Abstract
Claviceps purpurea is an important pathogen of grasses and source of novel chemical compounds. Three groups within this species (G1, G2, and G3) have been recognized based on habitat association, sclerotia and conidia morphology, and alkaloid production. These groups have further been supported by RAPD and AFLP markers, suggesting this species may be more accurately described as a species complex. However, all divergent ecotypes can coexist in sympatric populations with no obvious physical barriers to prevent gene flow. In this study, we used both phylogenetic and population genetic analyses to test for speciation within C. purpurea using DNA sequences from ITS, a RAS-like locus, and a portion of beta-tubulin. The G1 types are significantly divergent from the G2/G3 types based on each of the three loci and the combined dataset, whereas the G2/G3 types are more integrated with one another. Although the G2 and G3 lineages have not diverged as much as the G1 lineage based on DNA sequence data, the use of three DNA loci does reliably separate the G2 and G3 lineages. However, the population genetic analyses strongly suggest little to no gene flow occurring between the different ecotypes and we argue that this process is driven by adaptations to ecological habitats; G1 isolates are associated with terrestrial grasses, G2 isolates are found in wet and shady environments, and G3 isolates are found in salt marsh habitats.
Keywords: Claviceps, fungi, genealogy, genetic structure, speciation
Introduction
Species concepts have been intensely debated in the literature and no one concept is generally accepted among biologists. Traditional species concepts for the classification of fungi are based on morphology and reproductive biology, but phylogenetic approaches have become increasingly popular in recent years to resolve species identification (Taylor et al. 2000; Harrington & Rizzo 1999). Moreover, phylogenetic analyses have challenged morphological species concepts and have been especially helpful in delineating fungal species with few morphological characters. Phylogenetic approaches have also been sought for the many fungi in which sexual reproduction is not known to occur, making biological species concepts impossible to implement. Phylogenetic studies have routinely identified cryptic species within morphological species in various fungal genera including Fusarium (Skovgaard et al. 2002; O’Donnell et al., 2000), Stachybotrys (Cruse et al. 2002), Tricholoma (Horton 2002), Coccidioides and close relatives (Koufopanou et al. 2001), Cenococcum (Douhan & Rizzo 2005), Neurospora (Dettman et al. 2003), and lichenized genera such as Physcia (Myllys et al. 2001) and Letharia (Kroken & Taylor 2001).
What has not been determined in most of these studies is the mechanism or driving force behind the speciation process. In some instances, geographic isolation has been suggested, as in Coccidioides (Koufopanou et al. 2001) and Fusarium (O’Donnell et al. 2000), whereas in other cases these putative cryptic species may be found in the same geographic location and in some instances even isolated from the same soil core (Douhan & Rizzo 2005; Moyersoen et al. 2003; Skovgaard et al. 2002). Ecological theory predicts that the stable coexistence of identical competitors will not occur (Hardin 1960), suggesting that cryptic species occupying the same apparent niche may play different ecological roles. Therefore, ecological factors likely play a significant role in the speciation process of co-occurring organisms. This ‘ecological speciation’ process has received little attention in the fungi. We will adopt the terminology of Rundle and Nosil (2005) that defines ‘ecological speciation’ as the “process by which barriers to gene flow evolve between populations as a result of ecologically-based divergent selection.”
One group of fungi where ecological aspects may have influenced speciation is within the Claviceps purpurea complex. These are plant pathogenic fungi that infect the flowers of grasses and cause a disease known as ‘ergot.’ The infection results in a replacement of the grass floral tissue with fungal mycelium that grows to form a darkly pigmented survival structure called a sclerotium (Alexopolous et al. 1996). Sclerotia of C. purpurea contain potent alkaloids that are toxic to humans and other animals (Hudler, 1998). This has been important during human history because sclerotia have been unwittingly mixed with grain and the fungal toxins have contaminated bread flour. The consumption of bread tainted with C. purpurea causes the debilitating disease known as ‘Holy Fire’ or ‘St. Anothony’s Fire,’ which includes a range of symptoms such as itching, headache, hallucinations, gangrene, seizures, and even death (Hudler, 1998). Despite the negative impacts of C. purpurea alkaloids, interesting and beneficial chemical compounds have also been derived from these fungi, including the infamous lysergic acid diethylamide (LSD) and pharmaceuticals used to treat both postpartum bleeding and migraine headaches (Hart 1999, Rehacek & Sajdl 1993)
Pazoutová et al. (2000) synthesized previous research on C. purpurea morphology, alkaloid chemistry and genetics, and identified three distinct groups within the species, describing them as ‘chemoraces.’ Rather than species delimitation based on host range, which was historically common in C. purpurea taxonomy, intraspecific groups were defined based on habitat specialization. The largest group, G1, is associated with land grasses. G2 isolates are associated with grasses in ‘wet and shady’ environments, whereas G3 isolates are found only on grasses in salt marsh habitats. G3 isolates are synonymous with C. purpurea var. spartinae (Duncan et al. 2002) and are referred to herein as G3. Sclerotia of both G2 and G3 C. purpurea float in water while sclerotia from terrestrial C. purpurea (G1) sink, clarifying the significance of earlier reports that sclerotia of some isolates float, and the association of this trait with host habitat (Stager 1922).
Our objective in this study was to investigate speciation within the C. purpurea complex using phylogenetic and population genetic approaches based on representative samples. Isolates included in this study were selected as representative of the variation within C. purpurea sensu lato and chosen to cover a large geographic area, a wide host range within the Poaceae, and all three habitat types. The group identity of each isolate used in this study was determined previously by RAPD and AFLP (Fisher et al. 2005b; 2005c; Pazoutová et al. 2000). RAPD and AFLP analyses of these, and additional isolates, supports the recognition of three discrete groups within C. purpurea and revealed high genetic variability between groups, with less than 2% of polymorphic markers shared across all isolates. Similarly, analysis of molecular variation (AMOVA) revealed that genetic variability was mainly due to variations between groups rather than within groups or populations, revealing a high probability of phylogenetic substructure (Fisher et al. 2005b; 2005c). Determining if the C. purpurea complex is made up of one or multiple species is important because it is common practice in wide-ranging studies to make no distinction between the three different C. purpurea lineages (Tooley et al. 2001; Stensrud et al. 2005; Scheffer & Tudzynski 2006; Dabkevicius & Mikaliunaite 2006; Scheffer et al. 2005).
