Abstract
Objective
To apply gene expression profiling to the study of peripheral blood mononuclear cells from patients with inflammatory myopathies, in order to provide insight into disease pathogenesis and identify potential biomarkers associated with disease activity.
Methods
We used Affymetrix whole-genome microarrays to measure the expression of ~38,500 genes in 65 blood and 15 muscle samples from 44 patients with dermatomyositis (DM), polymyositis (PM), inclusion body myositis (IBM), myasthenia gravis, or genetically determined myopathies and from 12 healthy volunteers. In 9 patients, 2 samples were obtained at different time points, when disease was either active or improving, and these paired blood samples were also compared. Bioinformatics techniques were used to identify genes with significant differential expression among diagnostic categories and in relation to disease activity. We corroborated the microarray data with quantitative real-time reverse transcriptase–polymerase chain reaction.
Results
Most patients with active DM or PM, but not patients with IBM, had significant and high up-regulation of the type I interferon-α/β (IFNα/β)–inducible genes in blood. Furthermore, the up-regulation of these genes correlated with disease activity in DM and PM, with down-regulation occurring when disease was controlled with treatment.
Conclusion
DM and PM are diseases characterized by the systemic overexpression of IFNα/β-inducible genes. The magnitude of the overexpression of these genes is higher in DM and correlates with disease activity in both disorders. Although PM and IBM have been modeled as having similar immunologic processes occurring within muscle, there are substantial differences in the expression of IFNα/β-inducible genes in blood in these diseases.
Dermatomyositis (DM) is an autoimmune inflammatory myopathy characterized clinically by subacute or chronic progressive proximal muscle weakness and characteristic skin changes. Although DM has been modeled as a disease attributable to an antibody-directed attack against endothelial antigens and resulting ischemia of muscle (1), no well-characterized pathogenic antibodies or endothelial antigens have been identified (2).
Gene expression profiling of muscle in patients with adult DM compared with that in patients with other inflammatory myopathies and normal healthy control subjects has revealed a gene transcriptional signature that is dominated by the up-regulation of interferon-α/β (IFNα/β)–inducible genes (3). Plasmacytoid dendritic cells (PDCs), which are natural IFNα/β-producing cells, are present in DM muscle. The IFNα/β-induced protein myxovirus resistance A (MxA) is expressed in perifascicular myofibers and capillaries. These observations suggest that tissue damage in DM derives from a self-destructive overactivation of the innate immune system (3).
In the current study, we performed large-scale gene expression studies using microarrays on blood samples from patients with inflammatory myopathies to determine whether distinct gene expression patterns that we previously observed in muscle (4) were present in peripheral blood mononuclear cells (PBMCs). We additionally looked at how disease activity affected the blood gene expression profile and how this changed during treatment-induced improvement in patients with inflammatory myopathies.
PATIENTS AND METHODS
Study subjects
We performed 65 microarray experiments on blood samples from a total of 56 prospectively enrolled patients, 36 of whom had inflammatory myopathies (12 with DM, 11 with polymyositis [PM], and 13 with inclusion body myositis [IBM]). For additional control groups, we studied 5 patients with myasthenia gravis (MG; a noninflammatory autoimmune myopathy), 3 patients with genetically determined myopathies (2 with myotonic dystrophy type 2 and 1 with mitochondrial myopathy), and 12 healthy volunteer subjects. Six patients with DM and 2 with PM provided blood samples for microarray experiments at 2 different time points, one when disease was active, the other when disease was improving; 1 patient with refractory DM provided 2 samples at different time points, both when disease was active. All patients met research criteria for definite or probable DM or PM (5) and definite or possible IBM (6). Patients with systemic lupus erythematosus (SLE) were excluded.
The clinical features of the patients with DM (mean age 47 years) and those with PM (mean age 56 years) are outlined in Table 1. Six patients, 2 with DM and 4 with PM, had interstitial lung disease. Of these, the 2 DM patients and 1 of the PM patients additionally had anti–histidyl–transfer RNA (anti–Jo-1) antibodies. None of the patients with IBM (6 men and 7 women, average age 69 years) was receiving immunomodulatory medication. At the time of recruitment, healthy volunteers had not had any serious illness in the last 6 months, had not started any new medications in the last 6 months, and had had no serious cold, flu, or other infection in the previous 2 months. The volunteers consisted of 5 men and 7 women and had an average age of 46 years (range 30–62 years). An internal review board approved the study. Written informed consent was obtained from all participating patients and healthy volunteers.
Table 1.
