Abstract
Eighteen histone deacetylases (HDACs) are present in humans, categorized into two groups: zinc-dependent enzymes (HDAC1–11) and NAD+-dependent enzymes (sirtuins 1–7). Among zinc-dependent HDACs, HDAC6 is unique. It has a cytoplasmic localization, two catalytic sites, a ubiquitin-binding site, and it selectively deacetylases α-tubulin and Hsp90. Here, we report the discovery that the redox regulatory proteins, peroxiredoxin (Prx) I and Prx II are specific targets of HDAC6. Prx are antioxidants enzymes whose main function is H2O2 reduction. Prx are elevated in many cancers and neurodegenerative diseases. The acetylated form of Prx accumulates in the absence of an active HDAC6. Acetylation of Prx increases its reducing activity, its resistance to superoxidation, and its resistance to transition to high-molecular-mass complexes. Thus, HDAC6 and Prx are targets for modulating intracellular redox status in therapeutic strategies for disorders as disparate as cancers and neurodegenerative diseases.
Keywords: acetylation, hydrogen peroxide, histone deacetylase inhibitors
Eighteen histone deacetylases (HDACs) have been identified in humans and classified based on homologies to yeast HDACs (1, 2). Class I (HDACs 1, 2, 3, and 8), class II (HDACs 4, 5, 7, and 9), class IIB (HDACs 6 and 10), and class IV (HDAC11) are zinc-dependent deacetylases. Class III HDACs (sirtuins 1–7) are not zinc-dependent deacetylases and have an absolute requirement for NAD+ for their activity (3).
HDAC6 is unique among the zinc-dependent HDACs (4–12). It has a primary cytoplasmic localization, full duplication of its two catalytic sites, and a ubiquitin-binding domain at the C terminus. Inhibition of HDAC6 activity by the specific inhibitor, tubacin, or its down-regulation by siRNA, can increase accumulation of acetylated α-tubulin (6, 7) and can alter cellular mobility and can increase acetylated Hsp90 (8–10), inducing client protein degradation. The ubiquitin-binding activity of HDAC6 mediates the recruitment of autophagic material to aggresomes, decreasing the cytotoxic effects of these aggregates (11, 12). Thus, HDAC6 functions in various cellular processes that are dependent and independent of its catalytic activity and affects cell growth, migration, and cell death.
In this work, we made the discovery that HDAC6 has an important role in redox regulation and cellular stress response. We found that the redox regulatory proteins peroxiredoxin I (Prx I) and II (Prx II) are specific targets of HDAC6 deacetylase. Acetylated Prx I and Prx II accumulate in cells lacking HDAC6 deacetylase activity. We found that the prostate cancer cell (LAPC4) does not express HDAC6 protein. This finding was confirmed by siRNA knockdown of HDAC6 and by the specific inhibition of HDAC6 with tubacin (13) in a cell line, which expresses HDAC6 protein (LNCaP). Prx I and Prx II are highly homologous 2-cysteine members of the Prx protein family that function as antioxidants at low resting H2O2 levels (14, 15). At higher levels of H2O2, the cysteine residue can be oxidized to sulfonic acid, with transformation of these proteins to high-molecular-mass protein complexes. Prx I and Prx II are reported to be elevated in many cancers and in various neurodegenerative disorders (16–23). In cancer cells, Prx I and Prx II can confer resistance to chemotherapy and radiation therapy (15–20). Stress-related cellular dysfunction caused by reactive oxygen species (ROS) appears to be involved in the development of various neurodegenerative diseases (22, 23).
We found that acetylation of Prx I and Prx II proteins increases their activity in reducing H2O2 and increases their relative resistance to superoxidation and to transition to high-molecular-mass complexes. At high levels of H2O2, Prx are transformed to large-molecular-mass complexes (14, 15). HDAC6 and its specific targets Prx I and Prx II are shown to play an important role in modulating response to H2O2-induced cellular stress. This understanding of the specific deacetylase function of HDAC6 with Prx I and Prx II suggests that manipulating HDAC6 activity and, in turn, the redox activities of the 2-Cys Prx proteins, Prx I and Prx II, have an important potential in therapeutic strategies for cancers, neurodegenerative diseases, and other disorders that may involve cellular apoptosis.
Results
α-Tubulin and 50- to 22-kDa Proteins are HDAC6 Targets.