Materials and methods
Molecular analyses
Fungal isolates & DNA extraction
Claviceps purpurea isolates used in this study and their origins are listed in Table 1. For field-collected isolates, sclerotia were collected during the fall of 2000, 2001 and 2002 and brought to University of California, Davis for culturing. Sclerotia were surface sterilized, cultured, dried and refrigerated as described in Fisher et al. (2005c). Numbered isolates were obtained as pure cultures from S. Paloutová (Institute of Microbiology, Czech Republic) including: G1 isolates 165, 204, 428 and G2 isolates 236 and 434 (Fisher et al. 2005c; Paloutová et al., 2000).
Table 1.
Claviceps purpurea isolates used in this study.
| Code | Group | Location | Year collected | Host |
|---|---|---|---|---|
| 165 | G1 | Zubri, Czech Republic | 1994 | Poa pratensis |
| 204 | G1 | Lauderdale, Alabama | 1996 | Festuca arudinaceae |
| 428 | G1 | Hohenheim, Germany | ? | Secale cereale |
| NGE1 | G1 | Newfoundland, Canada | 2001 | Leymus mollis |
| WFA | G1 | Nahcotta, Washington | 2002 | Festuca arundinaceae |
| WLS | G1 | Nahcotta, Washington | 2002 | Lolium spp. |
| 236 | G2 | Vlei Pole u Bousova, Czech Republic | ? | Molinia coerulea |
| 434 | G2 | Phillipsreuth, Germany | 1998 | Dactylis spp |
| WAB-1 | G2 | Long Beach, Washington | 2002 | Ammophila breviligulata |
| WCN2 | G2 | Willapa Bay, Washington | 2002 | Calamagrostis nutkaensis |
| WDG | G2 | Leadbetter State Park, Washington | 2002 | Dactylis glomerata |
| WDS | G2 | Willapa River, Washington | 2002 | Deschampsia caespitosa |
| WHS | G2 | Long Beach, Washington | 2002 | Holcus lanatus |
| ARG1 | G3 | Argentina Celpa Marsh, Argentina | 2002 | Spartina densiflora |
| CDE1 | G3 | Point Reyes, CA | 2002 | Spartina foliosa |
| CMD1 | G3 | MacDoel, CA | 2002 | Secale cereale |
| CPE10 | G3 | Palo Alto, CA | 2001 | Spartina foliosa |
| FSA1 | G3 | St. Augustine, Florida | 2000 | Spartina alterniflora |
| IRE12 | G3 | Dublin, Ireland | 2001 | Spartina anglica |
| RH2 | G3 | Rhode Island | 2001 | Spartina alterniflora |
| WDI-1 | G3 | Willapa River, Washington | 2002 | Distichlous spicata |
Tissue for DNA extraction was obtained by growing isolates on cellophane overlaid on PDA as described by Fisher et al. (2005c) or by scraping fungal mycelium directly from PDA plates. Total DNA was extracted using the methods described by Daehler et al. (1999) or slightly modified from Gardes and Bruns (1993). After DNA was recovered in solution, 75 μl of supernatant was withdrawn and mixed with 200 μl Tris-EDTA buffer (10 mM Tris, 1 mM EDTA (pH 7.8). DNA concentration was quantified by spectrophotometry.
PCR, Sequencing and phylogeny construction
PCR was performed in 40 μl reaction mixtures containing 2 μl of a 1:10 to 1:25 dilution of template DNA, 1X PCR buffer (Invitrogen, Carlsbad, CA), 2.5 mM MgCl2, 0.2 mM each dNTP (Invitrogen), 7.5 μM of each primer, and 0.5 U of Taq polymerase (Invitrogen). Three loci were amplified and sequenced; ITS rDNA using the primers ITS1F and ITS 4 (Gardes & Bruns 1993; White et al. 1990), a portion of a beta-tubulin gene using the primers described in Annis and Panaccione (1998), and a putative RAS-like protein using primers described by Carbone and Kohn (1999). Thermocycling conditions consisted of an initial hold at 94°C (4 min), followed by 30 cycles with a denaturing step of 94°C (30 sec), annealing temperature of 50°to 55°C (30 sec), and an extension temperature of 72°C (1 min). All amplifications were performed in a PE-9700 thermocycler (Perkin Elmer Corp., Norwalk, CT) or a MyCycler (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Each locus was sequenced in both directions using Big Dye® Terminator v3.1 chemistry (Applied Biosystems, Foster City, CA, USA). The PCR products were cleaned using ExoSapit (USB Amersham, Uppsala, Sweden) following the manufactures instructions and sequenced at the Core Instrumentation Facility (CIF) of the University of California’s Institute of Integrative Genome Biology at UC Riverside. The sequences were edited using Sequencher (version 4.1.2, Gene Codes Corporation, Ann Arbor, MI), aligned using Clustal X (version 1.81) (Chenna et al. 2003) and manually edited in MacClade version 4 (Maddison & Maddison 2001). Maximum Parsimony (MP) analysis using the heuristic search procedure with 1000 random-addition-sequence replicates and tree-bisection-reconnection branch swapping were conducted using PAUP* version 4.0 beta 10 (Swofford 2002). Confidence in tree topology was examined using bootstrap with 1,000 replicates under the heuristic option. All trees were midway rooted.
Population genetic analyses
We investigated the genetic separation of the three ecotypes (G1, G2, & G3) using the program MIGRATE-N (Version 2.3.3: Beerli and Felsenstein 1999, 2001, Beerli 2006) that is based on coalescence theory (cf Kingman 2000). The three ecotypes were treated as independent genetic units where only migrants and mutation could import new alleles into a unit. We compared two different models: one single population versus 3 populations with 6 different migration rates between all populations. This allowed us to test whether the 3 loci were powerful enough to reject if these three ecotypes were generated by a single random mating population.
MIGRATE-N does not take into account potential splitting of the ancestral population. Under such a model, the different ecotypes should show low migration rates if there was no recent divergence or no ongoing gene flow. We tested whether high gene flow between the ecotypes could be rejected using a Bayes Factor approach (Kass and Raftery, 1995) by running the Bayesian module of MIGRATE-N for both models and compared the marginal likelihoods.