Clinical features of the 12 patients with DM and 11 patients with PM*
| Disease, patient/age/sex† | Perifascicular atrophy |
Disease duration, months |
Treatment | Treatment duration, months |
CK level, units/liter | Symptoms | MMT score, 0–150 | MMT score Δ | Meds Δ | MITAX score, 0–72‡ | Other diagnoses |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Active DM | |||||||||||
| BGE10/38/F§ | No | 108 | Pred., IVIG | 4 | 948 | ↑ | 142 | ↓ | ↑ | 12 | – |
| BGE19/61/F§ | Yes | 12 | Pred., MMF | 24 | 165 | ↑ | 139 | ↓ | ↑ | 13 | Breast cancer |
| BGE46/25/F§ | Yes | 8 | Pred. | 0.1 | 393 | ↑ | 128 | ↓ | ↑ | 12 | – |
| BGE79/46/F§ | No | 72 | None | 0 | 740 | ↑ | 133 | ↓ | ↑ | 12 | Calcinosis |
| BGE92/27/F§ | Yes | 17 | Pred. | 11 | 1,140 | ↑ | 146 | ↓ | ↑ | 13 | ILD, anti–Jo-1 |
| BGE95/53/M§ | Yes | 25 | MTX | 23 | 6,416 | ↑ | 131 | ↓ | ↑ | 13 | Diabetes |
| BGE99/21/M§ | Yes | 3 | None | 0 | 352 | ↑ | 148 | ↓ | ↑ | 12 | – |
| BGE110/54/F | Yes | 6 | Pred. | 2 | 1,140 | ↑ | 146 | ↓ | ↑ | 13 | ILD, anti–Jo-1 |
| Improving DM | |||||||||||
| BGE15/62/F | Yes | 36 | Pred., MMF | 12 | 80 | ↓ | 150 | Full¶ | ↓ | 2 | – |
| BGE17/70/F | No | 24 | Pred., IVIG, MMF | 6 | 19 | ↓ | 123 | ↑ | ↓ | 2 | – |
| BGE36/59/F | No | 12 | Pred., MMF | 1 | 1,652 | ↓ | 140 | ↓ | ↓ | 2 | – |
| BGE80/44/F | No | 1 | Pred. | 1 | 71 | ↓ | 140 | ↓ | ↓ | 2 | – |
| Active PM | |||||||||||
| BGE3/55/F | No | 26 | Pred., MMF | 20 | 2,100 | ↑ | 140 | ↓ | ↑ | – | – |
| BGE32/72/F | No | 3 | Pred., IVIG | 2 | 1,219 | ↑ | 100 | ↓ | ↑ | – | – |
| BGE47/72/F | No | 11 | Pred., MMF | 9 | 3,027 | ↑ | 118 | ↓ | ↑ | – | – |
| BGE98/38/F | No | 16 | Pred. | 16 | 129 | ↑ | 139.66 | ↓ | ↑ | – | MCTD |
| BGE106/59/F§ | No | 2 | None | 0 | 4,720 | ↑ | 128.65 | ↓ | ↑ | – | – |
| BGE119/47/F§ | No | 5 | None | 0 | 1,256 | ↑ | 135 | ↓ | ↑ | – | ILD, MCTD |
| BGE121/67/M | No | 12 | None | 0 | 1,102 | ↑ | 137 | ↓ | ↑ | – | ILD |
| Improving PM | |||||||||||
| BGE11/65/F | No | 81 | AZA, IVIG | 48 | 472 | ↓ | 149 | ↑ | ↓ | – | ILD, anti–Jo-1 |
| BGE26/71/F | No | 29 | Pred., AZA, IVIG | 29 | 29 | ↓ | 149 | ↑ | ↓ | – | ILD |
| BGE50/40/F | No | 2 | Pred., IVIG | 2 | 663 | ↓ | 138 | ↑ | ↓ | – | – |
| BGE58/30/M | No | 25 | Pred., MTX, IVIG | 25 | 1,743 | ↓ | 119 | ↑ | ↓ | – | – |
DM = dermatomyositis; PM = polymyositis; CK = creatine kinase; MMT = manual muscle testing; MITAX = Myositis Intention to Treat Activity Index; Meds = medications; Pred. = prednisone; IVIG = intravenous immunoglobulin; ↑ = increase; ↓ = decrease; MMF = mycophenolate mofetil; ILD = interstitial lung disease; MTX = methotrexate; MCTD = mixed connective tissue disease; AZA = azathioprine.
Patient numbers correspond to those shown in Figure 1.
DM patients only.
Paired blood specimens from these patients were studied, yielding a total of 32 blood samples from patients with DM or PM.
The patient’s MMT score was 150 and was therefore at full (normal) strength.
Assessment of disease activity
We classified the DM and PM patients as those with active disease and those with improving disease. Those patients who met 3 of the following 4 criteria were classified as having active disease: 1) they had increasing symptoms, 2) they had increasing objective weakness on manual muscle testing, 3) they had an elevated and (if more than 1 measurement was available) an increasing serum level of creatine kinase (CK), and 4) the treating physician increased the patient’s immunotherapy. Similar features have been previously used to define active disease in myositis (7). DM and PM patients were classified as having active or improving disease prospectively, prior to analysis of gene expression data. Manual muscle testing based on the Medical Research Council scale (8) was used to assess strength; a composite score for 30 different muscle groups was calculated, giving a maximum score of 150. We used the Myositis Intention to Treat Activity Index (MITAX) (9), as proposed by the International Myositis Assessment and Clinical Studies group, as an additional measure of disease activity. The MITAX is a multisystem assessment tool used to look at the muscle, mucocutaneous, gastrointestinal, respiratory, and musculoskeletal systems. Good interrater reliability has been reported for this measure of disease activity. In the 9 patients with paired samples, we saw an average reduction in the MITAX score of 8.5 between samples obtained during active disease and those obtained during improving disease. Scores for samples obtained during active disease ranged from 12 to 13, and samples obtained during improving disease had a score of 2.
PBMC collection, muscle tissue collection, and RNA extraction
We collected 10 ml of blood from patients and volunteers into EDTA-containing tubes (57 samples), or in some cases (8 samples) directly into PAXgene RNA tubes (Qiagen, Valencia, CA). For the EDTA-containing tubes, after centrifugation, we aspirated the plasma (upper layer) down to 1 mm from the red blood cells, and we then carefully aspirated 500 μl of buffy coat into cryostat storage tubes already filled with 1.2 ml of solution of RNAlater (Ambion, Austin, TX). We froze the combined buffy coat and RNAlater at −20°C. Using RiboPure (Ambion), RNA was extracted from the buffy coat and from PAXgene RNA tubes. The RNA concentration was measured using a spectrophotometer, and RNA quality was evaluated by running 1 μg of RNA on 1% agarose gels.
RNA was extracted as previously described (4) from muscle biopsy samples weighing 70–120 mg. Muscle biopsy tissue was obtained at the time of active disease from 15 patients (5 with DM, 5 with PM, and 5 with IBM), all of whom also had blood microarray studies performed at time points of active or improving disease, and from 5 patients without neuromuscular disease who were undergoing diagnostic biopsies. Muscle RNA extraction was done with RiboPure in a manner similar to PBMC RNA extraction. Of these 15 microarray studies of muscle with inflammatory myopathy, 9 (3 of DM muscle, 2 of PM muscle, and 4 of IBM muscle) were previously performed with portions of the data used in publication, and reanalyzed in this study, and 6 were newly performed specifically for these studies.