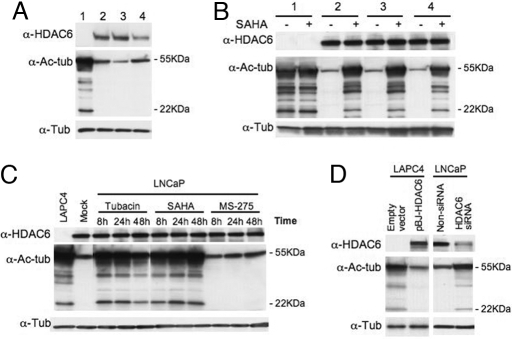
In studies (24) that determined the expression of the zinc-dependent HDACs in transformed and normal cells, we found that a human prostate cancer cell, LAPC4, did not express HDAC6 protein (Fig. 1A, lane 1). All other zinc-dependent HDACs of class I, II, and IV were expressed in both the normal and transformed cells examined (24).
Fig. 1.
Lack of inhibition of HDAC6 proteins induces accumulation of aetylated 50- to 22-kDa proteins. (A) Western blot analysis of lysates of prostate cancer cells: lane 1, LAPC4; lane 2, Du145; lane 3, LNCaP; lane 4, PC3. α-HDAC6, HDAC6; α-Ac-Tub, acetylated α-tubulin; α-Tub, α-tubulin as loading control. (B) Western blot analysis of lysates of prostate cell lines as in A were cultured without (−) or with (+) 5 μM SAHA for 24 h. (C) Western blot analysis of LNCaP cells cultured with tubacin, SAHA, or MS-275 (5 μM each) for the times indicated. (D) Western blot analysis of LAPC4 cells transfected with HDAC6 gene (Left) or LNCaP cells transfected with HDAC6 siRNA oligonucleotides (Right).
To determine the basis of LAPC4 lack of expression of HDAC6 protein, Northern blot analyses were performed on lysates of these cells. No transcript of this protein was detected (data not shown). By using RT-PCR, HDAC6 transcript was detectable with two pairs of primers designed on the 3′ origin [supporting information (SI) Fig. S1A]. Using primers covering the entire transcript, we could not amplify the 5′ end of the transcript corresponding to the CDS1 and CDS2 fragments compared with LNCaP cells (Fig. S1A). This finding is consistent with expression of a truncated HDAC6 transcript that could not be translated into a HDAC6 protein in LAPC4 cells.
Using an anti-acetylated α-tubulin antibody, we found that LAPC4 cells (Fig. 1A, lane 1) accumulated acetylated α-tubulin and other acetylated proteins, ranging in size from ≈50 to 22 kDa. In three human prostate cancer cell lines that express HDAC6, Du145, LNCaP, and PC3, there was a low level of acetylated α-tubulin but no detectable acetylated proteins corresponding to 50–22 kDa (Fig. 1A). Noteworthy, SIRT2, an α-tubulin deacetylase (25), is present in LAPC4 cells but apparently does not deacetylate the 50- to 22-kDa proteins (data not shown).
We next determined whether these acetylated proteins were deacetylase targets of HDACs by using suberoylanilide hydroxamic acid (SAHA, vorinostat), a paninhibitor of zinc-dependent HDACs (26). LAPC4, Du145, LNCaP, and PC3 were cultured with or without SAHA. Lysates of these prostate cancer cells cultured with the HDAC inhibitor showed a similar pattern of acetylated proteins present in LAPC4 cells (Fig. 1B). Thus, inhibition of HDACs, including HDAC6, is associated with the accumulation of acetylated proteins. Cells cultured without SAHA had low levels of acetylated α-tubulin and no detectable acetylated proteins corresponding to 50–22 kDa.
The following experiments were performed to determine whether the 50- to 22-kDa proteins are specific substrates of HDAC6. First, LNCaP cells were cultured with either SAHA, with tubacin, a specific HDAC6 inhibitor (13), or with MS-275, a HDACi that inhibits class I HDACs but not HDAC6 (27). LNCaP cells cultured with SAHA or tubacin, but not cells cultured with MS-275, accumulated acetylated α-tubulin and the 50- to 22-kDa proteins (Fig. 1C). LNCaP cells cultured with SAHA or MS-275 but not cells cultured with tubacin-accumulated acetylated histones (data not shown). Second, LAPC4 cells transfected with HDAC6 gene, inducing expression of HDAC6 protein, had low levels of acetylated α-tubulin and no detectable 50- to 22-kDa acetylated proteins (Fig. 1D). Third, LNCaP cells transfected with HDAC6 siRNA, resulting in decreased expression of HDAC6 protein, accumulated acetylated 50- to 22-kDa proteins (Fig. 1D). These findings are consistent with the 50- to 22-kDa proteins being specific HDAC6 nonhistone protein substrates.