We used the default settings for MIGRATE-N, except for the following run-options: we used the Bayesian inference module; one single long run using heating with temperatures of 1.0, 1.5, 3.0, and 10,000, totaling 50,100,000 visited parameter and genealogy changes in the cold (1.0) chain; sampling 10 replicates of each 50,000 in intervals of 100; after discarding the first 100,000 visits. We used uniform prior distributions with ranges from 0 to 0.1 for the mutation scaled population size Θ, that is the effective population size Ne times the mutation rate μ per site and generation, and ranges from 0 to 5000 for M, that is the mutation scaled immigration rate m/μ. The mutation model used was the F84 model (Felsenstein 2004) with a transition-transversion ratio set to 2. We report the mode and median, and the 95% credibility set of the posterior distribution for all estimated parameters of the two models.
Estimates of the average number of nucleotide differences, and shared, fixed and unique number of mutations between the G1, G2, and G3 ecotypes were calculated in DNAsp 4.0 (Rozas et al. 2003). Fst values were also calculated in DNAsp 4.0, with permutation tests used to test significance using 1000 randomizations.
Results
Phylogenetic inference
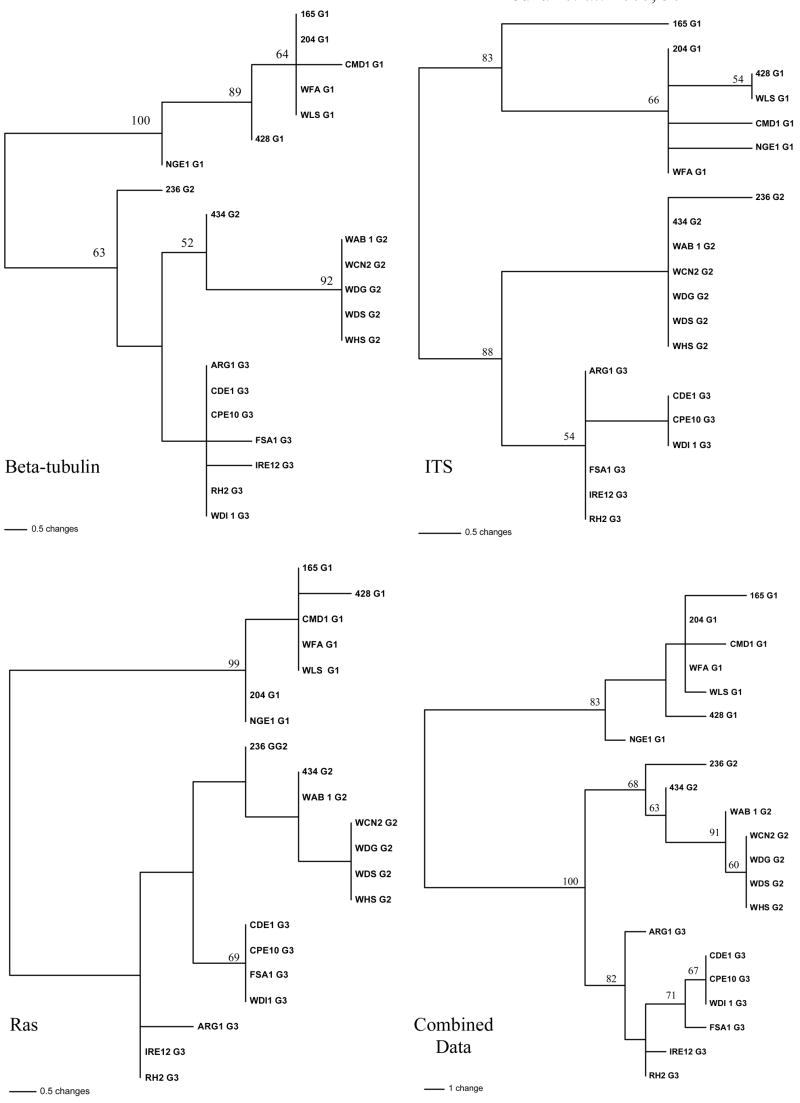
For beta-tubulin, 426 characters were analyzed with 5 parsimony un-informative and 13 parsimony informative characters. Figure 1 shows one of the 29 most parsimonious trees that were found with a length of 19. Consistency index, retention index, and the rescaled consistency index were 0.947, 0.987, and 0.935, respectively. All G2 isolates except 236 had two copies of the repeat AACTG starting at position 113 in the alignment whereas all G1 and G3 isolates had a single copy of the AACTG repeat. All G1 isolates clustered in a clade with a bootstrap support of 100. The G2 and G3 isolates clustered together into one weakly supported clade. Although all G3 isolates clustered together with weak boostrap support, G2 isolates were more variable with two isolates, 434 and 236, notably different from the rest (Fig. 1).
Fig. 1.
Maximum parsimony analyses of three individual loci (ITS, Betat-tbulin, Ras) and the combined data set from G1, G2, G3, isolates of Claviceps purpurea. Bootstrap support of 50% and above are indicated above nodes based on 1,000 replicates. Sequences have been deposited in GenBank under accession numbers (XXXXX-XXXXXX).
For ITS, 586 characters were analyzed with 4 parsimony un-informative and 8 parsimony informative characters. Figure 1 shows the single most parsimonious tree with a length of 14. Consistency index, retention index, and the rescaled consistency index were 0.929, 0.977, and 0.907, respectively. The G1 clade was supported with a bootstrap score of 83. The G2 and G3 isolates clustered together with a bootstrap value of 88. Although G2 and G3 isolates clustered separately into there own subclades, these subclades had weak bootstrap support.
For RAS, 375 characters were analyzed with 5 parsimony un-informative and 13 parsimony informative characters. Figure 1 shows one of a total of 354 most parsimonious trees that were found with a length of 15. Consistency index, retention index, and the rescaled consistency index were 0.800, 0.955, and 0.764, respectively. The G1 isolates clustered together with a bootstrap support of 99. The G2 and G3 isolates clustered together with poor bootstrap support for any sub-groupings.
In the combined dataset including all three genes, 1294 characters were analyzed with 8 parsimony un-informative and 32 parsimony informative characters. The consistency index, retention index, and the rescaled consistency index were 0.840, 0.956, and 0.803, respectively. The G1, G2, and G3 clades were supported by bootstrap values of 83, 68, and 82, respectively. The G2/3 clade was supported with a bootstrap value of 100. No obvious substructing based on geography was found for any of the three loci or the combined dataset.
