pathway (14, 16, 23, 26). Thus, no function is recorded when either of the paired arginines in UhpT and GlpT are replaced by lysine (14, 23; unpublished), and in the same fashion, NtrA displays only low residual activity when either of its paired arginine residues are substituted by lysine (26). In OxlT, it is also clear that arginine and lysine are poor substitutes for one another (Table 1), and this likely explains the low functional recovery after MTSEA treatment of R272C, despite that this target appears fully modified (Fig. 2). This line of reasoning also applies in the case of K355C, which shows little or no functional recovery when non-lysine-like side chains are generated by exposure to MTS-linked probes, even those that introduce positive charge (Fig. 1D).
These considerations prompted us to ask whether OxlT substrate specificity was altered on restoration of function by MTSEA in the R272C and K355C mutants. If so, we suggest this would be strong direct evidence that these positions are involved in substrate recognition. Indeed, we find that altering side-chain architecture modulates selectivity among three potential substrates: malonate, formate, and nitrate (Table 3, Fig. 4) and offer a simple speculation to may partly account for such results. Thus, while side-chain length is approximately the same for lysine and arginine, the side chain generated by MTSEA modification (-CH2SSCH2CH2NH3+) would extend an additional 1.85 Å into the binding domain. Comparisons of the GlpT x-ray structure (14) and the OxlT homology model (17) suggests this domain is normally a thin discoid about 3 Å × 10 Å, so intrusion of the larger side-chain would reduce the (putative) active-site cross-sectional area and volume. It follows that this might introduce bias against larger substrates, as is observed for the comparison of malonate and oxalate (Fig. 4). One might also expect that binding of smaller substrates could be enhanced. A highly developed argument shows how geometric aspects of a permeation pathway contribute to selectivity in ion channels (28), and while application of such considerations to OxlT remains speculative, the observed changes in substrate specificity do give strong support to the hypothesis that R272 and K355 contribute to ligand-binding in OxlT.
Acknowledgments
We thank The Johns Hopkins A.B. Mass Spectrometry/Proteomic Facility for providing training in MALDI-TOF and for access to the necessary equipment.
The abbreviations are
- MALDI-TOF
matrix-assisted laser desorption/ionisation-time of flight
- MFS
Major Facilitator Superfamily
- MTS
methanethiosulfonate
- MTSEA
methanethiosulfonate ethylamine
- TM
transmembrane helix
Footnotes
This work was supported by grants from the National Science Foundation (MCB-0235305) and the National Institutes of Health (R46 GM24196-30). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact
References
- 1.Anantharam V, Allison MJ, Maloney PC. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J Biol Chem. 1989;264:7244–7250. [PubMed] [Google Scholar]
- 2.Ruan ZS, Anantharam V, Crawford IT, Ambudkar SV, Rhee SY, Allison MJ, Maloney PC. Measurement of the substrate dissociation constant of a solubilized membrane carrier. J Biol Chem. 1992;267:10537–10543. [PubMed] [Google Scholar]
- 3.Abe K, Hayashi H, Maloney PC. Exchange of aspartate and alanine. Mechanism for development of a proton-motive force in bacteria. J Biol Chem. 1996;271:3079–3084. doi: 10.1074/jbc.271.6.3079. [DOI] [PubMed] [Google Scholar]
- 4.Lolkema JS, Poolman B, Konings WN. Bacterial solute uptake and efflux systems. Curr Opin Microbiol. 1998;1:248–253. doi: 10.1016/s1369-5274(98)80018-0. [DOI] [PubMed] [Google Scholar]
- 5.Abe K, Ohnishi F, Yagi K, Nakajima T, Higuchi T, Sano M, Machida M, Sarker RI, Maloney PC. Plasmid-encoded asp operon confers a proton motive metabolic cycle catalyzed by an aspartate-alanine exchange reaction. J Bacteriol. 2002;11:2906–2913. doi: 10.1128/JB.184.11.2906-2913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pao SS, Paulsen IT, Maier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saier MH, Jr, Busch W. The transporter classification (TC) system, (2002) Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 8.Heymann JA, Sarker R, Hirai T, Shi D, Milne JL, Maloney PC, Subramaniam S. Projection structure and molecular architecture of OxlT, a bacterial membrane transporter. EMBO J. 2001;20:4408–4413. doi: 10.1093/emboj/20.16.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirai T, Heymann J, Shi D, Sarker RI, Maloney PC, Subramaniam S. Three-dimensional structure of a bacterial oxalate transporter. Nat Struct Biol. 2002;9:597–600. doi: 10.1038/nsb821. [DOI] [PubMed] [Google Scholar]
