Abstract
Protein N-glycosylation is widespread among biological systems, and the fundamental process of transferring a lipid-linked glycan to suitable asparagine residues of newly synthesized proteins occurs in both prokaryotes and eukaryotes. The core reaction is mediated by Stt3p family members, and in many organisms this component alone is sufficient to constitute the so called oligosaccharyltransferase (OST). However, eukaryotes typically have a more elaborate OST with several additional subunits of poorly defined function. In the mammalian OST complex one such subunit, ribophorin I, is proposed to facilitate the N-glycosylation of certain precursors during their biogenesis at the endoplasmic reticulum. Here, we use cell culture models to show that ribophorin I depletion results in substrate-specific defects in N-glycosylation, clearly establishing a defined physiological role for ribophorin I. To address the molecular mechanism of ribophorin I function, a cross-linking approach was used to explore the environment of nascent glycoproteins during the N-glycosylation reaction. We show for the first time that ribophorin I can regulate the delivery of precursor proteins to the OST complex by capturing substrates and presenting them to the catalytic core.
Keywords: cross-linking, endoplasmic reticulum, N-glycosylation, RNAi, STT3
In eukaryotic cells, N-glycosylation is typically the most common protein modification that occurs in the ER lumen. N-glycosylation is facilitated by a large heterologous protein complex called the oligosaccharyltransferase (OST), which catalyses the attachment of a high mannose oligosaccharide en bloc onto suitable free asparagine residues of newly synthesized nascent chains during translocation into the ER lumen (1). The addition of an N-glycan provides a molecular tag that can promote the folding, maturation, and quality control of a precursor. Defects in the process of N-glycosylation, for example, congenital disorders, can result in devastating physiological consequences (2).
Our current understanding of the OST stems from research that utilizes a number of systems, including simple models such as prokaryotes and more complex and diverse eukaryotes. The mammalian equivalents of many, if not all Saccharomyces cerevisiae (S. cerevisiae) OST subunits, are known and comprise: ribophorin I (Ost1p), ribophorin II (Swp1p), OST48 (Wbp1p), STT3A and STT3B (Stt3), N33 and IAP (Ost3p and Ost6p), and Dad1 (Ost2p) (3–5). In addition, two other putative mammalian OST subunits, DC2 and KCP2, have been identified but to date have no known function (5).
There is now compelling evidence that the catalytic Center of the OST complex lies with the STT3 subunit(s) (6–9). In mammals, two STT3 isoforms exist, STT3A and STT3B; these share 70% sequence similarity and are expressed at varying levels depending upon tissue or cell-type (4). Despite their similarity, these two STT3 isoforms are not functionally equivalent when analyzed in vitro (10). In many prokaryotes, a single STT3 homologue is sufficient to mediate the N-glycosylation reaction (11–13). This observation raises the interesting question of why most eukaryotic OSTs contain multiple distinct subunits and what precise role these other subunits play during N-glycosylation.
Mammalian ribophorin I and ribophorin II are abundant ER membrane proteins (14, 15) that were coisolated as part of an enzyme complex showing in vitro OST activity (16). Although subsequent studies suggested that ribophorin I and its S. cerevisiae equivalent (Ost1p) could form part or all of the active site of the OST complex (17, 18), the case for STT3 playing this role is now compelling (11). In a previous study, we showed that a subset of newly synthesized membrane proteins transiently associated with ribophorin I immediately after their departure from the Sec61 translocon (19). Based on these data, we proposed that ribophorin I may function to retain potential substrates in close proximity to the catalytic subunit of the OST, thereby improving the efficiency of their N-glycosylation (19).
Our subsequent in vitro analysis of ribophorin I function showed that this OST subunit dramatically enhances the N-glycosylation of selected substrates but is apparently dispensable for the efficient N-glycosylation of many others (10). Here, we show for the first time that ribophorin I depletion has a selective effect upon the N-glycosylation of endogenous substrates expressed in cultured mammalian cells. By using a novel combination of siRNA-mediated depletion and in vitro cross-linking, we find that ribophorin I acts to selectively regulate the delivery of substrates to the catalytic core of the OST complex.
