Abstract
Bacteria and their viruses (phages) are locked in an evolutionary contest, with each side producing constantly changing mechanisms of attack and defense that are aimed to increase the odds of survival. As a result, phages play central roles in a great variety of genetic processes and drive rapid evolution of their bacterial hosts, which might ultimately work to the benefit of the host in a long run.
Introduction
Bacterial viruses (bacteriophages or phages) are the most abundant life form on earth [1, 2*], and as such they exert a profound impact on their hosts. During infection, various components of the cellular enzymatic machinery are modified by phage-encoded functions or are altogether inactivated to be replaced with phage components. The diversity of mechanisms that phages employ to deal with the cellular machinery provides invaluable insights into molecular details of its function and regulation.
Apart from simply “eating” (Greek phagein) bacteria, phages constitute a major evolutionary force that drives changes in host genome by fueling co-evolution of arms-race mechanisms, promoting lateral transfer of genetic information, facilitating genomic rearrangements, etc. Here, we briefly summarize recent advances in our understanding of the effects of dsDNA phages on their bacterial hosts.
Effect of phages on host macromolecular machinery
DNA replication
While some phages, such as λ, use individual components of the host replication apparatus (for example, 3), or inhibit them [4, 5], most analyzed dsDNA phages encode elements of their own replication machinery [6**] and do not rely on regulation of host replication enzymes. Thus, recent identification of Staphylococcus aureus phage proteins that specifically interact with the host replication apparatus is important [7**]. The authors systematically determined the ability of individual plasmid-expressed ORFs from 26 S. aureus phages to inhibit the growth of their host. Several phage proteins were found to interact with components of host replication machinery. Such pairwise interactions between phage-encoded polypeptides and their host replication apparatus targets were then used for development of high-throughput screening assays to find small molecules that disrupt these interactions and may thus serve as lead compounds for development of antibiotics that target bacterial replication [7**]. The most recent work focused on the inhibition of the host DnaN sliding clamp by two polypeptides from phages Twort and G1 [8]. Both polypeptides were shown to bind the sliding clamp and prevent its loading onto DNA and its interaction with DNA polymerase. Since neither protein has any effect on DNA replication of the related species, Streptococcus pyogenes, they must interact with evolutionary variable regions of the sliding clamp. Presently, it remains unclear whether inhibition of host replication is the main biological function of these polypeptides or indeed whether host replication inhibition is functionally important during phage infection.
DNA transcription
RNA polymerase (RNAP) is the most highly regulated enzyme in the cell and, not surprisingly, is commonly targeted by phage during infection. Interactions between phage factors and host RNAPs have long served as a rich source of paradigms for transcriptional regulation [reviewed in 9]. Recent advent of genomic sequences of novel phages has continued to reveal novel mechanisms. Three phages are noteworthy in this regard. Xp10, a siphovirus (long non-contractile tail virus) morphologically similar to phage ⌊, was shown to encode its own single-subunit RNAP [10]. Previously, only short-tailed podoviruses similar to phage T7 were known to encode their own RNAPs and use it for late genes expression [9]. Using a combination of genomic, computational, and modeling approaches Djordjevic et al [11] found that the gene regulatory strategy of Xp10 effectively combines the apparently orthogonal strategies of T7 and ⌊. Key to this strategy is Xp10 transcription regulator p7 which i) blocks transcription from most cellular and early viral promoters by host RNAP and thus stimulates late gene transcription by viral RNAP and ii) stimulates late gene transcription by antiterminating host RNAP transcription that initiates from a p7-resistant promoter. Identification of the site of p7 interaction on host RNAP provided a structural explanation for the unusual dual function of p7 [12].
Most recently, E. coli phage phiEcoM-GJ1, a myovirus (contractile tail virus) morphologically similar to phage T4, was also found to encode a single-subunit RNAP [13], suggesting that the tailed phages of every morphological type can rely on their own RNAPs for transcription of viral genes. The computational and modeling approach developed in [11] is broadly applicable to phages encoding their own RNAP and, if applied to phiEcoM-GJ1, is expected to reveal the regulatory strategy of its multiplication cycle and will likely identify novel transcription regulators.
The third example of a phage with a novel transcription regulation strategy is the YuA phage of P. aeruginosa [14*]. Expression of YuA late genes requires host ⎛54, a minor RNAP ⎛ factor that is unrelated to major ⎛70 class factors and that involves a complex set of ATP-dependent regulators for transcription. Biological implications and mechanistic details of this currently unique case of ⎛54 involvement in phage gene expression require further investigation.