HDAC6 has two catalytic domains, and the second domain has been generally reported to be dominant for the deacetylation of α-tubulin (7, 13, 28). A mutant HDAC6 with an inactive second tubulin catalytic domain (H611A) was transfected into LAPC4 cells. In these transfected cells, the expression of acetylated α-tubulin and the 50- to 22-kDa proteins was similar to that of nontransfected LAPC4 cells lacking HDAC6 proteins (data not shown). This finding suggests that the second catalytic domain of HDAC6 is required for the deacetylation of the 50- to 22-kDa proteins.
Identification of 22-kDa Proteins: Prx I and Prx II.
To identify the 50- to 22-kDa proteins, acetylated proteins were immunoprecipitated from cell lysates prepared from LAPC4 cells by using an anti-acetylated tubulin antibody. The immunoprecipitate was subjected to SDS/PAGE and stained with Coomassie blue. The band corresponding to the 22-kDa protein was recovered and analyzed by mass spectrometry (MS/MS) (29). Two proteins were found, Prx I and Prx II, and we confirmed their presence in the immunoprecipitate from LAPC4 lysates by Western blotting using specific antibodies (anti-Prx I and anti-Prx II) (Fig. 2A). In cells cultured up to 48 h with SAHA or MS-275, the total expression of Prx I or Prx II was unchanged (data not shown).
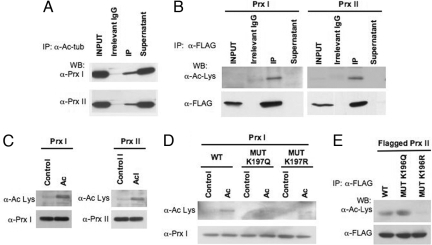
Fig. 2.
Identification of acetylated peroxiredoxins. (A) Detection of Prx I and Prx II in LAPC4 immunoprecipitate (IP) generated with anti-acetylated tubulin (α-Ac-tub) antibody. (B) Flagged Prx I and Prx II overexpressed in LAPC4 cells were immunoprecipitated with FLAG antibody and analyzed by Western blotting (WB) with the anti-acetylated lysine antibody (α-Ac-Lys). (C) Prx I and Prx II recombinant proteins were acetylated in vitro, fractionated in SDS gel, and probed with anti-acetylated lysine antibody. (D) Recombinant Prx I wild type (WT) and mutants (K197Q and K197R) were acetylated in vitro. (E) Flagged Prx II proteins overexpressed in LAPC4 cells were immunoprecipitated and probed with anti-acetylated lysine antibody.
To confirm that the 22-kDa proteins were acetylated Prx I and Prx II, their corresponding coding regions were cloned into a FLAG plasmid and overexpressed ectopically in LAPC4 cells. FLAG-immunoprecipitated proteins from lysates of transfected LAPC4 cells were analyzed by Western blotting with an anti-acetylated lysine antibody. The anti-acetylated lysine antibody specifically recognized Prx I and Prx II (Fig. 2B).
Using recombinant Prx proteins, acetylated Prx I and Prx II were prepared by reacting them with histone acetyltransferase (HAT) and acetylcoenzyme A (acetyl-CoA). The acetylation of Prx I and Prx II was confirmed by Western blotting, using anti-acetylated lysine antibody (Fig. 2C).
The intact positive charge of Lys191 in the C terminus of yeast Prx is important for the reducing activity and resistance of the protein to superoxidation by H2O2 (15, 30, 31). We next tested whether the lysine residues in human Prx (Lys197 in Prx I; Lys196 in Prx II), corresponding to Lys191 of yeast Prx, were acetylation sites. Mutants of recombinant Prx I were generated: K197Q (Gln is substituted for Lys and mimics acetylated lysine) and K197R (Arg is substituted for Lys and is a nonacetylated mimic) and reacted with HAT and acetyl-CoA (Fig. 2D). A weak signal was detected in Prx mutant K197Q and in K197R, compared with wild-type Prx I. These findings indicate that the Prx I Lys197 is an acetylation site. We also generated Prx II mutants (K196Q and K196R) cloned in a FLAG plasmid and transfected them into LAPC4 cells. The immunoprecipitates were analyzed by Western blotting using an anti-acetylated lysine antibody. The wild-type Prx II and the K196Q mutant presented strong positive signals, whereas the K196R mutant presented no detectable signal (Fig. 2E). These results confirmed that Prx II Lys196 is also a site of acetylation. The present results do not rule out that there may be other lysines in each protein that are acetylated.
Acetylation of Prx Increases Reducing Activity.