Target preparation, hybridization, and signal detection
Microarray studies were performed for muscle as previously described, using Affymetrix HG-U133A microarrays (Affymetrix, Santa Clara, CA) (4). PBMC samples were processed using Affymetrix HG-U133A plus 2.0 microarrays and GeneChip Operating System version 1.3.
Data processing
The Affymetrix HG-U133A plus 2.0 GeneChip has 54,675 probe sets, including 63 control probe sets. Probe set annotations (HG-U133_Plus_2 Annotations file, 3/9/2007) were obtained from the NetAffx Analysis Center (https://www.affymetrix.com/analysis/netaffx/index.affx). The expression levels were calculated using GC-Content Robust Multichip Analysis (GCRMA), which was implemented in the Bioconductor GCRMA package (available at http://www.bioconductor.org/download/oldrelease/bioc1.6/popular/gcrma.html). This algorithm produces an improved expression measurement by accounting for GC-content–based bias and optical noise behavior from all the arrays in an experiment (10). Quality control was performed by visual inspection of scanned and reconstructed images to identify gross artifacts and by careful assessment of the quality assessment parameters including control probe sets. All blood and muscle microarray data were analyzed together with GCRMA in this study.
Data analysis and visualization
The average and 90% confidence intervals (90% CIs) of fold changes were calculated for each disease group compared with the control group, and the P values for 2-group comparisons were determined by Welch t-test (Table 2). We applied stringent criteria to select genes as significantly up-regulated, requiring a P value < 0.0001 and the lower limit of 90% CIs ≥ 3.0. Genes were identified as IFNα/β induced through searches of literature (11–13) and molecular databases.
Table 2.
Evidence that IFNα/β-inducible genes are the most highly overexpressed genes in patients with active DM and patients with PM, but not in other control groups*
| P, active DM vs. normal subjects | Fold expression vs. normal subjects
|
Fold expression, improving vs. active disease
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene symbol | Gene title | Active DM (90% CI) | Active PM | IBM | MG | MYO | DM | PM | |
| IFI27 | Interferon alpha–inducible protein 27 | 7.94 × 10−6 | 130 (28–277) | 21.65 | 2.79 | 1.01 | 0.94 | −2.34 | 1.31 |
| IFI44L | Interferon-induced protein 44-like | 2.84 × 10−7 | 104 (39–194) | 64.23 | 7.01 | 0.68 | 4.09 | −9.65 | −5.16 |
| RSAD2 | Radical S-adenosyl domain/CIG5 | 2.20 × 10−7 | 66 (26–154) | 42.04 | 4.63 | 1.09 | 2.98 | −9.76 | −6.00 |
| IFI44 | Interferon-induced protein 44 | 1.61 × 10−6 | 57 (17–104) | 27.28 | 2.94 | 0.52 | 1.46 | −21.26 | −7.11 |
| LOC129607 | Hypothetical protein LOC129607 | 4.74 × 10−9 | 47 (20–114) | 29.34 | 3.48 | 1.20 | 2.51 | −16.42 | −5.68 |
| OAS1 | 2′, 5′-oligoadenylate synthetase 1 | 3.80 × 10−9 | 36 (13–84) | 15.13 | 2.05 | 0.35 | 1.04 | −18.22 | −3.90 |
| EPSTI1 | Epithelial stromal interaction 1 | 4.27 × 10−7 | 33 (15–57) | 16.24 | 3.09 | 0.73 | 3.50 | −17.93 | −2.21 |
| BIRC4BP | X1AP-associated factor 1 | 7.99 × 10−6 | 24 (12–39) | 17.38 | 4.18 | 0.78 | 1.76 | −13.25 | −3.45 |
| IFIT5 | Interferon-induced tetratricopeptide 5 | 6.81 × 10−6 | 20 (10–32) | 14.65 | 2.63 | 0.89 | 1.98 | −13.24 | −3.65 |
| OASL | 2′-5′-oligoadenylate synthetase-like | 3.23 × 10−8 | 19 (9–50) | 9.49 | 2.42 | 0.38 | 0.84 | −11.07 | −3.92 |
| OAS3 | 2′-5′-oligoadenylate synthetase 3 | 7.74 × 10−6 | 19 (8–34) | 12.68 | 1.94 | 0.60 | 1.38 | −13.18 | −5.02 |
| IFIT1 | Interferon-induced tetratricopeptide 1 | 2.87 × 10−6 | 18 (8–47) | 15.50 | 1.92 | 0.37 | 1.45 | −8.93 | −7.83 |
| PLSCR1 | Phospholipid scramblase 1 | 5.49 × 10−7 | 16 (9–25) | 14.97 | 3.57 | 5.68 | 2.69 | −4.19 | −2.35 |
| HERC5 | Hect domain and RLD 5 | 1.21 × 10−6 | 15 (5–30) | 11.62 | 1.91 | 0.49 | 1.13 | −13.58 | −4.14 |
| EIF2AK2 | Interferon-inducible protein kinase | 2.78 × 10−6 | 13 (7–24) | 12.18 | 4.59 | 1.52 | 3.16 | −6.23 | −1.76 |
| TNFSF10 | TNF superfamily, member 10 | 6.42 × 10−5 | 13 (7–21) | 10.79 | 3.58 | 2.69 | 3.26 | −5.21 | −2.56 |
| GBP1 | Guanylate binding protein 1 | 3.26 × 10−5 | 13 (6–21) | 6.73 | 3.83 | 0.54 | 3.32 | −9.79 | −1.75 |
| TNFAIP6 | TNF alpha-induced-protein 6 | 4.83 × 10−6 | 11 (6–17) | 8.05 | 1.76 | 3.06 | 1.65 | −4.44 | −2.00 |
| IFIT3 | Interferon-induced tetratricopeptide 3 | 5.46 × 10−8 | 9 (6–14) | 7.41 | 1.84 | 0.59 | 1.82 | −5.45 | −2.95 |
| SAMD9L | Sterile alpha motif domain 9-like | 1.59 × 10−5 | 9 (5–17) | 13.96 | 7.58 | 0.39 | 2.10 | −13.71 | 1.31 |
| CHMP5 | Chromatin-modifying protein 5 | 4.21 × 10−5 | 9 (5–13) | 6.42 | 1.60 | 1.90 | 1.88 | −4.21 | −2.44 |
| ISG15 | ISG15 ubiquitin-like modifier | 5.24 × 10−5 | 9 (4–15) | 4.86 | 1.04 | 0.19 | 0.55 | −5.05 | −4.62 |
| OAS2 | 2′-5′-oligoadenylate synthetase 2 | 5.94 × 10−5 | 8 (4–14) | 4.15 | 0.94 | 0.38 | 1.23 | −9.74 | −6.46 |
| IFIH1 | Interferon-induced helicase C domain 1 | 1.50 × 10−5 | 7 (4–10) | 7.40 | 1.86 | 0.50 | 1.74 | −8.09 | −2.97 |