Results and Discussion
Ribophorin I Function in Cultured Cells.
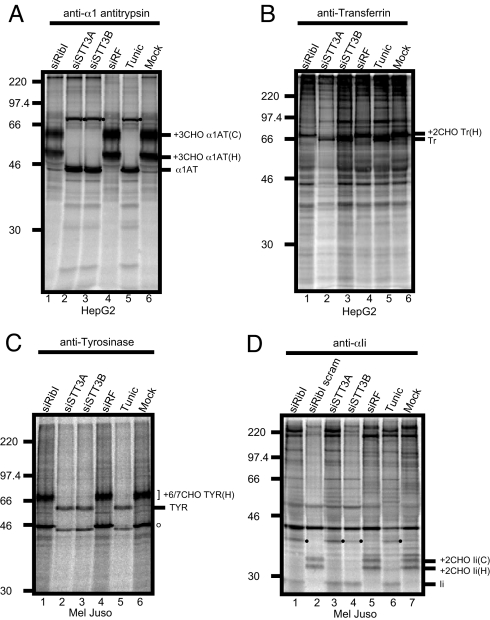
HepG2 cells express a number of secretory glycoproteins (20) and hence were selected to study the effect of ribophorin I depletion by siRNA. Importantly, our previous work has established that the integrity of the remaining OST complex is not disrupted after ribophorin I depletion (10), minimizing the possibility of indirect or pleiotropic effects after ribophorin I knockdown. For comparison, the two mammalian STT3 isoforms were also depleted [supporting information (SI) Fig. S1A], and cells were treated with the antibiotic tunicamycin that inhibits assembly of the lipid-linked oligosaccharide precursor necessary for N-glycosylation to occur. Both of these treatments were proven to be effective at inhibiting protein N-glycosylation in vitro (10). α1-antitrypsin (α1AT) is a secretory protein with three sites for N-glycosylation. Whole cell extracts were analyzed after metabolic labeling and immunoprecipitation; two major intracellular forms of α1AT were observed (Fig. 1A). The faster migrating species has a high mannose form of N-linked glycan added [Fig. 1A, lane 6, +3CHO α1AT(H)] and is sensitive to Endo-H treatment (Fig. S1B). The second, slower migrating species has more complex N-linked glycan typically acquired during transit through the Golgi complex and is insensitive to Endo-H treatment [Fig. 1A, lane 6, +3CHO α1AT(C) and Fig. S1B]. Tunicamycin treatment resulted in a complete loss of N-glycosylation of α1AT (Fig. 1A, lane 5). Likewise, knocking down either of the two STT3 isoforms, STT3A or STT3B, also resulted in complete inhibition of N-glycosylation (Fig. 1A, lanes 2 and 3). In contrast, N-glycosylation of α1AT appears completely unperturbed by ribophorin I depletion (Fig. 1A, lane 1), although Western blot analysis confirmed a substantial reduction of >80% in the cellular levels of the target protein after siRNA treatment (Fig. S1A) (cf. ref. 10). A second endogenous HepG2 secretory glycoprotein, transferrin, was also analyzed for the effects of OST subunit depletion. Once again, tunicamycin treatment and siRNA-mediated depletion of STT3A or STT3B resulted in a complete loss of N-glycosylated transferrin (Fig. 1B, lanes 2, 3, 5, and 6, see Tr), whereas ribophorin I knockdown did not affect the N-glycosylation status of transferrin. However, less total material was recovered from the ribophorin I knockdown compared with the control cells [Fig. 1B, lanes 1, 5, and 6, see +2CHOTr(H)].
Fig. 1.