Population Genetic analyses
3-population model (Each ecotype is treated as a population): Results on migration rates are all low between all populations with large credibility intervals, that for individual loci always include zero; the combined estimates exclude zero migration rates. The posterior distribution for the scaled migration rates are all heavily skewed to the right, and peak all near zero compared to the whole distribution. We are confident that the prior distribution, a uniform distribution, had little influence on our results. The population sizes Θ reveal that the ecotype G3 has more variability and a larger population than the other types, G1 and G2. Table 2 gives credibility intervals and mode and medians. Typical values for the number of immigrants Nm (ΘM/4) are 1 immigrant every four generations.
Table 2.
Combined estimates over three loci of mutation scaled population size Θ and mutation scaled migration rates M. The estimates were found using the Bayesian inference module in MIGRATE-N.
| Model | Parameter | Mode | Median | 95% Credibility interval |
|---|---|---|---|---|
| 3-population | ΘG1 | 0.00062 | 0.001 | 0.00008 – 0.00356 |
| ΘG2 | 0.00049 | 0.00087 | 0.00008 – 0.00302 | |
| ΘG3 | 0.00353 | 0.00478 | 0.00137 – 0.00923 | |
| MG2 to G1 | 543 | 868 | 25 – 2370 | |
| MG3 to G1 | 508 | 848 | 5 – 2405 | |
| MG1 to G2 | 663 | 948 | 30 – 2530 | |
| MG3 to G2 | 688 | 982 | 10 – 2695 | |
| MG1 to G3 | 333 | 528 | 0 – 1490 | |
| MG2 to G3 | 283 | 548 | 5 – 1560 | |
| 1-population | ΘG1+ G2+ G3 | 0.01125 | 0.01185 | 0.00670 – 0.01760 |
1-population model (All ecotypes are pooled into one population): the population is much larger than the combined population size of the 3-population model suggesting that, because we ignored migration, the length of the genealogies are lengthened which results in an overestimate of the population size (Excoffier, 2004). We compared our results from the 3-population model with 1-population model using a Log-Bayes factor (BF). Using the reported marginal likelihoods from the two MIGRATE-N analyses: log L(3-population model) = −2073.887689; log L(1-population model) = −2204.2369. Then the Log(BF) is log L(3) − log L(1) = 130.34. Values higher than 10 suggest that we should strongly prefer the first model, in our case the three-population model (Kass and Raftery, 1995).
Table 3 shows the summary statistics in the comparison between the G1, G2, and G3 isolates of C. purpurea. In the G2–G3 comparisons, the average number of nucleotide differences and nucleotide diversity were consistently lower than in the comparisons of G1–G3 and G1–G2. This is also consistent with lower Fst values for G2–G3 comparisons as compared with G1–G3 and G1–G2 comparisons, except for the ITS locus in which the lowest Fst value was for the G1–G3 comparison. All Fst comparisons were significant (P < 0.001), suggesting limited gene flow between the three C. purpurea lineages from different habitats, which was consistent with the coalescence analyses.
Table 3.
DNA divergence between G1, G2, and G3 Claviceps purpurea isolates in the three analyzed loci and the combined data set measured as number of segregating sites, fixed and shared polymorphic sites, average number of nucleotide differences (k), average number of nucleotide substitutions per site between habitat types (π), and population differentiation values between habitat types (Fst).
| No. of polymorphic sites | Fixed differences | No. of polymorphic compared to monomorphic* | No. of Polymorphic compared to monomorphic† | Shared differences | k | π | Fst | |
|---|---|---|---|---|---|---|---|---|
| Beta-tubulin | ||||||||
| G1–G2 | 15 | 0 | 10 | 3 | 2 | 6.385 | 0.0152 | 0.8216 |
| G2–G3 | 9 | 1 | 2 | 6 | 0 | 3.121 | 0.0083 | 0.6875 |
| G1–G3 | 13 | 7 | 4 | 2 | 0 | 5.747 | 0.0137 | 0.9034 |
| ITS | ||||||||
| G1–G2 | 11 | 5 | 1 | 6 | 0 | 4.121 | 0.0072 | 0.8369 |
| G2–G3 | 5 | 3 | 1 | 1 | 0 | 2.121 | 0.0037 | 0.8800 |
| G1–G3 | 9 | 3 | 1 | 6 | 0 | 3.495 | 0.0061 | 0.7719 |
| Ras | ||||||||
| G1–G2 | 7 | 4 | 1 | 2 | 0 | 3.648 | 0.0134 | 0.9091 |
| G2–G3 | 4 | 0 | 2 | 3 | 0 | 1.802 | 0.0065 | 0.5172 |
| G1–G3 | 7 | 5 | 0 | 2 | 1 | 3.692 | 0.0135 | 0.8600 |
| Combined | ||||||||
| G1–G2 | 33 | 14 | 11 | 8 | 0 | 14.08 | 0.0115 | 0.8504 |
| G2–G3 | 17 | 4 | 6 | 8 | 0 | 6.516 | 0.0053 | 0.7042 |
| G1–G3 | 29 | 15 | 10 | 5 | 1 | 12.93 | 0.0102 | 0.8576 |
Number of sites that are polymorphic in the first species compared and monomorphic in the second species
Number of sites that are polymorphic in the second species compared and monomorphic in the first species
Discussion
In this study, phylogenetic and population genetic analyses showed marked genetic differences among the different ecotypes and suggest little or no gene flow among the different ecotypes. We can also definitely reject models that assume random mating between the different ecotypes based on the analyses. The G1 types are significantly divergent from the G2/G3 habitat types based on each of the three loci and the combined dataset, whereas the G2/G3 types are more integrated with one another. However, although the G2 and G3 lineages have not diverged as much as the G1 lineage based on DNA sequence data, the use of three DNA loci does reliably separate the G2 and G3 lineages. The fact that the G2 and G3 lineages are more closely related to each other than to the G1 lineage is strongly supported by the fact that sclerotia from G2 and G3 isolates float in water while those of G1 isolates sink (Pazoutová et al. 2000). Results from this study are in agreement with previous conclusions based on AFLP and RAPD data, in which only 2% of genetic markers were shared among the three lineages of C. purpurea. Perhaps because of their high rate of polymorphism or because of their assessment of variation across the entire genome, AFLP and RAPD data were more informative than DNA sequences for separating the different ecotypes. A similar situation was recently shown in two species complexes of mycoparasites, Hypomyces microspermus and H. chrysospermus (Douhan & Rizzo 2003). In this case, AFLP clearly differentiated cryptic lineages within both H. microspermus and H. chrysospermus, whereas ITS rDNA sequence data recovered the same cryptic lineages, but in some cases with weak bootstrap support (Douhan & Rizzo 2003).