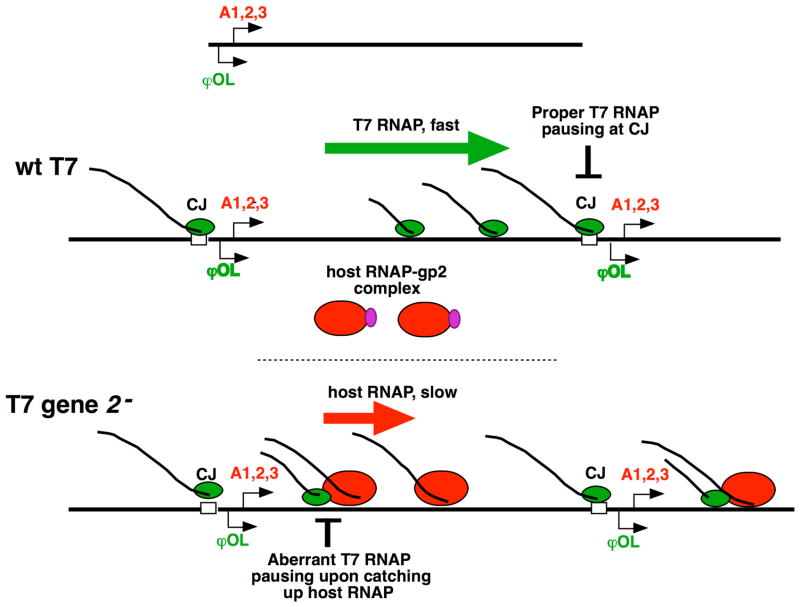
While the spirit of new phage biology is the exploration of diversity, classical phages of E. coli offer important advantages because a large number of genetic tools to manipulate their host are readily available. Two recent additions are the Keio collection of knockouts in dispensable E. coli genes and the ASKA library of cells containing 85% of E. coli ORFs under inducible promoters on high-copy plasmids. These collections enable systematic analysis of the effects of deletion/overexpression of nonessential E. coli genes on the process of phage infection. The Richardson group used these libraries to study T7 infection [15]. In addition to genes whose products are known to influence the growth of T7, several novel host genes whose importance for T7 development was previously unrecognized were identified. One such gene, udk, coding for a cytosine/thymidine kinase, impaired T7 growth when overexpressed [15]. T7 mutants that are able to grow on Udk-overproducing strains were mapped [16*] and found to contain lesions in early promoters, substitutions in the T7 lysozyme, which decrease its ability to bind and inhibit T7 RNAP, or mutations in gene 2, whose product, gp2, binds to E. coli RNAP and prevents promoter recognition [17]. Early genes of T7 phage, including the T7 RNAP gene, are transcribed by the E. coli RNAP; T7 RNAP transcribes phage middle genes, including gene 2, and late genes. Host RNAP inhibition by gp2 in vitro and the timing of its expression in vivo suggest that gp2 may orchestrate a switch from host to viral RNAP transcription. However, T7 infection in the absence of gp2 fails not at the stage when the switch from host to viral RNAP transcription occurs, but does much later, during T7 DNA packaging [18]. During packaging, concatemeric products of T7 genome replication are cleaved into unit-length genomes that are inserted into virion proheads, right genome end first. In the absence of gp2, processing of the genome left end proceeds aberrantly. T7 gene 2 mutants that overcome the defects caused by udk overexpression encode gp2 that binds host RNAP better than the wild-type gp2 [16*]. Qimron et al propose a model of gp2 function that is consistent with the available data (Fig. 1). They posit that normal processing of T7 concatemers requires that the transcribing T7 RNAP pauses at a specific site at the concatemer junction and recruits the processing and packaging factors gp18 and gp19. This pausing is enhanced by T7 lysozyme. In the absence of gp2, transcription by slow-moving host RNAP from strong rightward early promoters presents a roadblock to transcription by the much faster T7 RNAP, causing it to pause, which in turn generates undue recruitment of gp18 and gp19, and aberrant packaging. Decreasing the strength of T7 lysozyme interaction with viral RNAP, removing the early phage promoters, or increasing the strength of gp2 interaction with host RNAP should decrease aberrant T7 RNAP stalling. Udk may alter the elongation rate by T7 or host RNAP by changing the concentration of NTPs in the cell. Alternatively, Udk may bind gp2 and inactivate it.