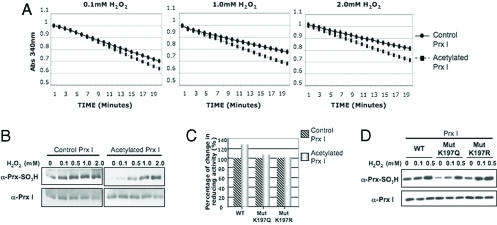
We next evaluated the effect of acetylation of Prx I on its H2O2-reducing activity, using the peroxidase reaction with the thioredoxin (Trx) system as an electron donor, Trx reductase, and NADPH (32–34). The activity of recombinant Prx I protein in reducing H2O2 was assayed by measuring NADPH oxidation (absorbance at 340 nm) (30, 34). The in vitro acetylated Prx I was more active than the nonacetylated Prx I in reducing H2O2 over a range of 0.1–2.0 mM (Fig. 3A).
Fig. 3.
Acetylation of Prx increases its reducing activity and resistance to overoxidation. (A) Activity of recombinant Prx I, control (●), and acetylated forms (■) was assayed for H2O2 reduction by measuring NADPH oxidation (A340 nm) (21). (B) Acetylated Prx I is more resistant to being overoxidized (anti-Prx-SO3H antibody). (C) H2O2-reducing activity of acetylated recombinant Prx I, WT, and mutants (K197Q and K197R). The percentage change in 340 mM, O.D., per minute, per amount of protein in 20 min is shown. Activity of the nonacetylated protein in 0.1 mM H2O2 was taken as 100%. (D) Western blot analysis of oxidized Prx I after 1-h exposure to H2O2 in a peroxidase reaction in WT, MUTK197Q, and MUTK197R.
Prx I and Prx II both regulate intracellular H2O2 levels while at the same time are regulated by H2O2. Under normal redox conditions, Prx I and Prx II scavenge H2O2 by building disulfide bridges that can be reduced via the thioredoxin system (30, 32–36). At high levels of H2O2, Prx-reducing activity is lost through overoxidation of the active sulfhydryl site to sulfinic or sulfonic acid (33–35). Upon overoxidation, Prx undergoes a transition to a high-molecular-mass complex (14). Using an anti-Prx-SO3H antibody that specifically recognizes both sulfinic and sulfonic forms of overoxidized cysteine in Prx, we found that the acetylated Prx I was more resistant to inactivation by H2O2 than the nonacetylated form of the protein (Fig. 3B).
To test whether acetylation of Lys197 was critical in determining of Prx H2O2-reducing activity, the effect of in vitro acetylation on the reducing activity of mutant Prx I proteins, K197Q and K197R, was assayed. Acetylation of the mutant proteins did not increase their reducing activity as it did in the wild-type protein (Fig. 3C). The acetylated mimic mutant (K197Q) had a lower level of oxidized protein (assayed by reaction with anti-Prx-SO3H antibody) compared with the wild-type and K197R mutant proteins (Fig. 3D). These findings indicate that acetylation of Lys197 is involved in determining Prx-reducing activity and protection against overoxidation.
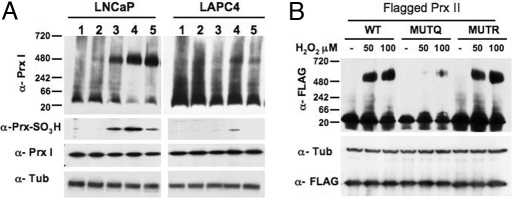
Overoxidation of Prx I or Prx II causes the loss of H2O2-reducing activity and the transition to high-molecular-mass complex protein (14, 15). LNCaP cells exposed to concentrations of 50 and 100 μM H2O2 for 20 min accumulate high-molecular-mass complexes of ≈480 kDa (Fig. 4A). By comparison, in LAPC4 cells, which have acetylated Prx, the high-molecular-mass complexes accumulate only when they were exposed to 100 μM H2O2, but not 50 μM H2O2 (Fig. 4A). Similar results were found for Prx II (data not shown). The level of overoxidized Prx (anti-Prx-SO3H) was lower in LAPC4 cells compared with LNCaP cells (Fig. 4A), which is consistent with acetylated Prx in LAPC4 cells being more active in H2O2-reducing activity and being more resistant to overoxidation and transition to the high-molecular-mass complexes.
Fig. 4.