| MX1 | Myxovirus resistance 1 | 5.21 × 10−6 | 6 (3–13) | 6.04 | 1.69 | 0.21 | 0.83 | −4.54 | −4.65 |
The 25 genes with the highest differential expression in patients with active dermatomyositis (DM) are listed in descending order of their fold expression (with 90% confidence intervals [90% CIs]) in patients with active DM compared with normal subjects. The symbols of well-established interferon-α/β (IFNα/β)–inducible genes are in boldface. The marked up-regulation in patients with active DM (n = 8) compared with expression in normal subjects (n = 12) is also seen in patients with active polymyositis (PM) (n = 7), but not in patients with active inclusion body myositis (IBM) (n = 13), myasthenia gravis (MG) (n = 5), or genetically determined myopathies (MYO) (n = 3). We applied stringent criteria to select genes as significantly up-regulated, requiring a P value of < 0.0001 and the lower limit of 90% CIs ≥ 3.0. The comparison of groups with improving and active disease shows the down-regulation of gene expression that occurs with treatment-related improvement. XIAP = X-linked inhibitor of apoptosis; TNF = tumor necrosis factor.
Group fold changes and 90% CIs were calculated comparing blood and muscle specimens from 8 patients with active DM, 11 patients with improving DM, 7 patients with active PM, 6 patients with improving PM, 13 patients with IBM, 5 patients with MG, 3 patients with genetically determined myopathies, and 12 normal subjects. Additionally, 9 patients (7 with DM, 2 with PM) with paired samples (18 samples) were analyzed pairwise for treatment-associated changes in gene signatures. Blood and muscle expression data were compared for 13,398 genes common to both HG-U133A and HG-U133A plus 2.0 microarray chips mapped according to Affymetrix probe set identifications.
Quantitative real-time reverse transcriptase–polymerase chain reaction (RT-PCR)
We performed quantitative real-time RT-PCR for 2 IFN-inducible genes, IFIT1 and Mx1 (MxA), on 18 samples (from 4 patients with active DM, 5 patients with improving DM, 4 patients with IBM, and 5 healthy volunteers) using primers designed with Primer3 software (Whitehead Institute, Cambridge, MA) and purchased commercially (Operon Biotechnologies, Huntsville, AL). Primers used were as follows: for MxA, 5′-CGGCTAACGGATAAGCAGAG-3′ (forward) and 5′-ACCTACAGCTGGCTCCTGAA-3′ (reverse); for IFIT1, 5′-AAAAGCCCACATTTGAGGTG-3′ (forward) and 5′-GAAATTCCTGAAACCGACCA-3′ (reverse).
RNA (1 μg) was reverse-transcribed to complementary DNA (cDNA) with oligo(dT)20 and Ready-to-Go reverse transcription kit from Amersham Biosciences (Piscataway, NJ). SYBR Green I–based real-time RT-PCR was carried out on an Opticon Monitor (MJ Research, Waltham, MA) with cDNA templates (1/100 of the RT reaction) using Taq polymerase (Promega, Madison, WI) and buffer (2 mM MgCl2, 400 mM dNTP [Roche, Basel, Switzerland], 0.5 × SYBR Green I, and 0.8 mM of each PCR primer [Operon Biotechnologies]) in a 25-ml final reaction volume. The samples were loaded into wells of Low Profile 96-well microplates (Abgene, Epsom, UK). After an initial denaturation step at 95°C for 5 minutes, conditions for cycling were 40 cycles of denaturation (at 95°C for 30 seconds), annealing (at 57°C for 30 seconds), and extension (at 72°C for 1 minute). The fluorescence signal was measured immediately after incubation at 79°C for 5 seconds following each extension step, eliminating possible primer dimer detection. At the end of PCRs, a melting curve was generated to confirm the specificity of the PCR product. For each run, serial dilutions of human GAPDH plasmids were used as standards for quantitative measurement of the amount of amplified cDNA. All PCRs were run in triplicate.
The comparative threshold method was used to quantify the amplified transcripts. Mean fold ratios of amplified transcripts were calculated comparing samples from patients with improving DM with samples from patients with active DM, samples from patients with active DM with samples from normal subjects, samples from patients with improving DM with samples from normal subjects, and samples from patients with IBM with samples from normal subjects.
Immunohistochemistry
Frozen muscle sections from 15 patients (5 with DM, 5 with PM, and 5 with IBM) whose muscle underwent microarray studies were stained with anti-MxA antibodies (courtesy of Dr. Otto Haller, Department of Virology, University of Freiburg, Freiburg, Germany) as previously described (3) and examined for correlation with the results of transcript studies.
RESULTS
Blood IFNα/β-inducible gene transcripts are the most up-regulated of all genes in PBMCs from patients with active DM and to a lesser extent from patients with active PM
A comparison of transcript expression levels for patients with active DM and active PM with those for healthy controls revealed that genes induced by IFNα/β had the largest fold changes and the highest statistical significance among the ~38,500 measured transcripts (P values less than 0.0001) (Table 2).