Figure 1. A model for the role of the host RNAP inhibitor gp2 during T7 infection (after [16*]).
At the top, the linear genome of the T7 phage is shown. Arrows indicate three strong early promoters recognized by host RNAP (A1,2,3, red) and phage RNAP promoter ⎞OL (green). Other phage RNAP promoters are omitted for clarity. Below, the concatemeric intermediate of T7 genome replication is shown (open boxes labeled CJ are concatemeric junctions). During the wild-type T7 infection, gp2, the product of phage gene 2 binds to and inhibits host RNAP. T7 RNAP transcribes most of the T7 genome and pauses at CJ, recruiting the processing and packaging factors gp18 and gp19. In the absence of gp2 (bottom), host RNAP transcription proceeds throughout the infection. The transcription elongation rate of host RNAP is much lower than that of T7 RNAP, leading to T7 RNAP stalling behind transcribing host RNAP, and inducing aberrant pauses followed by recruitment of gp18/19 and incorrect processing at the left end of the genome.
mRNA translation
The cell usually contains a sufficient supply of ribosomes to produce extra proteins on short notice. Thus, many phages utilize unmodified host translation machinery to translate their mRNA. However, many diverse and unrelated phages encode tRNA. According to one recent work, phage tRNAs tend to recognize codons that are abundant in phage ORFs but are underrepresented in host ORFs [19**]. Such phage-encoded tRNAs should therefore stimulate preferential translation of viral mRNAs. However, in contrast to the observations made in [19**], codon specificities of tRNAs encoded by phages of the T4 group do not correlate with codon composition of viral genes [20]. Nevertheless, the remarkably high level of sequence conservation of tRNA genes among phages of this group indicates that they may play an important role that remains to be identified.
Another work [21] found that Thermus phage ⎞YS40 might control the expression of its genes through a novel mechanism that directs the cellular translation apparatus towards the leaderless mRNAs of phage middle and late genes. To accomplish this, the phage appears to specifically upregulate the levels of IF2, a translation factor that has been previously implicated in translation of leaderless RNA [22].
Effects of phages on host genomes
Phages can profoundly influence genomes of their bacterial hosts through various mechanisms. Infection of Campylobacter jejuni with its virulent bacteriophage CP34 leads to major genomic rearrangements in surviving bacteria [23**]. These rearrangements occur through recombination at the sites of integration of Mu-like prophages, with the largest rearrangement being a 590 kb inversion. These rearrangements are associated with the acquisition of phage resistance but, importantly, also reduce C. jejuni capability to colonize chicken intestine. Curiously, resistance through chromosomal rearrangements was acquired only in clones isolated from phage-treated infected chickens: spontaneous phage-resistant mutants obtained in vitro did not contain these rearrangements. This work demonstrates that in vitro plating experiments might not always be good models for naturally occurring phage resistance in bacterial populations. Another observation of this work is that, when reintroduced into chickens in the absence of the phage, formerly phage-resistant Campylobacter isolates undergo further rearrangements and again become both sensitive to the phage and proficient in colonization of chickens [23**]. These observations reveal a remarkable genomic plasticity that bacteria can show in response to bacteriophage. Molecular mechanisms as well as the prevalence of genomic rearrangements in natural C. jejuni populations remain to be determined.
Another recent work studied the coexistence of a host bacterium (Pseudomonas fluorescens SBW25) and its phage SBW25√2 over ~200 bacterial generations [24**]. Among 36 populations independently maintained in the presence of the phage, 9 evolved mutator phenotypes, with 10–100 fold increases in mutation rates due to mutations in mismatch repair genes mutL and mutS. None of the 36 control populations maintained in the absence of the phage had acquired mutator phenotypes. The mutS mutants gained a selective advantage over the wild-type bacteria when grown in the presence (but not in the absence) of the phage, indicating that the mutator phenotype itself rather than possible associated mutations imparts growth advantage [24**]. While the emergence of mutator phenotypes has been shown to occur in several systems (reviewed in 25), the phage-induced mutator phenotype shown in [24**] is novel. Applying the “evolutionary throttle”, afforded by the acquisition of mutator phenotype, might bring a temporary advantage in the phage-host race to the bacterial population.