Effect of OST subunit knockdown on the N-glycosylation of endogenously expressed proteins in HepG2 and Mel Juso cells. (A and B) HepG2 cells were transfected with siRNAs targeting ribophorin I (lane 1), STT3A (lane 2), STT3B (lane 3), a nonfunctional control siRNA (siRF) (lane 4), or mock treated (lane 6). To control for loss of N-glycosylation, HepG2 cells were incubated with 2 μg/ml tunicamycin for 12 h before isolation on day 2 (lane 5). Cells were pulse labeled with [35S] methionine/cysteine for 45 min, solubilized in IP buffer and specific products recovered by immunoprecipitating with antisera recognizing α1-antitrypsin (α1AT, see A) or transferrin (Tr, see B). In all cases, the presence and number (x) of N-linked glycans present on a particular protein is indicated by the suffix xCHO. For α1AT (A), two populations of glycoproteins reflecting products with both high mannose type N-linked glycans [+3CHOα1AT(H)] and complex N-glycans [+3CHOα1AT(C)] were detected. With Tr (B), only high mannose type glycans were observed. (C and D) Mel Juso cells were treated with siRNAs or tunicamycin as above. Additionally, a scrambled ribophorin I duplex (siRibI scram) was used during the analysis of the invariant chain (D, lane 2). Cells were labeled with [35S] methionine/cysteine for 30 min, solubilized in IP buffer and immunoprecipitated with antisera recognizing tyrosinase (TYR, see C) or invariant chain (Ii, see D). Two closely migrating forms of tyrosinase were detected representing chains with 6 or 7 high mannose type N-glycans attached [+6/7 CHO TYR(H)]. Two quite distinct forms of N-glycosylated Ii were detected; these reflect discrete populations of proteins with high mannose [+2CHO Ii(H)] or complex type [+2CHO Ii(C)] N-linked glycans as described above for α1AT. The appearance of higher molecular weight species after inhibiting N-glycosylation of α1AT and Ii (A and D, filled circles) may reflect aberrant dimerization of the nonglycosylated proteins (compare with ref. 34). In the case of TYR, Mel Juso cells express a cross-reacting glycoprotein of ∼46 kDa (C, open circle) that behaves in a similar fashion to authentic TYR after treatment with the various siRNAs or tunicamycin. This product is most likely a splice variant of tyrosinase (35).
Our previous in vitro studies (10) suggested that the N-glycosylation of secretory proteins was unaffected by ribophorin I depletion, and our analysis of endogenously expressed glycoproteins in HepG2 cells is entirely consistent with this notion. We next addressed the substrate specificity of ribophorin I by using Mel Juso cells, because these express the invariant chain of the MHC class II complex (Ii) (21), a glycoprotein that we identified as a ribophorin I dependent substrate in vitro (10). We observed that cellular levels of ribophorin I, STT3A and STT3B, in Mel Juso cells were specifically and efficiently knocked down by using siRNA (Fig. S1D). We also analyzed the effect on the N-glycosylation of two endogenously expressed membrane proteins, tyrosinase and Ii. After metabolic labeling and immunoprecipitation, tyrosinase was found to migrate as a 70-kDa doublet corresponding to products with 6 and 7 N-linked glycans as previously described (22) (Fig. 1C, lane 6). Both products are sensitive to Endo-H treatment (Fig. S1E). Depletion of STT3A and STT3B results in the complete loss of tyrosinase N-glycosylation (Fig. 1C, lanes 2, 3, 5, and 6), whereas ribophorin I knockdown has no effect (Fig. 1C, lanes 1, 5, and 6). When we examined the Ii expressed in Mel Juso cells by using the same approach, two distinct species of ∼35 kDa were observed after immunoprecipitation (Fig. 1D, lane 7). The faster migrating product has high mannose type N-linked glycans [Fig. 1D, lane 7, +2CHO Ii(H)] that are Endo-H sensitive (Fig. S1F). The slower migrating species bears complex N-linked glycans [Fig. 1D, lane 7, +2CHO Ii(C)] that are insensitive to Endo-H treatment (Fig. S1F). In contrast to our analysis of tyrosinase, we find that Ii requires all three OST subunits, ribophorin I, STT3A, and STT3B, for its efficient N-glycosylation in Mel Juso cells (Fig. 1D, lanes 1, 3, 4, 6, and 7). Normal levels of N-glycosylation are observed with a control siRNA and a scrambled version of the ribophorin I specific duplex (Fig. 1D, lanes 2, 5, and 7). Based on data obtained by using siRNA mediated depletion of ribophorin I, we conclude that its normal physiological role is substrate specific. Hence, after a ribophorin I knockdown, N-glycosylation is either completely inhibited (Ii) or unaffected (α1AT, transferrin and tyrosinase). This analysis establishes the cellular role of ribophorin I and provides the first direct evidence that both STT3 isoforms are required for efficient N-glycosylation in mammalian cells for the four precursors analyzed here.