High-molecular-mass complexes in LAPC4 and LNCaP cells. (A) Native gels showing Prx I high-molecular-mass complexes induced by exposure to H2O2 for 20 min in LNCaP and LAPC4 cells. Concentration of H2O2 (lanes): 1, none; 2, 25 μM; 3, 50 μM; 4, 100 μM; 5, 100 μM + fluid change and recovery for 1 h. (Lower) Denaturing gels showing the levels of oxidized Prx (α-Prx-SO3H), Prx I (α-Prx I), and α-tubulin (α-Tub) in the LNCaP and LAPC4 lysates. (B) Flagged Prx II high-molecular-mass complexes in LNCaP cells treated with the indicated concentrations of H2O2; WT, and transfected with MUTQ (K196Q) or MUTR (K196R) are shown (see Results).
To confirm that the lower level of oxidized Prx in LAPC4 H2O2-treated cells was associated with Prx acetylation, Prx II-flagged proteins (wild type and mutants) were overexpressed in LNCaP cells and then treated with H2O2. The Prx II K196Q acetylated mimic formed high-molecular-mass complexes only at 100 μM H2O2 whereas in the wild type and K196R mutant the complexes formed at 50 and 100 μM H2O2 (Fig. 4B). Acetylation of Prx is a major determinant in its protection from overoxidation.
Sensitivity to H2O2-Induced Cell Death of LAPC4, LNCaP, and Normal Human Foreskin (HFS) Cells.
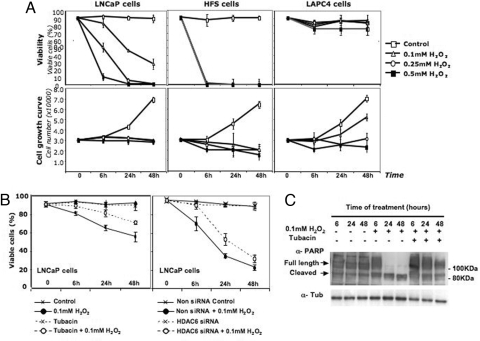
ROS are generated in cells in response to several types of environmental stress that lead to apoptosis and cell death (37). Prx I and Prx II play a role in modulating cellular response to ROS (30, 31). LAPC4 cells, in which there is an accumulation of acetylated Prx I, H2O2 concentrations (0.5 mM) that induce 100% of LNCaP and HFS cells to undergo apoptosis and cell death (Fig. 5A) induces <10% LAPC4 cell death. LNCaP (Fig. 1A) and HFS cells (data not shown) lack detectable acetylated Prx (22-kDa) proteins. When LAPC4 cells were transiently transfected with HDAC6 gene and expressed HDAC6 protein (Fig. 1D), H2O2 (0.5 mM) induced >40% of the cells to undergo cell death (data not shown). When HDAC6 was inhibited in LNCaP cells by tubacin or transient transfection with HDAC6 siRNA, acetylated Prx accumulated (Fig. 1 C and D). LNCaP cells were significantly more resistant to H2O2-induced cell death when HDAC6 activity was inhibited by tubacin (at 24 h, P = 0.004) (Fig. 5C Upper) or by HDAC6 siRNA compared with control cells (at 24 h, P = 0.014) (Fig. 5C Lower). The loss of viability was greater in cultures of cells transfected with siRNA than cells exposed to tubacin, which may reflect the manipulation required for siRNA transfection. In both studies, inhibiting HDAC6 was associated with significant increased resistance to H2O2.
Fig. 5.
HDAC6 inhibition reduces sensitivity to H2O2-induced cell death. (A) Cell growth (Lower) and viability (Upper) of LNCaP, HFS, and LAPC4 cells cultured with H2O2 at the concentrations and times indicated. (B) (Left) LNCaP cells were precultured with 8 μM tubacin for 4 h and then exposed to H2O2 for times indicated. (Right) LNCaP cells were transfected with non-silencing RNA (non-siRNA) or HDAC6 siRNA 48 h before H2O2 treatment for times indicated. (C) Analysis of PARP degradation in lysates of LNCaP cells: no additions, treated with 0.1 mM H2O2, and precultured with 8 μM tubacin, and exposed to 0.1 mM H2O2 for the times indicated.
Poly(ADP-ribose) polymerase (PARP) degradation is a marker of cellular apoptosis (37). PARP was assayed in lysates of LNCaP cells cultured with and without tubacin for 4 h and then exposed to 0.1 mM H2O2 for 48 h. Cells cultured with tubacin plus H2O2 had nondegraded PARP (Fig. 5D), similar to the pattern observed in cells not exposed to H2O2. LNCaP cells exposed to H2O2 without prior culture with tubacin had degraded PARP (Fig. 5D). Thus, inhibition of HDAC6 activity is associated with resistance to H2O2-induced apoptotic cell death.