Figure 1 shows the blood IFNα/β-inducible gene expression signature in inflammatory myopathies. Of the 25 most highly up-regulated genes, at least 21 (84%) are known to be IFNα/β inducible. None of these genes were significantly up-regulated in patients with IBM, MG, or genetically determined myopathies. The magnitude of up-regulation was generally higher in DM than in PM. Quantitative RT-PCR showed that the IFN-inducible genes Mx1 and IFIT1 were highly up-regulated in blood from patients with active DM, supporting our observations from the microarray data. The average coefficient of variance of triplicate samples was 0.15, with a high correlation between triplicate runs of 0.99. Correlation of the RT-PCR data with microarray data was excellent (for Mx1, R2 = 0.9889; for IFIT1, R2 = 0.9978). Overall, 8 of 8 patients with active DM and 5 of 7 patients with active PM had levels of overexpression of IFNα/β-inducible genes that exceeded those of all other 50 blood specimens studied.
Figure 1.
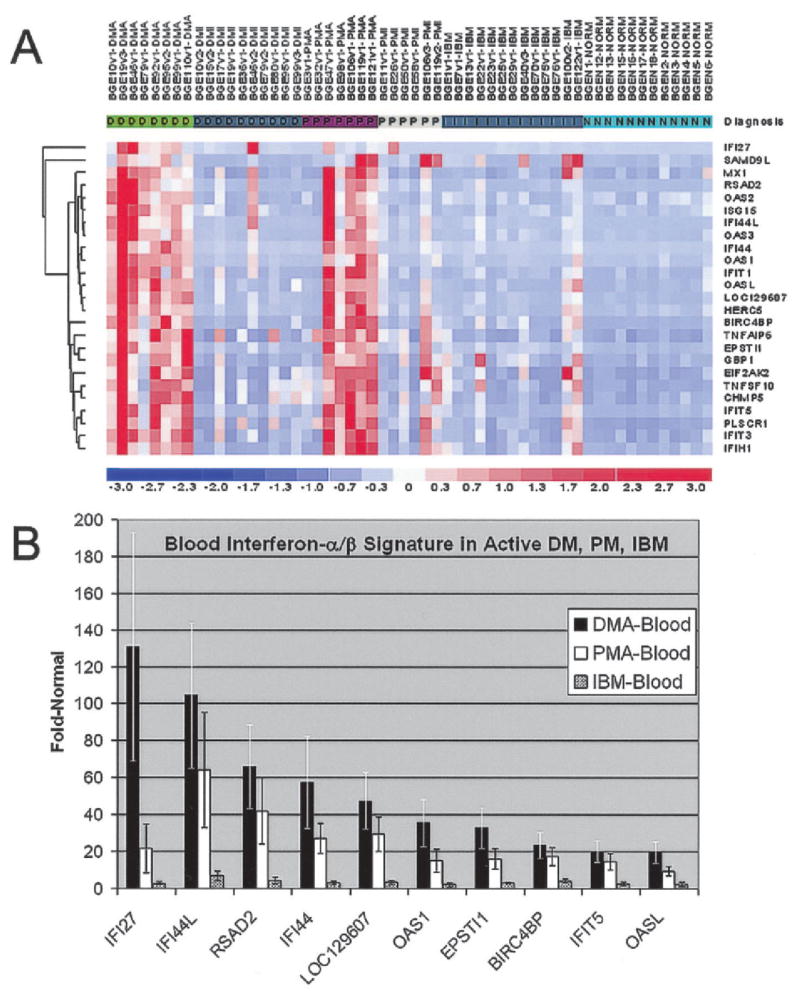
Blood interferon-α/β (IFNα/β)–inducible gene expression signature in inflammatory myopathies. A, Hierarchical clustering and visual representation of the magnitude of expression of the 25 most highly expressed genes in active dermatomyositis (DMA) compared with that in normal subjects (NORM) (see Table 2). At least 84% of these are known IFNα/β-inducible genes. Expression levels are higher in active dermatomyositis and active polymyositis (PMA) than in improving dermatomyositis (DMI), improving polymyositis (PMI), and active inclusion body myositis (IBM) compared with those in normal subjects. Values for individual patients are in columns. Values for genes are in rows. Red represents the highest expression levels. Blue represents the lowest expression levels. B, Comparison of the magnitude of the type I IFN signature for the 10 most up-regulated genes in active dermatomyositis with that in active polymyositis and active IBM. Values are the mean ± SEM for each gene.
Down-regulation of IFNα/β-inducible genes with clinical improvement in DM and PM
We compared transcript profiles of 8 samples from patients with active DM with those of 11 samples from patients with improving DM, and we separately compared transcript profiles of 7 samples from patients with active PM with those of 6 samples from patients with improving PM. The genes most highly down-regulated with improvement in disease were predominantly IFNα/β inducible (Table 2, last 2 columns at right). Quantitative RT-PCR similarly confirmed improvement in DM patients. In paired samples from the same patients with DM (n = 6) or PM (n = 2) who had active and improving disease at 2 different time points, the type I IFN–inducible genes were again the most down-regulated of all genes (Figure 2). For the 1 patient with refractory DM, little overall change for many type I IFN–inducible genes was observed in the paired specimens.
Figure 2.