Coevolution of genes: marine phages
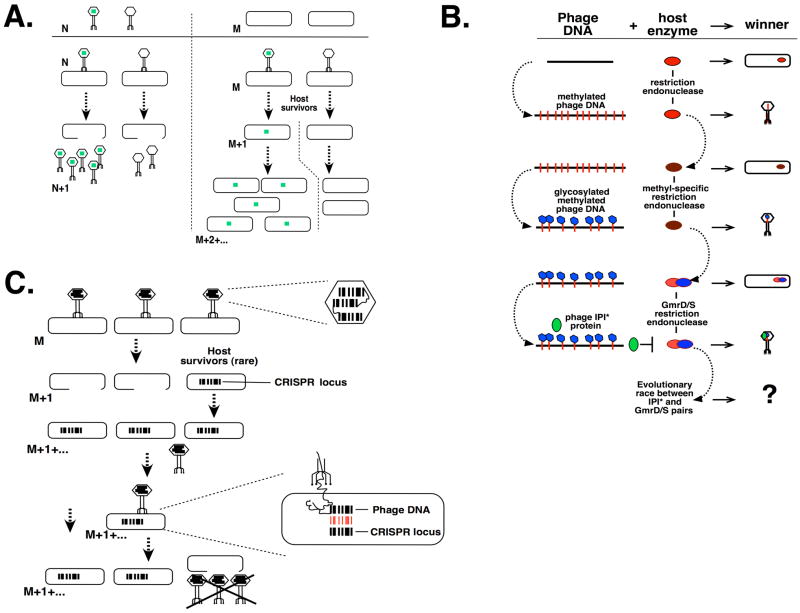
Phage P-SSP7 and its host, a marine cyanobacterium Prochlorococcus MED4, demonstrate a remarkable relationship [26**]. P-SSP7 is related to the T7 phage. Unlike T7, the P-SSP7 genome contains several bacterial metabolism genes, including the psba gene encoding photosystem II D1. Exchange of metabolism genes between marine viruses and their cyanobacterial hosts appears to be a widespread phenomenon [27]. By carrying photosynthesis-related genes in their genomes, phages might provide a boost of host metabolism during infection and in this way speedup the infection of slow-growing cells living in resource-poor ocean water environment, acquiring a selective advantage. Consistent with their role in infection, P-SPP7 bacterial metabolism genes are expressed in a coordinated manner despite their scattered location in the phage genome [26**]. The authors hypothesize that these “higher-fitness” host genes have a faster rate of evolution while borne by the virus than they would have if they remained in the host. “Outsourcing” of gene evolution to the phage might therefore provide a competitive advantage to bacteria that later reacquire these genes and thus be advantageous to bacteria in a long run (Fig. 2A). Currently, the reverse transfer of metabolism genes remains hypothetical.
Figure 2. Examples of genetic impact of phage on the host.
The host bacteria are shown as rounded rectangles. M and N represent generation numbers for host and phage, respectively; their absolute values are arbitrary. M/N+1 indicate progeny generations. A. Co-evolution. A variant of a metabolism gene that specifies a beneficial trait to the host is shown as a green square. Left. Phage that carries the beneficial trait enables faster host metabolism during the course of infection, resulting in a more productive infection. Right. Host bacteria that survive the infection and acquire the beneficial trait have more efficient metabolism and thus have a growth advantage in competition with their predecessors. B. Arms race. An example of a contest between phage DNA protective modifications and host restriction system is shown (based on a situation encountered in T4 phage and its relatives). Each genetic interaction (infection) is shown in the same horizontal level, with the outcome of each interaction listed on the right. Host as the winner indicates that the restriction system has degraded the phage DNA; phage as the winner indicates that the phage DNA is resistant to the host’s restriction system. Evolutionary arms race steps undertaken by host or virus are indicated with bent vertical arrows, alongside the changing side. Adapted from [30]. C. CRISPR. Acquired immunity. Phage infection results in the demise of most cells, with an exception of rare survivors that incorporate portions of phage genome (shown as bar codes) into their CRISPR loci. The newly acquired CRISPR locus blocks propagation of the phage if there is a match between phage sequences and sequences of the CRISPR locus (the match is shown in red).