Should three distinct species within the C. purpurea complex be recognized taxonomically? As previously mentioned, traditional species concepts for the classification of fungi are based on morphology and reproductive biology, but phylogenetic approaches may be more powerful to accurately resolve fungal lineages (Taylor et al. 2000; Harrington & Rizzo 1999). During the speciation process, reproductive barriers arise between groups of individuals. The two groups at first share allelic polymorphisms until one of the two groups become fixed for certain alleles whereas the other group remains polymorphic (Avise 1994; Geiser et al. 1998). Therefore, based on previous AFLP and RAPD data, it would appear that the justification for species recognition would be warranted. However, Taylor et al. (2000) advocate identifying phylogenetic species of fungi by analyzing evolutionary relationships among multiple genes, which they call the Genealogical Concordance Phylogenetic Species Recognition (GCPSR). They suggest the use of multiple genes to determine the transition from concordance to conflict among taxa, which can be used to determine species boundaries. The conflict is thought to be due to recombination occurring between individuals. Based on the GCPSR criteria we would reject that there are three species within the C. purpurea complex and conclude that two species exist, representing the G1 and G2/G3 lineages. However, for more recently derived taxa, a marker system such as AFLP may be more powerful than DNA sequence loci to determine species boundaries because methods such as AFLP screen many loci across a genome. Despite the strengths of markers such as AFLP, inferring phylogenies based on randomly amplified fragments can be problematic, and there are arguments both for (Buntjer et al. 2002; Després et al. 2002; Kardolus et al. 1998) and against this approach (Seberg & Peterson 1998).
The overall evidence from our phylogenetic and population genetic analyses and previous studies suggest that the three lineages (G1, G2, and G3) should be recognized as unique species, or at least as varieties. Duncan et al. (2002) proposed renaming the G3 lineage as C. purpurea var. spartinae based on a combination of host identity (Spartina spp.), sclerotia ecology (floatation) and conidial morphology. However, since the variety status was proposed, G3 isolates were found infecting a non-Spartina host in nature (Distichlis spicata – Fisher et al. 2005b). Therefore, the named variety based on host association does not seem warranted. Regardless of what these fungi are called, it is important that some distinction be made since C. purpurea sensu lato is widely studied as a model plant pathogen of economic importance (Tudzynski & Scheffer 2004). Several lines of evidence suggest three divergent lineages within C. purpurea and these lineages likely have important biological differences relevant for studies in the wider scientific community. Currently, it is common practice in wide-ranging studies of everything from pathogenicity of C. purpurea on various hosts to molecular studies of virulence genes to phylogenetic assessments of the genus Claviceps to make no distinction between the three different C. purpurea lineages (Stensrud et al. 2005; Scheffer & Tudzynski 2006; Dabkevicius & Mikaliunaite 2006; Scheffer et al. 2005).
Some authors have argued that species concepts should have an ecological basis as well as a genetic one (Harrington & Rizzo 1999), and we suggest that ecological factors have driven the speciation process in C. purpurea. In estuarine habitats, the transition from salt-water tolerant species such as Spartina spp. and Distichlis spicata, to riparian and terrestrial grasses, is often gradual, with no physical barrier between habitat types (Mitsch & Gosselink 1993). The boundaries of the high tide and the slope of the terrain delimit the change from one habitat to the next, and this will differ by site. For example, in Willapa Bay, Washington (USA) the G1, G2 and G3 lineages coexist within a distance of less than 100 meters, with no physical barriers to spore dispersal by rain splash or insect vectors (Fisher et al. 2005b). It is not known if G1, G2 and G3 C. purpurea diverged sympatrically in a habitat such as a coastal estuary, or allopatrically with geographic barriers to gene flow. However, since there was no phylogeographic structure found in this diverse sampling of isolates and host grass species, it seems likely that ecological factors were more important in the speciation process of these fungi. The genus Claviceps is thought to have a Gondwanan orgin and most species in the genus are tropical or subtropical. It has been hypothesized that species close to C. purpurea migrated from South America to North America after the formation of the Panama land bridge, and they later spread to Europe and Africa (Pazoutová 2003). It is only these species that developed the ability to deal with cold winters and semi-arid conditions, which is potentially more evidence suggesting that ecological habitat helped shaped this species complex.
Although it remains to be determined if the three habitat-associated lineages are reproductively isolated, results from our phylogenetic analysis suggest that reproduction among G2/3 isolates would be more likely than between either G1 and G2 or G1 and G3 isolates. However, the population genetic analyses strongly suggest little to no gene flow occurring between the different ecotypes. In order for reproduction to occur, theses different ecotypes must be able to infect the same hosts. The results of a host range study showed that G3 isolates can infect both riparian and terrestrial grasses after artificial inoculation (Pazoutová et al. 2002), though its range in nature is so far limited to the C4 grasses Spartina spp. and Distichlis spicata. Similarly, Pazoutová et al. (2000; 2002) reported that G2 isolates can infect both riparian and terrestrial grasses in the greenhouse but this has not been documented under natural conditions. Currently we have not identified the barriers to gene flow in sympatric G1, G2 and G3 C. purpurea populations like in the Willapa Bay’s upper marsh, but possibilities include differences in flowering time or flowering duration among hosts, or differences in plant biochemistry which might preclude infection of C3 grasses by a pathogen adapted to C4 grass hosts. Thus, as in other fungi, there may be multiple habitat-related factors that drive the speciation process. For example, it has been suggested that the behavior of insect vectors or physiological barriers to mating or infection may prevent gene flow among host races of the anther smut fungus Microbotryum violaceum in sympatric populations of three host plant species (Shykoff et al. 1999).
Regardless of whether the divergence among C. purpurea lineages occurred in allopatric or sympatric populations, selection pressures in maritime habitats are distinct from those necessary for dispersal and survival in terrestrial habitats and C. purpurea from Spartina and D. spicata (G3) exhibit characteristics uniquely suited for maritime environments. For example, sclerotia from G2 and G3 C. purpurea float on water (Pazoutová et al. 2000), presumably due to large intercellular spaces (Duncan et al. 2002), and flotation presumably aids both survival and dispersal in areas with flooding or tides. In addition, G3 sclerotia do not require a cold stratification prior to germination, a requirement common in C. purpurea from terrestrial habitats (G1) not likely to be met in coastal environments (Duncan et al. 2002). Although data is lacking, tolerance for highly salinated leaves, water, and soil is likely involved in directional selection leading to the isolation of the G3 lineage.