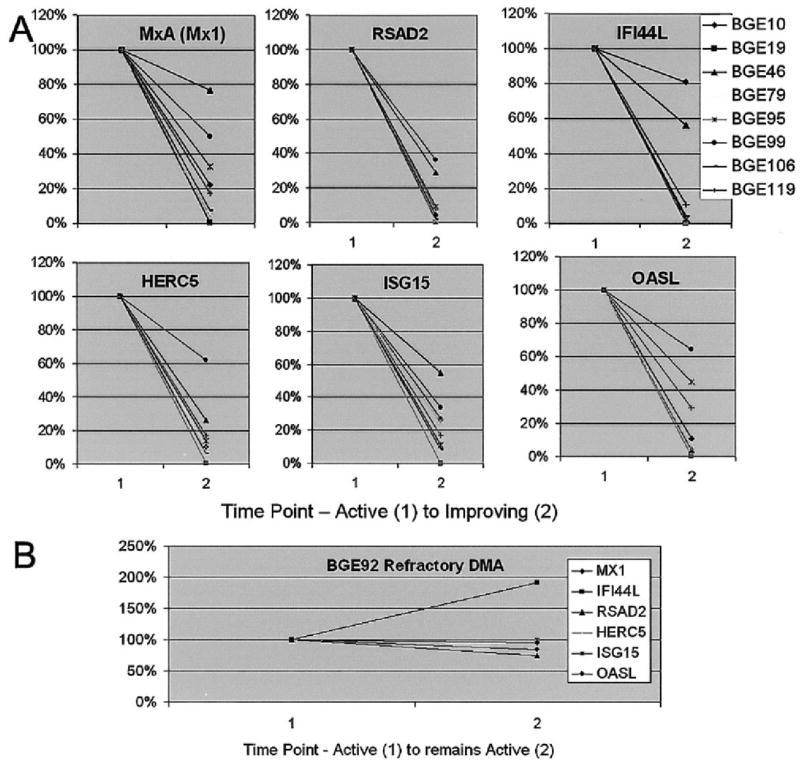
Down-regulation of 6 IFNα/β-inducible genes in 8 patients correlates with improvement in clinical disease from time point 1 (active) to time point 2 (improving). A, Gene expression studies and analysis of paired blood samples from 2 visits between which clinical improvement occurred, in 8 patients, showed marked down-regulation of multiple IFNα/β-inducible gene transcripts. Six of these genes are shown. B, In patient BGE92, with refractory active DM, expression of most of these 6 genes increased or remained unchanged in serial samples. See Figure 1 for definitions.
Up-regulation of IFNα/β-inducible genes is greater in muscle than in blood in DM, but not in PM or IBM
For 15 patients (5 each with DM, PM, or IBM), we compared the blood gene expression profiles with muscle gene expression using the 13,398 genes that are shared among both the U133A (used for muscle profiling) and U133A plus 2.0 (used for blood profiling) microarrays. In muscle of DM patients, there was marked up-regulation of the expression of the same IFN-inducible genes that we found to be highly up-regulated in blood (Figure 3). In contrast, in muscle of PM and IBM patients, only a modest increase of the IFNα/β-inducible gene transcription was present. This may have been due to infiltrating immune system cells that themselves express IFNα/β-inducible genes, such as MxA (Figure 4). Of particular interest in DM, there is a marked overexpression of certain IFN-inducible genes in muscle compared with that in blood (Figure 3). For example, expression of ISG15 in muscle of DM patients was ~570 times that in normal muscle and ~100-fold higher than that in blood of DM patients.
Figure 3.
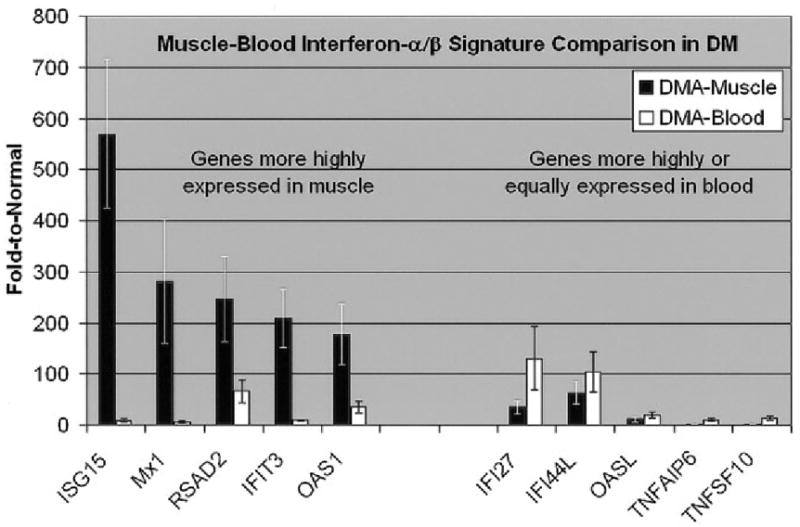
Comparison of microarray results in muscle and blood from patients with active DM. Shown are microarray data from 5 muscle samples and 8 blood samples. Values are the mean ± SEM for each gene. Although type I IFN–inducible genes are up-regulated in both blood and muscle from patients with DM, the degree of up-regulation is generally much greater in muscle than in blood and specifically greater for certain genes that accordingly may be more associated with direct mechanisms of tissue injury. Shown are fold increases for each gene transcript in muscle or blood from patients with active DM, compared with expression in muscle or blood, respectively, from normal subjects. See Figure 1 for definitions.
Figure 4.