Global analysis of steady-state levels of host transcripts during P-SSP7 infection revealed that while transcript levels of most genes are decreased, as expected, some genes are explicitly activated. While the mechanisms of this activation are unknown, the authors speculate that P-SPP7 evolved to use the products of some of the activated host genes for its own advantage. The authors cite as “the most compelling evidence for the co-evolutionary process” in their virus-host system the fact that many activated genes are found in the hypervariability islands and may be of phage origins [26**]. They do not exclude a simpler idea that host genes of viral origins are upregulated through a mechanism that may involve mobilization of resident prophages upon infection, and as such may have little to do with co-operation between P-SPP7 and its host.
Arms race: a case of T4 phage
Bacteria employ various strategies to thwart virus threats [reviewed in 28]. These strategies are readily countered by the phages. An interesting example of such an arms race is provided by recent analysis of T4, one of the best-studied phages. T4 genomic DNA contains hydroxymethylated cytidine residues (HMC) instead of cytidines found in the host DNA. In addition, phage HMC residues are glycosylated. A pathogenic E. coli CT596 strain excludes T4 phage that lacks the lp1 gene. CT596 encodes a two-gene restriction system (gmrS/gmrD carried on a prophage) that specifically recognizes and cleaves glycosylated HMC DNA but has no effect on unglycosylated DNA [29]. The product of ip1, internal protein I (IPI), is targeted inside the viral capsid by the N-terminal capsid targeting sequence that is subsequently removed, resulting in mature processed IP1 (called IPI*). IPI* specifically binds to and inactivates the GmrS/GmrD complex. Several hundred copies of IPI* are injected into the host along with T4 DNA. Evidently, the sole function of IPI* is to counter GmrS/GmrD defense mechanism [30*], suggesting that the phage and its host have evolved the corresponding systems specifically to counteract each other (Fig. 2B). The latest work revealed a subsequent step in this arms race, demonstrating that another E. coli strain restricts T4 by degrading glycosylated HMC-DNA despite the presence of IPI*. This immunity to IPI* arose through acquisition of an altered gmrS/gmrD system that contains a fused GmrS-GmrD polypeptide with an overall 90% identity to the heterodimeric enzyme from E. coli CT596 [31]. The authors suggest that the altered system has evolved solely to counteract the inhibition by IPI*. Indeed, diverse bacterial species contain sequence homologs of gmrS/gmrD; at the same time, various T4 relatives contain genes whose genomic locations correspond to that of T4 lp1, but whose products share no sequence similarity with IPI, except for the capsid targeting sequence [30*, 31]. It is therefore possible that these proteins target additional, yet unknown host defense mechanisms. Thus, host restriction systems, phage DNA modifications, and phage-encoded inhibitors of host restriction endonucleases form a proprietary system whose evolution produces phage DNA modifications of increasing complexity and host restriction systems whose only function is to specifically counteract these modifications (Fig. 2B).
CRISPR: a bacterial immune system based on genetic memories of bad times?
Stretches of short (21–47bp) direct repeats interspaced with unrelated nonrepetitive sequences of similar size were found in ~50% of analyzed eubacterial and most of achaeal genomes and were called Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) [32–36*]. The number of repeats in a CRISPR locus varies from 2 to over 200 [36*]; the same genome could contain one or several different CRISPR loci. While the repeated sequences are very similar, they become more variable towards one end of the CRISPR locus. In addition to the repeated regions, a CRISPR locus usually contains an adjacent non-repeat leader region and several CRISPR-associated (CAS) genes [37].
Based on the observations that some sequences between repeats in CRISPR loci show similarity to phage and plasmid sequences, Makarova et al [38*] suggested that CRISPRs and CAS genes form a bacterial immune system directed against phages and other extrachromosomal genetic elements, and that this system may function through an RNAi-like mechanism. Indeed, recent works showed that CRISPR loci change in response to phage infection by acquiring nonrepetitive (spacer) sequences from the infecting phage, and that these changes are correlated with phage resistance [39**, 40*]. Furthermore, introduction of a phage sequence into a CRISPR spacer by genetic engineering introduced resistance to this particular phage in an otherwise naive, phage-sensitive strain. These results indicate that CRISPRs function by incorporating fragments of viral sequences into the spacers (Fig. 2C), and recent work confirms that directly [41*]. Fully consistent with these observations, CRISPR loci are among the most rapidly changing sequences in bacteria, are subject of active lateral gene transfer, and are involved in conferring phage resistance in natural populations [42*, 43*]. From a practical standpoint, CRISPR sequences effectively contain the history of a bacterial strain’s exposure to foreign DNA.