Host range within C. purpurea also suggests that ecology is more important than host in the evolution of these fungi, which is different from many plant-associated fungi. For example, within the plant pathogenic Magnaporthe grisea species complex there is strong evidence that speciation and genetic divergence are highly coupled with virulence on particular species or even varieties of host grasses. Although host switches have been documented, radiation events in this group of fungal pathogens appear to primarily follow the evolutionary history of hosts (Kohn 2005; Couch et al. 2005; Couch & Kohn 2002). A similar pattern of host-associated speciation has also been suggested for some symbiotic fungi, such as in the Pinus-associated ectomycorrhizal mushroom genus Suillus. In a study of disjunct Suillus species complexes from East Asia and Eastern North America, Wu et al. (2000) suggested that high host-fidelity for species or subgenera of Pinus has led to speciation through comigration.
“Ecological speciation” has previously been suggested as the mode of speciation among many diverse animal groups (Via 2001) including fishes (Rocha et al. 2005; Hatfield & Schluter 1999), lizards (Richmond & Reeder 2002; Ogden & Thorpe 2002; Rosenblum 2006), toads (Kruuk& Gilchrist 1997), brittle stars (Muths et al. 2006), snails (Cruz et al. 2004) and various phytophagous and non-phytophagous insects (Via et al. 2000; Runle & Nosil 2005). Similarly, at least six examples of putative ecological speciation have been inferred among angiosperm plants from diverse habitats on several continents (Wang et al. 1997; Rieseberg 2000; Lamont et al. 2003; Nagy 1997; Fine et al. 2005; Hall & Willis 2006). Ecological speciation in fungi has not received much attention and speciation is usually attributed to hosts for pathogenic fungi or due to geographic vicariance, both of which may or may not have ecological similarities. However, as far as we know, Claviceps purpurea is only the second specific example of a fungus where ecological speciation is the main proposed mode of speciation. The other example of possible ecological speciation in fungi is in the insect-pathogenic fungus Metarhizium anisopliae. In a study of M. anisopliae in Ontario (Canada), Bidochka et al. (2001) showed that this fungus was clearly segregated into two genetic groups, one common in agricultural areas and the other in forested habitats. The authors did not find consistent differences in host preference between the cryptic species, but they detected distinct differences in tolerance to UV light and temperature sensitivity. Although these are the only examples that we know of where ecological speciation has been indicated in fungi specifically, we expect that this is a product of the lack of study and we expect that many more cases of ecological speciation will be found among fungi in the future.
Acknowledgments
Financial support of the Agricultural Experiment Station, University of California Riverside, for GD is gratefully acknowledged. PB is supported by the joint NSF/NIGMS Mathematical Biology program under NIH grant R01 GM 078985. We also thank Sylvia Pazoutová for cultures of C. purpurea.
References
- Alexopolous CJ, Mims C, Blackwell M. Introductory Mycology. John Wiley and Sons, Inc; New York, USA: 1996. [Google Scholar]
- Annis SL, Panaccione DG. Accumulation of peptide synthetase gene transcripts and ergopeptines in cultures of ergopeptine-producing fungi. Canadian Journal of Microbiology. 1998;44:80–86. doi: 10.1139/w97-130. [DOI] [PubMed] [Google Scholar]
- Avise JC. Molecular Markers, Natural History and Evolution. Chapman & Hall; New York, NY: 1994. [Google Scholar]
- Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–73. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P. Effect of unsampled populations on the estimation of population sizes and migration rates between sampled populations. Molecular Ecology. 2004;13:827–836. doi: 10.1111/j.1365-294x.2004.02101.x. [DOI] [PubMed] [Google Scholar]
- Beerli P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics. 2006;22:341–345. doi: 10.1093/bioinformatics/bti803. [DOI] [PubMed] [Google Scholar]
- Bidochka MJ, Kamp AM, Lavender TM, Dekoning J, De Croos JNM. Habitat association in two genetic groups of the insect-pathogenic fungus Metarhzium anisoplaie: Uncovering cryptic species? Applied and Environmental Microbiology. 2001;67:1335–1342. doi: 10.1128/AEM.67.3.1335-1342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntjer JB, Otsen M, Nijman IJ, Kuiper MTR, Lenstra JA. Phylogeny of bovine species based on AFLP fingerprinting. Heredity. 2002;88:46–51. doi: 10.1038/sj.hdy.6800007. [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- Chenna R, Sugawara H, Koide T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch BC, Fudal I, Lebrun M-H, Tharreau D, Valent B, van Kim P, Nottéghem J-L, Kohn LM. Origins of host-specific populations of the blast pathogen, Magnaporthe oryzae, in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics. 2005;170:613–630. doi: 10.1534/genetics.105.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch BC, Kohn LM. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia. 2002;94:683–93. doi: 10.1080/15572536.2003.11833196. [DOI] [PubMed] [Google Scholar]
- Cruse M, Telerant R, Gallagher T, Lee T, Taylor JW. Cryptic species in Stachybotrys chartarum. Mycologia. 2002;94:814–822. [PubMed] [Google Scholar]
- Cruz R, Carballo M, Conde-Padin P, Rolan-Alvarez E. Testing alternative models for sexual isolation in natural populations of Littorina saxatilis: indirect support for by-product ecological speciation? Journal of Evolutionary Biology. 2004;17:288–293. doi: 10.1111/j.1420-9101.2003.00689.x. [DOI] [PubMed] [Google Scholar]
- Dabkevicius Z, Mikaliunaite R. The effect of fungicidal seed treatments on germination of rye ergot (Claviceps purpurea (Fr.) Tul.) sclerotia and on ascocarp formation. Crop Protection. 2006;25:677–683. [Google Scholar]
- Daehler CC, Anttila CK, Ayres DR, Strong DR, Bailey JP. Evolution of a new ecotype of Spartina alterniflora (Poaceae) in San Francisco Bay, California, USA. American Journal of Botany. 1999;86:543–546. [PubMed] [Google Scholar]