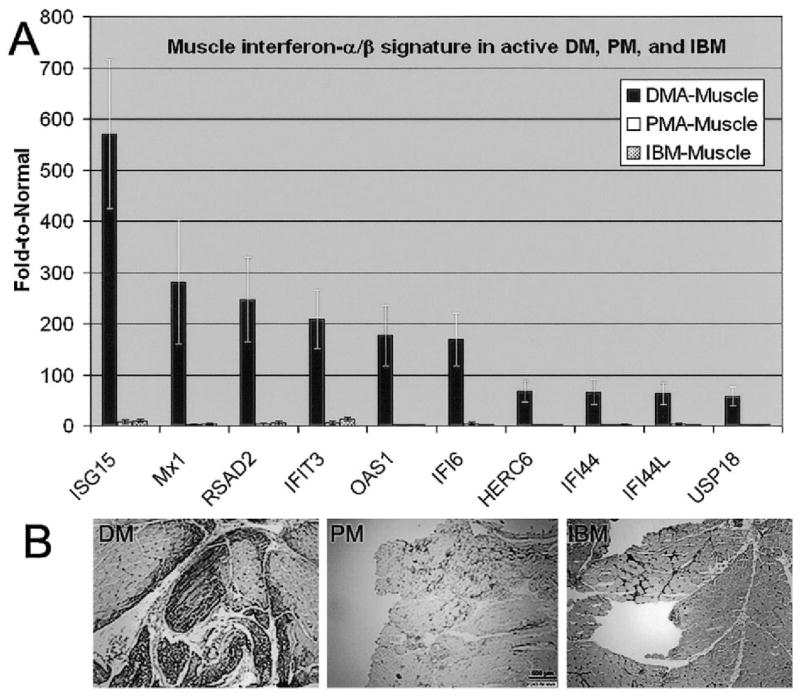
Distinct muscle expression of IFNα/β-inducible genes and protein in DM compared with that in PM and IBM. A, Muscle microarray data for 20 individuals (5 DM patients, 5 PM patients, 5 IBM patients, and 5 normal subjects). Values are the mean ± SEM for each group. The most highly differentially regulated genes in DM muscle are expressed at orders of magnitude greater than in PM and IBM. B, Immunohistochemistry for the IFNα/β-inducible protein myxovirus resistance A (MxA or Mx1) in DM, PM, and IBM. In DM, the protein is shown to be expressed by muscle fibers themselves, in contrast to PM and IBM, in which expression of MxA is limited to that by invading inflammatory cells. (Original magnification × 40.) See Figure 1 for other definitions.
Correlation of tissue pathology with up-regulation of IFNα/β-inducible MxA protein
As previously reported, 1 IFNα/β-inducible gene protein, MxA, is overexpressed in muscle in DM patients (3). In the current study, the MxA transcript level, although similarly elevated in blood from patients with active DM and in blood from patients with active PM (6.2-fold and 6.0-fold, respectively, compared with that in blood from normal subjects), was markedly higher by microarray studies in muscle from DM patients than in muscle from PM patients (281-fold and 2.5-fold, respectively, compared with that in muscle from normal subjects) (Figure 4). The marked enrichment of MxA transcript in muscle from DM patients was similarly accompanied by marked enrichment of MxA protein by immunohistochemistry in comparing muscle sections from DM and PM patients. In 4 of 5 DM patients, MxA staining was present intensely in many myofibers, particularly perifascicular myofibers, while in all 5 patients with PM and in all 5 patients with IBM, MxA staining was limited to infiltrating immune system cells (Figure 4). MxA staining is not present in normal muscle biopsy samples (3).
DISCUSSION
Our findings suggest that in most patients with DM or PM, but not in patients with IBM, there is a distinct blood gene expression profile characterized by marked overexpression of IFNα/β-inducible genes. Clinical improvement during immunosuppressive treatment is generally associated with a reduction in the overexpression of these genes toward normal levels. These findings, in relation to gene expression in muscle, have implications for hypotheses about pathogenicity and blood biomarkers of potential diagnostic use.
For DM, the IFNα/β gene signature in blood is highly correlated with the findings of microarray studies in muscle and supports the hypothesis that this disease may be driven by systemic and intramuscular overproduction of IFNα/β (3). Similar blood gene transcription signatures have been reported in SLE (11,14,15). Over-expression at the protein level for at least 1 of these genes (MxA) is present in DM muscle capillaries and perifascicular myofibers (3) and in DM skin (16,17). Additionally, PDCs, which are natural IFNα-producing cells, are abundant in DM muscle (3) and skin (17). Up-regulation of MxA transcript levels in blood have been observed in juvenile DM and may correlate with disease activity (18).
Although blood profiles exhibited similar levels of overexpression of IFNα/β-inducible genes in both DM and PM, in muscle some of these genes are more highly expressed by orders of magnitude only in DM (Figure 3). One explanation for this could be that although systemic activation of the innate immune system is present in both diseases, DM muscle is exposed to a greater amount of type I IFNs than PM muscle. This hypothesis is supported by previous findings of IFNα/β-secreting PDCs infiltrating DM muscle (3) in much greater numbers than seen in IBM and PM (19). Additionally, while in PM the expression of the IFNα/β-inducible protein MxA is confined to invading inflammatory cells, in DM MxA protein is present within myofibers (Figure 4).
The enrichment of such specific IFNα/β-inducible genes in muscle is likely an important clue to the nature of tissue injury in DM. Thus, the marked enrichment of ISG15 transcript in DM muscle suggests that of the various IFNα/β-inducible proteins up-regulated in DM blood and muscle, this particular molecule, a ubiquitin-like modifier, could be of greater relevance to the direct mechanisms of tissue injury in DM.
The distinct lack of highly up-regulated IFNα/β genes in IBM blood, compared with PM blood, contrasts with the otherwise similar nature of immunologic abnormalities that have previously been observed in muscle in these 2 diseases. These findings suggest a different magnitude of activation of the innate immune system in PM from that in IBM. Further study of this hypothesis would best be addressed in larger numbers of patients. Additionally, for many patients the diagnosis of IBM is delayed, recognized only after a previous diagnosis of glucocorticoid-resistant PM (20). Further characterization of the IFNα/β-inducible gene blood biomarkers in IBM and PM suggests the potential for future earlier diagnosis of IBM and avoidance of glucocorticoid treatment for such patients.
Our findings also suggest the utility of blood biomarkers of disease activity to supplement management of patients with DM or PM. In this study, we identified multiple blood biomarkers of active, medication-responsive myositis. Currently, there is a need for more specific tests to evaluate disease activity in DM or PM. The serum level of CK is generally reflective of disease activity in PM but may be normal in patients with active DM. The MITAX has been proposed as a clinical measure of disease activity. We calculated a MITAX score for our DM patients, which correlated well with our own assessment of disease activity. However, while the MITAX has been shown to be a good tool for disease activity assessment, intraclass correlation between assessors for muscle involvement was low, underlining the need for a more objective measure (9). An objective and inexpensive PCR-based blood test that correlates the expression of certain IFNα/β-inducible genes with disease activity could supplement the clinical management of DM and PM. Eventually, such tests might provide surrogate markers for treatment response in clinical trials.