Mechanisms of CRISPR function remain to be determined. Analysis of mutant phages that overcome CRISPR-based resistance shows that they contain alterations in sequences that became incorporated in the spacer. In addition, phages can overcome CRISPR-based immunity by changing a sequence in their genome that is immediately adjacent to the sequence found in the CRISPR spacer [41*]. It appears that phage sequences that are adjacent to (but are not part of) sequences found in CRISPR spacers are conserved, indicating that cellular machinery (a CAS protein?) recognizes some conserved motif before initiating the transfer of phage sequence into the CRISPR locus. Currently, nothing is known about the enzymology of CRISPR-mediated phage resistance. Transcription of CRISPR regions and processing of resultant transcripts has been experimentally demonstrated only in Archaea [44]. Transcription-mediated functioning of the CRISPR system in eubacteria remains to be proven and mechanisms of functioning of this exciting system remain to be identified. With tools for facile identification of CRISPR repeats becoming available [45, 46], rapid progress on elucidation of their structure and function is expected.
Conclusions
Despite the rapid progress, our understanding of even a few well-studied phages of E. coli is far from complete. New experimental approaches combining biochemistry with systematic genome-wide analysis of both the host and viral genes over the course of infection will continue to provide exciting results in this field for many years to come.
Acknowledgments
We thank E. P. Geiduschek for valuable advice. This article was supported in part by the Intramural Research Program of the US National Institutes of Health, National Institute of Environmental Health Sciences and an NIH RO1 grant GM59295 to KS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. A review that highlights the recent finding on contribution of viruses to marine ecosystems. The review covers both viruses of bacteria and eukaryotes and in that provides one of the most general views of viral dynamics. [DOI] [PubMed] [Google Scholar]
- 3.Taylor K, Wegrzyn G. Replication of coliphage lambda DNA. FEMS Microbiol Rev. 1995;17:109–119. doi: 10.1111/j.1574-6976.1995.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 4.Sergueev K, Court D, Reaves L, Austin S. E.coli cell-cycle regulation by bacteriophage lambda. J Mol Biol. 2002;324:297–307. doi: 10.1016/s0022-2836(02)01037-9. [DOI] [PubMed] [Google Scholar]
- 5.Datta I, Sau S, Sil AK, Mandal NC. The bacteriophage lambda DNA replication protein P inhibits the oriC DNA- and ATP-binding functions of the DNA replication initiator protein DnaA of Escherichia coli. J Biochem Mol Biol. 2005;38:97–103. doi: 10.5483/bmbrep.2005.38.1.097. [DOI] [PubMed] [Google Scholar]
- 6**.Weigel C, Seitz H. Bacteriophage replication modules. FEMS Microbiol Rev. 2006;30:321–81. doi: 10.1111/j.1574-6976.2006.00015.x. One of the most comprehensive reviews that covers essentially every enzyme involved in prokaryotic replication. A must-read for anyone in the field. A supplementary compendium of replication enzymes might be also very useful for reference. [DOI] [PubMed] [Google Scholar]
- 7**.Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M. Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol. 2004;22:185–191. doi: 10.1038/nbt932. To the best of our knowledge, this work shows the first examples of bacteriophage-encoded inhibitors of host replicase other than inhibitors of replication initiation. Based on this work, it is likely that additional phage-derived inhibitors will be identified and their cellular targets and mechanisms of action will be described. In addition, the work has practical application, suggesting strategies for designing antibacterial drugs. [DOI] [PubMed] [Google Scholar]
- 8.Belley A, Callejo M, Arhin F, Dehbi M, Fadhil I, Liu J, McKay G, Srikumar R, Bauda P, Bergeron D, Ha N, Dubow M, Gros P, Pelletier J, Moeck G. Competition of bacteriophage polypeptides with native replicase proteins for binding to the DNA sliding clamp reveals a novel mechanism for DNA replication arrest in Staphylococcus aureus. Mol Microbiol. 2006;62:1132–1143. doi: 10.1111/j.1365-2958.2006.05427.x. [DOI] [PubMed] [Google Scholar]
- 9.Nechaev S, Severinov K. Bacteriophage-induced modifications of host RNA polymerase. Annu Rev Microbiol. 2003;57:301–322. doi: 10.1146/annurev.micro.57.030502.090942. [DOI] [PubMed] [Google Scholar]
- 10.Yuzenkova J, Nechaev S, Berlin J, Rogulja D, Kuznedelov K, Inman R, Mushegian A, Severinov K. Genome of Xanthomonas oryzae bacteriophage Xp10: an odd T-odd phage. J Mol Biol. 2003;330:735–748. doi: 10.1016/s0022-2836(03)00634-x. [DOI] [PubMed] [Google Scholar]