Discussion
This work describes the discovery of a function for HDAC6, namely the specific deacetylation of redox regulatory proteins Prx I and Prx II. Previous studies established that HDAC6 is the deacetylase for α-tubulin, cortactin, an actin binding protein and the chaperone protein Hsp90 (6–10). In addition to its deacetylase activity, HDAC6 has a ubiquitin binding site at its C terminus that can play a role in facilitating autophagic degradation of potentially noxious proteins (11, 12).
The present discovery that the redox regulatory proteins Prx I and Prx II, whose main function is cellular protection from free radical accumulation, are specific substrates of HDAC6 adds an important understanding of the functions of this deacetylase that has implications for diseases as disparate as cancers and neurodegenerative disorders.
The human prostate cancer cell LAPC4 lacks HDAC6 protein and accumulates the previously unrecognized substrates of HDCA6, acetylated Prx I and Prx II. Inhibition of HDAC6 with tubacin or its down-regulation with siRNA in both transformed cells (LNCaP, PC3, Du145) as well as normal cells (HFS) was associated with an accumulation of acetylated and Prx I and Prx II.
Prx I are elevated in many cancers including esophageal, pancreatic, melanoma, thyroid, and lung cancers (16–20). Elevated levels of Prx I and Prx II are associated with resistance to cancer therapy and promote aggressive survival phenotypes of cancer cells.
Prx have also been reported to be overexpressed and/or aberrantly expressed in several neurodegenerating disorders, including Alzheimer's, Pick's disease, and others associated with progressive aggregate formation (22, 23). The present work found that the acetylated form of Prx is more active in reducing H2O2 than the nonacetylated form. Thus, inhibition of HDAC6 deacetylase activity with a consequent accumulation of acetylated Prx could lead to a beneficial increase in antioxidant activity in neurodegenerative disorders. HDAC inhibitors such as SAHA, trichostatin A, and sodium butyrate have been shown to ameliorate disease progression in rodent models of Huntington's disease (38), spinal and bulbar muscular atrophy (39), amyotrophic lateral sclerosis (40), and Parkinson's disease (41). Although the mechanisms of the beneficial effects of the HDAC inhibitors in these neurodegenerative diseases are not known, inhibition of HDAC6 with consequent increase in Prx reducing activity may, in part, explain these effects.
However, in cancer cells, the increased reducing activity of Prx associated with inhibition of HDAC6 could contribute to resistance to therapy. This hypothesis would suggest that as part of an anticancer therapeutic regimen, inactivating Prx activity might be beneficial (42–45).
In summary, the discovery that HDAC6 is a specific deacetylase for the redox regulatory proteins Prx I and Prx II whose activity is regulated, in part, by acetylation, suggests that the activity of this deacetylase and of the redox proteins can be useful targets for therapeutic strategies in these disparate disorders. The redox proteins, Prx, have a role in both the resistance of certain cancers to therapy and quite a different role in possibly slowing the progression of neurodegenerative diseases. Developing inhibitors of Prx acetylation may be a useful therapeutic strategy for cancers. However, an agent such as a specific HDAC6 inhibitor that enhances the accumulation of acetylated Prx and protects these proteins from overoxidation may be useful in treating neurodegenerative disorders that involve apoptotic cell death.
Methods
Cell Lines, Reagents, and Antibodies.
LNCaP, Du145, PC3, and HFS were obtained from American Type Culture Collection. LAPC4 cells were kindly provided by Charles Sawyers (Memorial Sloan–Kettering Cancer Center). Antibodies used were: anti-acetylated tubulin (Sigma), anti-HDAC6 (Santa Cruz Biotechnology), anti-tubulin, (Calbiochem), anti-acetylated lysine (Cell Signaling), anti-Prx I and Prx II (Upstate), anti-Prx-SO3H (Abcam), anti-PARP (BD Biosciences). MS-275 was obtained from Calbiochem. Tubacin and SAHA were kindly provided by Stuart Schreiber (Harvard University, Cambridge, MA) and Ronald Breslow (Columbia University, New York), respectively.
Western Blotting and Immunoprecipitation.
Western blotting was performed as published in ref. 24. Immunoprecipitation was performed either by using the anti-acetylated tubulin antibody, which was covalently bound to CNBr-activated Sepharose 4B beads according to the manufacturer's instructions (Amersham), or by using anti-FLAG beads (Sigma). Cell lysate and beads were incubated overnight at 4°C. Bound proteins were eluted by competition with 100 μg/ml specific peptide. For protein identification, different elution fractions were fractionated in SDS/polyacrylamide gels. Gels were stained with Coomassie blue R-250, and visible bands were cut and submitted to mass spectrometry analyses (29).