Acknowledgments
Dr. Walsh’s work was supported by the Muscular Dystrophy Association (Career Development grant MDA3949). Dr. Beggs’ work was supported by the Muscular Dystrophy Association (grant MDA3971) and the NIH (grant AR-044345 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases). Dr. Greenberg’s work was supported by the Muscular Dystrophy Association (grant MDA3878). Microarray data were generated by the Children’s Hospital Boston Gene Expression Core, supported by the NIH (grant NS-40828).
Footnotes
Drs. Walsh and Kong contributed equally to this work.
Drs. Yao, Jallal, and Kiener own stock or stock options in MedImmune. Dr. Amato has received consulting fees (less than $10,000) from MedImmune. Dr. Greenberg has received consulting fees and speaking fees (less than $10,000) from MedImmune.
AUTHOR CONTRIBUTIONS
Dr. Greenberg had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study design. Walsh, Pinkus, Beggs, Amato, Greenberg.
Acquisition of data. Walsh, Pinkus, Amato, Greenberg.
Analysis and interpretation of data. Walsh, Kong, Yao, Jallal, Kiener, Pinkus, Beggs, Amato, Greenberg.
Manuscript preparation. Walsh, Yao, Jallal, Kiener, Pinkus, Beggs, Amato, Greenberg.
Statistical analysis. Walsh, Kong, Greenberg.
References
- 1.Dalakas MC, Hohlfeld R. Polymyositis and dermatomyositis. Lancet. 2003;362:971–82. doi: 10.1016/S0140-6736(03)14368-1. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg SA, Amato AA. Uncertainties in the pathogenesis of adult dermatomyositis. Curr Opin Neurol. 2004;17:359–64. doi: 10.1097/00019052-200406000-00018. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R, et al. Interferon-α/β-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57:664–78. doi: 10.1002/ana.20464. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg SA, Sanoudou D, Haslett JN, Kohane IS, Kunkel LM, Beggs AH, et al. Molecular profiles of inflammatory myopathies. Neurology. 2002;59:1170–82. doi: 10.1212/wnl.59.8.1170. [DOI] [PubMed] [Google Scholar]
- 5.Hoogendijk JE, Amato AA, Lecky BR, Choy EH, Lundberg IE, Rose MR, et al. 119th ENMC international workshop: trial design in adult idiopathic inflammatory myopathies, with the exception of inclusion body myositis, 10–12 October 2003, Naarden, The Netherlands. Neuromuscul Disord. 2004;14:337–45. doi: 10.1016/j.nmd.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Griggs RC, Askanas V, DiMauro S, Engel A, Karpati G, Mendell JR, et al. Inclusion body myositis and myopathies. Ann Neurol. 1995;38:705–13. doi: 10.1002/ana.410380504. [DOI] [PubMed] [Google Scholar]
- 7.Nagy G, Pallinger E, Antal-Szalmas P, Aleksza M, Marschalko M, Brozik M, et al. Measurement of intracellular interferon-γ and interleukin-4 in whole blood T lymphocytes from patients with systemic lupus erythematosus. Immunol Lett. 2000;74:207–10. doi: 10.1016/s0165-2478(00)00265-0. [DOI] [PubMed] [Google Scholar]
- 8.Griggs RC, Moxley RT, 3rd, Mendell JR, Fenichel GM, Brooke MH, Pestronk A, et al. for the Clinical Investigation of Duchenne Dystrophy Group. Prednisone in Duchenne dystrophy: a randomized, controlled trial defining the time course and dose response. Arch Neurol. 1991;48:383–8. doi: 10.1001/archneur.1991.00530160047012. [DOI] [PubMed] [Google Scholar]
- 9.Isenberg DA, Allen E, Farewell V, Ehrenstein MR, Hanna MG, Lundberg IE, et al. International consensus outcome measures for patients with idiopathic inflammatory myopathies: development and initial validation of myositis activity and damage indices in patients with adult onset disease. Rheumatology (Oxford) 2004;43:49–54. doi: 10.1093/rheumatology/keg427. [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12:882–93. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- 11.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–5. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–20. [PubMed] [Google Scholar]
- 13.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–8. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–23. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han GM, Chen SL, Shen N, Ye S, Bao CD, Gu YY. Analysis of gene expression profiles in human systemic lupus erythematosus using oligonucleotide microarray. Genes Immun. 2003;4:177–86. doi: 10.1038/sj.gene.6363966. [DOI] [PubMed] [Google Scholar]
- 16.Wenzel J, Scheler M, Bieber T, Tuting T. Evidence for a role of type I interferons in the pathogenesis of dermatomyositis. Br J Dermatol. 2005;153:462–3. doi: 10.1111/j.1365-2133.2005.06786.x. [DOI] [PubMed] [Google Scholar]
- 17.Wenzel J, Schmidt R, Proelss J, Zahn S, Bieber T, Tuting T. Type I interferon-associated skin recruitment of CXCR3+ lymphocytes in dermatomyositis. Clin Exp Dermatol. 2006;31:576–82. doi: 10.1111/j.1365-2230.2006.02150.x. [DOI] [PubMed] [Google Scholar]
- 18.O’Connor KA, Abbott KA, Sabin B, Kuroda M, Pachman LM. MxA gene expression in juvenile dermatomyositis peripheral blood mononuclear cells: association with muscle involvement. Clin Immunol. 2006;120:319–25. doi: 10.1016/j.clim.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg SA, Pinkus GS, Amato AA, Pinkus JL. Myeloid dendritic cells in inclusion-body myositis and polymyositis. Muscle Nerve. 2007;35:17–23. doi: 10.1002/mus.20649. [DOI] [PubMed] [Google Scholar]
- 20.Amato AA, Gronseth GS, Jackson CE, Wolfe GI, Katz JS, Bryan WW, et al. Inclusion body myositis: clinical and pathological boundaries. Ann Neurol. 1996;40:581–6. doi: 10.1002/ana.410400407. [DOI] [PubMed] [Google Scholar]