- 11.Djordjevic M, Semenova E, Shraiman B, Severinov K. Quantitative analysis of bacteriophage Xp10 transcription strategy. Virology. 2006;354:240–251. doi: 10.1016/j.virol.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 12.Yuzenkova Y, Zenkin N, Severinov K. Mapping of RNA polymerase residues that interact with bacteriophage Xp10 transcription antitermination factor p7. J Mol Biol. 2008;375:29–35. doi: 10.1016/j.jmb.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamalludeen N, Kropinski AM, Johnson RP, Lingohr E, Harel J, Gyles CL. Complete Genomic Sequence of Bacteriophage {phi}EcoM-GJ1: a Novel Phage that has Myovirus Morphology and a Podovirus-like RNA polymerase. Appl Environ Microbiol. 2008;74:516–525. doi: 10.1128/AEM.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Ceyssens PJ, Mesyanzhinov V, Sykilinda N, Briers Y, Roucourt B, Lavigne R, Robben J, Domashin A, Miroshnikov K, Volckaert G, Hertveldt K. The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J Bacteriol. 2008;190:1429–1435. doi: 10.1128/JB.01441-07. This work is of particular interest to those studying mechanisms of transcription, as it identified a potentially a novel strategy of phage transcription regulation, which involves a ⎛54-llike transcription factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qimron U, Marintcheva B, Tabor S, Richardson CC. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc Natl Acad Sci U S A. 2006;103:19039–19044. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Qimron U, Kulczyk AW, Hamdan SM, Tabor S, Richardson CC. Inadequate inhibition of host RNA polymerase restricts T7 bacteriophage growth on hosts overexpressing udk. Mol Microbiol. 2008;67:448–457. doi: 10.1111/j.1365-2958.2007.06058.x. This article demonstrates a novel relationship between host RNA polymerase, its phage-encoded inhibitor gp2 and a metabolism gene udk, and by this prompts us to revisit our thinking about the long-standing question of the role of gp2 in T7 infection in particular, and of the roles of host RNAP at different stages of phage infection in general. [DOI] [PubMed] [Google Scholar]
- 17.Nechaev S, Severinov K. Inhibition of Escherichia coli RNA polymerase by bacteriophage T7 gene 2 protein. J Mol Biol. 1999;289:815–26. doi: 10.1006/jmbi.1999.2782. [DOI] [PubMed] [Google Scholar]
- 18.LeClerc JE, Richardson CC. Gene 2 protein of bacteriophage T7: purification and requirement for packaging of T7 DNA in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4852–4856. doi: 10.1073/pnas.76.10.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Bailly-Bechet M, Vergassola M, Rocha E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007;17:1486–1495. doi: 10.1101/gr.6649807. This paper addresses an long-observed but poorly understood phenomenon of ubiquitous presence of tRNA genes in genomes of various bacteriophages. Based on sequence analysis, the work posits an intriguing hypothesis on the role of these tRNA genes in phage infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolan JM, Petrov V, Bertrand C, Krisch HM, Karam JD. Genetic diversity among five T4-like bacteriophages. Virol J. 2006:23–30. doi: 10.1186/1743-422X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sevostyanova A, Djordjevic M, Kuznedelov K, Naryshkina T, Gelfand MS, Severinov K, Minakhin L. Temporal regulation of viral transcription during development of Thermus thermophilus bacteriophage phiYS40. J Mol Biol. 2007;366:420–435. doi: 10.1016/j.jmb.2006.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grill S, Moll I, Hasenöhrl D, Gualerzi CO, Bläsi U. Modulation of ribosomal recruitment to 5’-terminal start codons by translation initiation factors IF2 and IF3. FEBS Lett. 2001;495:167–171. doi: 10.1016/s0014-5793(01)02378-x. [DOI] [PubMed] [Google Scholar]
- 23**.Scott AE, Timms AR, Connerton PL, Loc Carrillo C, Adzfa Radzum K, Connerton IF. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 2007;3:e119. doi: 10.1371/journal.ppat.0030119. This outstanding paper not only demonstrates large-scale phage-induced genomic rearrangements in the host as a novel mechanism of dealing with virus threat, but also shows that these rearrangements do lead to phage resistance. Furthermore, the work shows that these rearrangements are reversible, and by this indicates that similar strategies might be used in other bacteria to evade virus threats. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Pal C, Maciá MD, Oliver A, Schachar I, Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–1081. doi: 10.1038/nature06350. This work shows that bacteria could respond to phage threat by acquiring the mutator phenotype, which confers a competitive advantage to the host specifically in the presence of viral threat. Findings of this work provide an important clue as to why mutation rates that are too low might be deleterious to organisms, and highlight that life span of a species might be determined by a fine compromise between fidelity of replication and survival in the presence of various threats. [DOI] [PubMed] [Google Scholar]