Transfections.
HDAC6 vectors were kindly provided by Stuart Schreiber. Prx I and Prx II coding region sequences were amplified by using cDNA from LAPC4 cells. The primers used for RT-PCR were: Prx I forward (5′-ggaagcttatgtcttcaggaaatg-3′), Prx I reverse (5′-ccgaattctcacttctgcttggag-3′), Prx II forward (5′-ggaagcttatggcctccggtaacg-3′) and Prx II reverse (5′-ccgaattcctaattgtgtttggag-3′). They were cloned into the pFLAG-CMV-4 vector (Sigma). Prx mutants were generated by using the QuikChange II site-directed mutagenesis kit (Stratagene). HDAC6 siRNA and nonsilencing oligonucleotides were obtained from Qiagen. Cells were transfected by using the Nucleofactor kit (Amaxa), following the manufacturer's instructions.
In Vitro Acetylation.
In vitro acetylation of Prx was performed by using 10 μg of Prx recombinant protein incubated at 30°C for 20 min with 2 μg of p300 HAT domain (Upstate) and 1.2 mM acetyl-CoA (Sigma) in a buffer containing 50 mM Tris·HCl (pH 8.0), 10% glycerol, 0.1 mM EDTA, and 3 mM DTT, in a 45-μl final volume.
Peroxiredoxin Activity.
Peroxidase activity was measured by monitoring the oxidation of NADPH by the decrease of absorbance at 340 nm (21). The reaction was carried out by using 50 mM Hepes-NaOH (pH 7.4), 3 μg of Trx reductase (Sigma), 4 μg of Trx (Sigma), 0.5 mM NADPH (Sigma), 5 μg of Prx I (BPS Bioscience), and different concentrations of H2O2 in a 150-μl final volume.
H2O2 Treatment and Prx High-Molecular-Mass Complex Detection.
Cells were seeded in 24-well plates at 5 × 104 cells per well and treated the day after, as follows. Chemical inhibition of HDAC6 activity was done by pretreatment with 8 μM tubacin for 4 h before H2O2 treatment. In the HDAC6 down-regulation experiment, cells were transiently transfected with HDAC6 siRNA or non-siRNA oligonucleotides and allowed to recover for 2 days and then treated with H2O2. Cell viability was evaluated by Trypan blue assay (45). Prx oligomers were identified after cells were treated with H2O2 for the appropriate times and concentrations. Cell lysates were fractionated in nondenaturing gels (Invitrogen) and analyzed by Western blotting as described in ref. 24.
Supplementary Material
Acknowledgments.
We are grateful to Dr. James Bradner and Stuart Schreiber, Harvard University, for providing the HDAC6 clones and tubacin. Studies reported here were supported in part by National Institutes of Health Grant P30CA08748-41, The Jack and Susan Rudin Foundation, David Koch Foundation, and Experimental Therapeutics Center at Memorial Sloan–Kettering Cancer Center.
Footnotes
Conflict of interest statement: Memorial Sloan–Kettering Cancer Center and Columbia jointly hold patents on SAHA and related compounds that were exclusively licensed to ATON Pharma, acquired by Merck in April 2004. P.A.M. was a founder of ATON and has a financial interest in the further development of SAHA (vorinostat) by Merck.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803749105/DCSupplemental
References
- 1.Lehrmann H, Pritchard LL, Harel-Bellan A. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv Cancer Res. 2002;86:41–65. doi: 10.1016/s0065-230x(02)86002-x. [DOI] [PubMed] [Google Scholar]
- 2.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: Overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 3.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75(1):435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 4.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdel A, et al. Active maintenance of mHDA2/mHDAC6 histone deacetylase in the cytoplasm. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 6.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. HDAC-6 interacts with and deacetylases tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bali P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs JJ, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 10.Aoyagi S, Archer TK. Modulating molecular chaperone Hsp90 functions through reversible acetylation. Trends Cell Biol. 2005;15:565–567. doi: 10.1016/j.tcb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 12.Pandey UB, Batlevi Y, Baehrecke EH, Taylor JP. HDAC6 at the intersection of autophagy, the ubiquitin-proteasome system and neurodegeneration. Autophagy. 2007;3:643–645. doi: 10.4161/auto.5050. [DOI] [PubMed] [Google Scholar]
- 13.Haggarty SJ, et al. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moon JC, et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- 15.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 16.Kinnula VL, et al. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196:316–323. doi: 10.1002/path.1042. [DOI] [PubMed] [Google Scholar]