- Després L, Gielly L, Redoutet B, Tabarlet P. Using AFLP to resolve phylogenetic relationships in a morphologically diversified plant species complex when nuclear and chloroplast sequences fail to reveal variability. Molecular Phylogenetics and Evolution. 2002;27:185–196. doi: 10.1016/s1055-7903(02)00445-1. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution. 2003;57:2721–2741. doi: 10.1111/j.0014-3820.2003.tb01515.x. [DOI] [PubMed] [Google Scholar]
- Douhan GW, Rizzo DM. Host-parasite relationships among bolete infecting Hypomyces species. Mycological Research. 2003;107:1342–1349. doi: 10.1017/s0953756203008542. [DOI] [PubMed] [Google Scholar]
- Douhan GW, Rizzo DM. Phylogenetic divergence in a local population of the ectomycorrhizal fungus Cenococcum geophilum. New Phytologist. 2005;166:263–271. doi: 10.1111/j.1469-8137.2004.01305.x. [DOI] [PubMed] [Google Scholar]
- Duncan RA, Sullivan R, Alderman SC, Spatafora JW, White JF. Claviceps purpurea var. spartinae var. nov.: an ergot adapted to the aquatic environment. Mycotaxon. 2002;81:11–25. [Google Scholar]
- Excoffier L. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Molecular Ecology. 2004;13(4):853–864. doi: 10.1046/j.1365-294x.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sinauer Associates; Sunderland MA: 2004. p. 664. [Google Scholar]
- Fine PVA, Daly DC, Munoz GV, Mesones I, Cameron KM. The contribution of edaphic heterogeneity to the evolution and diversity of Burseraceae trees in the western Amazon. Evolution. 2005;59:1464–1478. [PubMed] [Google Scholar]
- Fisher AJ, DiTomaso JM, Gordon TR. Conidial morphology and ecological characteristics as diagnostic tools for identifying Claviceps purpurea from salt-marsh habitats. Canadian Journal of Plant Pathology. 2005a;27:389–395. [Google Scholar]
- Fisher AJ, DiTomaso JM, Gordon TR. Sub-specific groups of Claviceps purpurea associated with grass species in Willapa Bay, Washington, and the prospects for biological control of invasive Spartina alterniflora. Biological Control. 2005b;34:170–179. [Google Scholar]
- Fisher AJ, Gordon TR, DiTomaso JM. Geographic distribution and diversity in Claviceps purpurea (Fr.) Tul from salt marsh habitats and characterization of Pacific Coast populations. Mycological Research. 2005c;109:439–446. doi: 10.1017/s0953756205002467. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for Basidiomycetes -application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Pitt JI, Taylor JW. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Willis JH. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution. 2006;60:2466–2477. [PubMed] [Google Scholar]
- Hardin G. The competitive exclusion principle. Science. 1960;131:1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- Harrington TC, Rizzo DM. Defining species in the fungi. Academic Publishers; London: 1999. [Google Scholar]
- Hart C. Forged in St. Anthony's Fire: drugs for migraine. Modern drug discovery. 1999;2:20–31. [Google Scholar]
- Hatfield T, Schluter D. Ecological speciation in sticklebacks: environment dependent hybrid fitness. Evolution. 1999;53:866–873. doi: 10.1111/j.1558-5646.1999.tb05380.x. [DOI] [PubMed] [Google Scholar]
- Horton TR. Molecular approaches to ectomycorrhizal diversity studies: variation in ITS at a local scale. Plant and Soil. 2002;244:29–39. [Google Scholar]
- Hudler GW. Magical Mushrooms, Mischievous Molds. Princeton University Press; 1998. [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. Journal of the American Statistical Association. 1995;90(430):773–795. [Google Scholar]
- Kardolus JP, van Eck HJ, van den Berg RG. The potential of AFLPs in biosystematics: a first application in Solanum taxonomy (Solanaceae) Plant Systematics and Evolution. 1998;210:87–103. [Google Scholar]
- Kingman JF. Origins of the coalescent. 1974–1982. Genetics. 2000;156(4):1461–1463. doi: 10.1093/genetics/156.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn LM. Mechanisms of fungal speciation. Annual Review of Phytopathology. 2005;43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- Koufopanou V, Burt A, Szaro T, Taylor JW. Gene genealogies, cryptic species, and molecular evolution in the human pathogen Coccidioides immitis and relatives (Ascomycota, Onygenales) Molecular Biology and Evolution. 2001;18:1246–1258. doi: 10.1093/oxfordjournals.molbev.a003910. [DOI] [PubMed] [Google Scholar]
- Kroken S, Taylor JW. A gene genealogical approach to recognize phylogenetic species boundaries in the lichenized fungus Letharia. Mycologia. 2001;93:38–53. [Google Scholar]
- Kruuk LEB, Gilchrist JS. Mechanisms maintaining species differentiation: predator mediated selection in a Bombina hybrid zone. Proceedings of the Royal Society of London B. 1997;264:105–110. [Google Scholar]
- Lamont BB, He T, Enright NJ, Krauss SL, Miller BP. Anthropogenic disturbance promotes hybridization between Banksia species by altering their biology. Journal of Evolutionary Biology. 2003;16:551–557. doi: 10.1046/j.1420-9101.2003.00548.x. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4: Analysis of phylogeny and character evolution. Version 4.03. Sinauer Associates; Sunderland, Massachusetts: 2001. [Google Scholar]
- Mitsch WJ, Gosselink JG. Wetlands. John Wiley & Sons, Inc; New York: 1993. [Google Scholar]
- Moyersoen B, Beever RE, Martin F. Genetic diversity of Pisolithus in New Zealand indicates multiple long-distance dispersal from Australia. New Phytologist. 2003;160:569–579. doi: 10.1046/j.1469-8137.2003.00908.x. [DOI] [PubMed] [Google Scholar]
- Muths D, Davoult D, Gentil F, Jollivet D. Incomplete cryptic speciation between intertidal and subtidal morphs of Acrocnida brachiata (Echinodermata: Ophiuroidea) in the Northeast Atlantic. Molecular Ecology. 2006;15:3303–3318. doi: 10.1111/j.1365-294X.2006.03000.x. [DOI] [PubMed] [Google Scholar]
- Myllys L, Lohtander K, Tehler A. Beta-tubulin, ITS and group I intron sequences challenge the species pair concept in Physcia aipolia and P. caesia. Mycologia. 2001;93:335–343. [Google Scholar]