- 25.Ferenci T. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 2003;11:457–461. doi: 10.1016/j.tim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 26**.Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, Kettler G, Sullivan MB, Steen R, Hess WR, Church GM, Chisholm SW. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–86. doi: 10.1038/nature06130. This work proposes that carrying host metabolism genes might be beneficial to the phage, which might provide a metabolism boost to the host during phage infection. The same trend might be also beneficial to the host itself. Thus, “outsourcing” evolution of metabolism genes to the phage might be a novel aspect of phage-host relationship. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 2006;4:e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoskisson PA, Smith MC. Hypervariation and phase variation in the bacteriophage ‘resistome’. Curr Opin Microbiol. 2007;10:396–400. doi: 10.1016/j.mib.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Bair CL, Black LW. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J Mol Biol. 2007;366:768–778. doi: 10.1016/j.jmb.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Bair CL, Rifat D, Black LW. Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI*. J Mol Biol. 2007;366:779–789. doi: 10.1016/j.jmb.2006.11.049. This work suggests the existence of an evolutionary arms race between the phage and host, through targeting the same interaction. Resulting from this race, both sides accumulate elaborate DNA modifications and proprietary enzymes evolved to counteract these modifications, both of which are gratuitous in other environments. These increasingly elaborate but ultimately futile arms races might be general phenomena pertinent to co-evolution of binary systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J Mol Biol. 2008;375:720–734. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakata A, Amemura M, Makino K. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol. 1989;171:3553–3356. doi: 10.1128/jb.171.6.3553-3556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mojica FJ, Ferrer C, Juez G, Rodríguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17:85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- 35.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 36*.Sorek R, Kunin V, Hugenholtz P. CRISPR - a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. This is a must-read review for anyone interested in the newly appreciated bacterial defense system. [DOI] [PubMed] [Google Scholar]
- 37.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. This work proposed an important functional role for CRISPR as an adaptable bacterial immune system. Many predictions of this work are finding experimental confirmation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. This outstanding work directly demonstrates the role of CRISPR loci in acquisition of immunity against phage. This is likely to be a general phenomenon and in that will open a new, exciting field of research. [DOI] [PubMed] [Google Scholar]
- 40*.Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. This important work provides an essential confirmation that acquisition of foreign sequences by CRISPR loci is indeed a general phenomenon. This work reinforces the notion that CRISPR loci are part of the bacterial immune system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. This important work provides experimental clues to the molecular mechanisms of CRISPR function. In particular, the work shows that generation of genetic “memory” in Streptococcus thermophilus is accomplished by incorporating foreign sequence fragments into the existing CRISPR locus. The work also indicates that incorporation of these sequences might rely on the recognition of certain motifs present in the phage immediately outside of the incorporated sequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. This important work provides evidence that CRISPR loci are rapidly changing and adapting to local environments in natural bacterial populations. [DOI] [PubMed] [Google Scholar]
- 43*.Kunin V, He S, Warnecke F, Peterson SB, Garcia Martin H, Haynes M, Ivanova N, Blackall LL, Breitbart M, Rohwer F, McMahon KD, Hugenholtz P. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008;18:293–297. doi: 10.1101/gr.6835308. This important work analyses the variability in bacterial genome caused primarily by specifics of the local environment, and independently shows that CRISPR loci are among the most highly variable genomic regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lillestøl RK, Redder P, Garrett RA, Brügger K. A putative viral defense mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]