- 17.Noh DY, et al. Overexpression of peroxiredoxin in human breast cancer. Anticancer Res. 2001;21:2085–2090. [PubMed] [Google Scholar]
- 18.Karihtala P, et al. Peroxiredoxins in breast carcinoma. Clin Cancer Res. 2003;9:3418–3424. [PubMed] [Google Scholar]
- 19.Chen WC, et al. Induction of radioprotective peroxiredoxin-I by ionizing irradiation. J Neurosci Res. 2002;70:794–798. doi: 10.1002/jnr.10435. [DOI] [PubMed] [Google Scholar]
- 20.Nonn L, Berggren M, Powis G. Increased expression of mitochondrial peroxiredoxin-3 (thioredoxin peroxidase-2) protects cancer cells against hypoxia and drug-induced hydrogen peroxide-dependent apoptosis. Mol Cancer Res. 2003;1:682–689. [PubMed] [Google Scholar]
- 21.Lee W, et al. Human peroxiredoxin 1 and 2 are not duplicate proteins: The unique presence of Cys-83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J Biol Chem. 2007;282:22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- 22.Multhaup G, et al. Reactive oxygen species and Alzheimer's disease. Biochem Pharmacol. 1997;54:533–539. doi: 10.1016/s0006-2952(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 23.Krapfenbauer K, et al. Aberrant expression of peroxiredoxin subtypes in neurodegenerative disorders. Brain Res. 2003;967:152–160. doi: 10.1016/s0006-8993(02)04243-9. [DOI] [PubMed] [Google Scholar]
- 24.Dokmanovic M, et al. Histone deacetylase inhibitors selectively suppress expression of HDAC7. Mol Cancer Ther. 2007;6:2525–2534. doi: 10.1158/1535-7163.MCT-07-0251. [DOI] [PubMed] [Google Scholar]
- 25.North BJ, et al. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 26.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotech. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 27.Dokmanovic M, Marks PA. Prospects: Histone deacetylase inhibitors. J Cell Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 28.Zou H, Wu Y, Navre M, Sang BC. Characterization of the two catalytic domains in histone deacetylase 6. Biochem Biophys Res Commun. 2006;341:45–50. doi: 10.1016/j.bbrc.2005.12.144. [DOI] [PubMed] [Google Scholar]
- 29.Winkler GS, et al. Isolation and mass spectrometry of transcription factor complexes. Methods. 2002;26:260–269. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- 30.Oktyabrsky ON, Smirnova GV. Redox regulation of cellular functions. Biochemistry (Mosc) 2007;72:132–145. doi: 10.1134/s0006297907020022. [DOI] [PubMed] [Google Scholar]
- 31.Koo KH, et al. Regulation of thioredoxin peroxidase activity by C-terminal truncation. Arch Biochem Biophys. 2002;397:312–318. doi: 10.1006/abbi.2001.2700. [DOI] [PubMed] [Google Scholar]
- 32.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Immenschuh S, Baumgart-Vogt E. Peroxiredoxins, oxidative stress, and cell proliferation. Antioxid Redox Signal. 2005;7:768–777. doi: 10.1089/ars.2005.7.768. [DOI] [PubMed] [Google Scholar]
- 34.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 35.Rabilloud T, et al. Proteomics analysis of cellular response to oxidative stress: Evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 36.Wagner E, et al. A method for detection of overoxidation of cysteines: Peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochem J. 2002;366:777–785. doi: 10.1042/BJ20020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 38.Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci USA. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minamiyama M, et al. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2004;13:1183–1192. doi: 10.1093/hmg/ddh131. [DOI] [PubMed] [Google Scholar]
- 40.Petri S, et al. Additive neuroprotective effects of a histone deacetylase inhibitor and a catalytic antioxidant in a transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2006;22:40–49. doi: 10.1016/j.nbd.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 41.Gardian G, et al. Neuroprotective effects of phenylbutyrate in the N171–82Q transgenic mouse model of Huntington's disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 42.Engel RH, Evens AM. Oxidative stress and apoptosis: A new treatment paradigm in cancer. Front Biosci. 2006;11:300–312. doi: 10.2741/1798. [DOI] [PubMed] [Google Scholar]
- 43.Fang J, Nakamura H, Iyer AK. Tumor-targeted induction of oxystress for cancer therapy. J Drug Target. 2007;15:475–486. doi: 10.1080/10611860701498286. [DOI] [PubMed] [Google Scholar]
- 44.Powis G, Mustacich D, Coon A. The role of the redox protein thioredoxin in cell growth and cancer. Free Radic Biol Med. 2000;29:312–322. doi: 10.1016/s0891-5849(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 45.Ungerstedt JS, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.