- Nagy ES. Selection for native characters in hybrids between two locally adapted plant subspecies. Evolution. 1997;51:1469–1480. doi: 10.1111/j.1558-5646.1997.tb01470.x. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Tack BK, Casper HH. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proceedings of the National Academy of Sciences. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden R, Thorpe RS. Molecular evidence for ecological speciation in tropical habitats. Proceedings of the National Academy of Sciences. 2002;99:13612–13615. doi: 10.1073/pnas.212248499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazoutová S, Olsovska J, Linka M, Kolínská R, Flieger M. Chemoraces and habitat specialization of Claviceps purpurea populations. Applied and Environmental Microbiology. 2000;66:5419–5425. doi: 10.1128/aem.66.12.5419-5425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazoutová S, Raybould AF, Honzátko A, Kolínská R. Specialized populations of Claviceps purpurea from salt marsh Spartina species. Mycological Research. 2002;106:210–214. [Google Scholar]
- Pazoutová S. Evolutionary strategy of Claviceps. In: White JF, Bacon CW, Hywel-Jones NL, Spatafora JW, editors. Clavicipitalean Fungi: Evolutionary Biology, Chemistry, Biocontrol and Cultural Impacts. Marcel Dekker; New York, Basel: 2003. pp. 329–354. [Google Scholar]
- Rehacek Z, Sajdl P. Ergot alkaloids: Chemistry, Biological Effects, Biotechnology. New York: Elsevier Science publishing; 1993. [Google Scholar]
- Richmond JQ, Reeder TW. Evidence for parallel ecological speciation in scincid lizards of the Eumeces skiltonianus species group (Squamata: Scincidae) Evolution. 2002;56:1498–1513. doi: 10.1111/j.0014-3820.2002.tb01461.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Crossing relationships among ancient and experimental sunflower hybrid lineages. Evolution. 2000;54:859–865. doi: 10.1111/j.0014-3820.2000.tb00086.x. [DOI] [PubMed] [Google Scholar]
- Rocha LA, Robertson DR, Roman J, Bowen BW. Ecological speciation in tropical reef fishes. Proceedings of the Royal Society of London B. 2005;272:573–579. doi: 10.1098/2004.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB. Convergent evolution and divergent selection: Lizards at the White Sands ecotone. American Naturalist. 2006;167:1–15. doi: 10.1086/498397. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Nosil P. Ecological speciation. Ecology Letters. 2005;8:336–352. [Google Scholar]
- Scheffer J, Tudzynski P. In vitro pathogenicity assay for the ergot fungus Claviceps purpurea. Mycological Research. 2006;110:465–470. doi: 10.1016/j.mycres.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Scheffer J, Chen CB, Heidrich P, Dickman MB, Tudzynski P. A CDC42 Homologue in Claviceps purpurea is involved in vegetative differentiation and is essential for pathogenicity. Eukaryotic Cell. 2005;4:1228–1238. doi: 10.1128/EC.4.7.1228-1238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seberg OG, Peterson G. Constructing phylogenies from discrete data-parsimony methods. In: Karp A, Isaac PG, Ingram DS, editors. Molecular Tools for Screening Biodiversity. Chapman & Hall; London: 1998. pp. 344–355. [Google Scholar]
- Shykoff JA, Meyhöfer A, Bucheli E. Genetic isolation among host races of the anther smut fungus Microbotryum violaceum on three host plant species. International Journal of Plant Science. 1999;160:907–916. doi: 10.1086/314179. [DOI] [PubMed] [Google Scholar]
- Skovgaard K, Bodker L, Rosendahl S. Population structure and pathogenicity of members of the Fusarium oxysporum complex isolated from soil and root necrosis of pea (Pisum sativum L.) FEMS Microbiology and Ecology. 2002;42:367–374. doi: 10.1111/j.1574-6941.2002.tb01026.x. [DOI] [PubMed] [Google Scholar]
- Stager R. Beitrag zur Verbreitungsbiologie der Claviceps-Sklerotien. Zentralblatt Fur Bakteriologie Parasitenkunde Infefektionskranheiten Und Hygiene. 1922;56:329–339. [Google Scholar]
- Stensrud O, Hywel-Jones NL, Schumacher T. Towards a phylogenetic classification of Cordyceps: ITS rDNA sequence data confirm divergent lineages and paraphyly. Mycological Research. 2005;109:41–56. doi: 10.1017/s095375620400139x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and other methods). Version 4.0b10. Sunderland, Massachusetts (USA): Sinauer Associates, Inc; 2002. [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Tooley PW, Goley ED, Carras MM, Frederick RD, Weber EL, Kuldauc GA. Characterization of Claviceps species pathogenic on sorghum by sequence analysis of the ß-tubulin gene intron 3 region and EF-1a gene intron 4. Mycologia. 2001;93:541–551. [Google Scholar]
- Tudzynski P, Scheffer J. Claviceps purpurea: molecular aspects of a unique pathogenic lifestyle. Molecular Plant Pathology. 2004;5:377–388. doi: 10.1111/j.1364-3703.2004.00237.x. [DOI] [PubMed] [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends in Ecology and Evolution. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. [DOI] [PubMed] [Google Scholar]
- Via S, Bouck AC, Skillman S. Reproductive isolation between divergent races of pea aphids on two hosts. II. Selection against migrants and hybrids in the parental environments. Evolution. 2000;54:1626–1637. doi: 10.1111/j.0014-3820.2000.tb00707.x. [DOI] [PubMed] [Google Scholar]
- Wang H, McArthur ED, Sanderson SC, Graham JH, Freeman DC. Narrow hybrid zone between two subspecies of big sagebrush (Artemisia tridentata: Asteraceae). IV. Reciprocal transplant experiments. Evolution. 1997;51:95–102. doi: 10.1111/j.1558-5646.1997.tb02391.x. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A guide to methods and applications. San Diego, USA: Academic Press; 1990. [Google Scholar]
- Wu QX, Mueller GM, Lutzoni FM, Huang YQ, Guo SY. Phylogenetic and biogeographic relationships of eastern Asian and eastern North American disjunct Suillus species (Fungi) as inferred from ITS sequences of nuclear ribosomal RNA ITS sequences. Molecular Phylogenetics and Evolution. 2000;17:37–47. doi: 10.1006/mpev.2000.0812. [DOI] [PubMed] [Google Scholar]