Analyzing Nascent Chain OST Interactions.
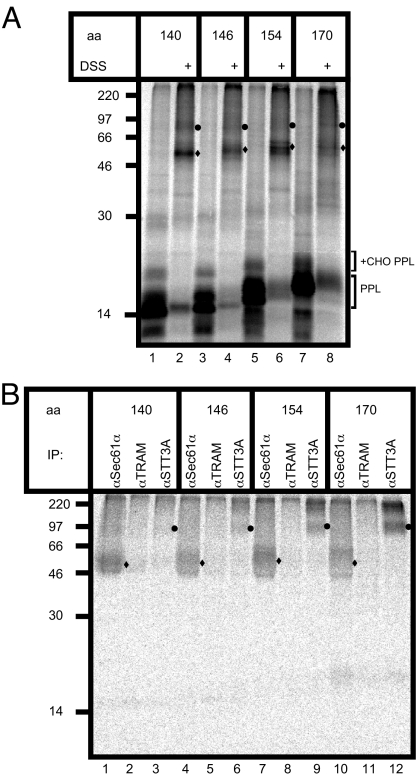
To decipher the molecular basis for ribophorin I function during the process of N-glycosylation we analyzed the association of ribophorin I−dependent and −independent substrates with the OST complex. We reasoned that this could be best achieved by combining an in vitro assay for OST function (10) with the use of cross-linking, so as to probe the environment of the nascent polypeptide chain during N-glycosylation. We initially turned to the model system first used to identify STT3A as the catalytic subunit of the mammalian OST (8). Hence, a derivative of the secretory protein pre-prolactin was constructed that contained a single consensus site for N-glycosylation together with a lone lysine residue located within this motif (denoted PPLNKT, cf. ref. 8). Whereas the original study exploited a photocross-linking approach (8), we were able to substitute this with the bifunctional amine-reactive reagent disulfo-succinimide (DSS). To maintain potential interactions between the nascent chain and subunits of the OST complex, the translocating polypeptide was artificially trapped at the ER translocon as a ribosome-bound peptidyl-tRNA (8). Under these conditions, discrete DSS-dependent cross-linking products were obtained with PPLNKT translocation intermediates of varying chain length (Fig. 2A). The various PPLNKT nascent chain lengths analyzed could all be N-glycosylated (Fig. 2A, lanes 1, 3, 5, and 7 +CHO PPL), and the major adducts observed (Fig. 2A, circles and diamonds in lanes 2, 4, 6, and 8) were consistent with cross-linking to subunits of both the ER translocon and the OST complex (8).
Fig. 2.
Analyzing nascent chain OST interactions. (A) PPLNKT translocation intermediates of increasing chain length (aa) were synthesized in rabbit reticulocyte lysate (RRL) supplemented with semipermeabilized HeLa cells and cross-linked by using DSS. The resulting glycosylated (+CHO PPL) and nonglycosylated PPL (PPL) chains are indicated together with two major DSS-dependent adducts that are visible in the total products (filled circles and diamonds). (B) Specific cross-linking products of the PPLNKT chains with Sec61α (filled diamonds) and STT3A (filled circles) were identified by immunoprecipitation.