- 10.Hirai T, Heymann JA, Maloney PC, Subramaniam S. Structural model for 12-helix transporters belonging to the major facilitator superfamily. J Bacteriol. 2003;185:1712–1718. doi: 10.1128/JB.185.5.1712-1718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abe K, Ruan ZS, Maloney PC. Cloning, sequencing, and expression in Escherichia coli of OxlT, the oxalate:formate exchange protein of Oxalobacter formigenes. J Biol Chem. 1996;271:6789–679. doi: 10.1074/jbc.271.12.6789. [DOI] [PubMed] [Google Scholar]
- 12.Foster DL, Boublik M, Kaback HR. Structure of the lac carrier protein of Escherichia coli. J Biol Chem. 1983;258:31–34. [PubMed] [Google Scholar]
- 13.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol 3-phosphate transporter of Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 15.Fu D, Maloney PC. Structure-function relationships in OxlT, the oxalate/formate transporter of Oxalobacter formigenes. Topological features of transmembrane helix 11 as visualized by site-directed fluorescent labeling. J Biol Chem. 1998;273:17962–17967. doi: 10.1074/jbc.273.28.17962. [DOI] [PubMed] [Google Scholar]
- 16.Fu D, Sarker RI, Abe K, Bolton E, Maloney PC. Structure/function relationships in OxlT, the oxalate-formate transporter of Oxalobacter formigenes. Assignment of transmembrane helix 11 to the translocation pathway. J Biol Chem. 2001;276:8753–8760. doi: 10.1074/jbc.M008417200. [DOI] [PubMed] [Google Scholar]
- 17.Yang Q, Wang X, Ye L, Mentrikoski M, Mohammadi E, Kim YM, Maloney PC. Experimental tests of a homology model for OxlT, the oxalate transporter of Oxalobacter formigenes. Proc Natl Acad Sci U S A. 2005;102:8513–8518. doi: 10.1073/pnas.0503533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu D, Maloney PC. Evaluation of secondary structure of OxlT, the oxalate transporter of Oxalobacter formigenes, by circular dichroism spectroscopy. J Biol Chem. 1997;272:2129–2135. doi: 10.1074/jbc.272.4.2129. [DOI] [PubMed] [Google Scholar]
- 19.Ambudkar SV, Maloney PC. Bacterial anion exchange. Use of osmolytes during solubilization and reconstitution of phosphate-linked antiport from Streptococcus lactis. J Biol Chem. 1986;261:10079–10086. [PubMed] [Google Scholar]
- 20.Cadene M, Chait BT. A robust, detergent-friendly method for mass spectrometric analysis of integral membrane proteins. Anal Chem. 2000;72:5655–5658. doi: 10.1021/ac000811l. [DOI] [PubMed] [Google Scholar]
- 21.Ye L, Jia Z, Jung T, Maloney PC. Topology of OxlT, the oxalate transporter of Oxalobacter formigenes, determined by site-directed fluorescence labeling. J Bacteriol. 2001;183:2490–2496. doi: 10.1128/JB.183.8.2490-2496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye L, Maloney PC. Structure/function relationships in OxlT, the oxalate/formate antiporter of Oxalobacter formigenes: assignment of transmembrane helix 2 to the translocation pathway. J Biol Chem. 2002;277:20372–20378. doi: 10.1074/jbc.M111140200. [DOI] [PubMed] [Google Scholar]
- 23.Dunten RL, Sahin-Toth M, Kaback HR. Role of the charge pair aspartic acid-237-lysine-358 in the lactose permease of Escherichia coli. Biochemistry. 1993;32:3139–3145. doi: 10.1021/bi00063a028. [DOI] [PubMed] [Google Scholar]
- 24.Sahin-Toth M, Kaback HR. Role of the charge pair aspartic acid-237-lysine-358 in the lactose permease of Escherichia coli. Biochemistry. 1993;32:10027–10035. doi: 10.1021/bi00063a028. [DOI] [PubMed] [Google Scholar]
- 25.Fann MC, Davies AH, Varadhachary A, Kuroda T, Sevier C, Tsuchiya T, Maloney PC. Identification of two essential arginine residues in UhpT, the sugar phosphate antiporter of Escherichia coli. J Membr Biol. 1998;164:187–195. doi: 10.1007/s002329900404. [DOI] [PubMed] [Google Scholar]
- 26.Unkles SE, Rouch DA, Wang Y, Siddiqi MY, Glass AD, Kinghorn JR. Two perfectly conserved arginine residues are required for substrate binding in a high-affinity nitrate transporter. Proc Natl Acad Sci U S A. 2004;101:17549–17554. doi: 10.1073/pnas.0405054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maloney PC, Anantharam V, Allison MJ. Measurement of the substrate dissociation constant of a solubilized membrane carrier. Substrate stabilization of OxlT, the anion exchange protein of Oxalobacter formigenes. J Biol Chem. 1992;267:10531–10536. [PubMed] [Google Scholar]
- 28.Hille B. Ionic Channels of Excitable Membranes. 2. Sinauer Associates, Inc; Sunderland, MA: 1992. pp. 347–376. [Google Scholar]