To conclusively identify the cross-linking partners of the PPLNKT polypeptides, the products were analyzed by immunoprecipitation. This confirmed that the PPLNKT nascent chains were efficiently cross-linked to the ER translocon component Sec61α (Fig. 2B, diamonds in lanes 1, 4, 7, and 10), and the OST subunit, STT3A (Fig. 2B, circles in lanes 3, 6, 9, and 12). In contrast, cross-linking to the ER translocon-associated component, TRAM, was extremely inefficient (Fig. 2B, lanes 2, 5, 8, and 11), consistent with the location of the lysine residue within the PPLNKT chains that is primarily responsible for adduct formation (23). This pattern of adducts confirms that N-glycosylation at the OST occurs while translocation across the ER membrane is in progress (8). The normally cotranslocational nature of N-glycosylation is further underlined by the loss of adduct formation with STT3A upon puromycin-mediated release of the trapped PPLNKT nascent chains from the ribosome (data not shown). PPLNKT cross-linking to STT3A is also strongly inhibited by a short tripeptide OST substrate, confirming that adduct formation reflects a biologically productive interaction between the nascent chain and the OST complex (Fig. S2).
Ribophorin I−Independent Substrates.
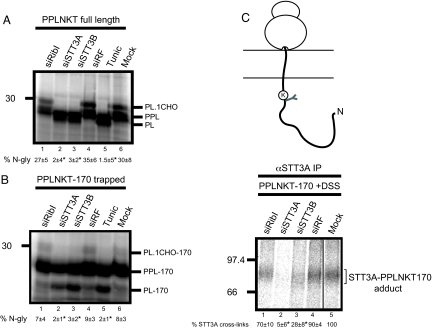
Having optimized the cross-linking of PPLNKT nascent chains to STT3A and established that in our system the longest translocation intermediate gave the clearest adducts (Fig. 2B, lanes 3, 6, 9, and 12; see also ref. 8), we analyzed the effect of siRNA mediated depletions upon the N-glycosylation of PPLNKT. When N-glycosylation of the full-length protein was analyzed by using an in vitro read out, depletion of STT3A and STT3B caused an almost complete loss of N-glycosylation compared with the ∼30% efficiency observed with control samples (Fig. 3A, lanes 2–6). In contrast, ribophorin I depletion had no significant effect on PPLNKT N-glycosylation (Fig. 3A, lanes 1, 4, and 6). We therefore conclude that PPLNKT is N-glycosylated independently of ribophorin I function, consistent with previous in vitro (10) and in vivo (this study) investigations showing ribophorin I has no detectable role during the N-glycosylation of four different secretory glycoproteins.
Fig. 3.
Molecular analysis of a ribophorin I−independent substrate. (A) Full-length PPLNKT was synthesized in vitro by using semipermeabilized HeLa cells prepared after siRNA mediated depletion of ribophorin I (lane 1), STT3A (lane 2), STT3B (lane 3), a nonfunctional control (siRF) (lane 4) or mock treatment (lane 6). Tunicamycin treatment served as a control (compare to Fig. 1). The resulting products, glycosylated (PL.CHO) and nonglycosylated (PL) prolactin and the precursor with intact signal sequence (PPL) are shown after SDS/PAGE. The relative proportion of glycosylated polypeptide was calculated for each sample and expressed as a percentage of the total protein recovered. The values below the lanes are the mean ± SEM of three independent experiments. Levels of N-glycosylation that differ from the mock treated control with a significance of P < 0.02 are indicated by asterisks. (B) A 170-residue PPLNKT translocation intermediate encoded by an mRNA lacking a stop codon was synthesized as before (compare to Fig. 2) and analyzed as described for A. (C) A proportion of the PPLNKT-170 translocation intermediates shown in B were treated with DSS and the resulting adducts with STT3A were recovered by immunoprecipitation. The amount of STT3A-PPLNKT-170 adduct for the mock sample was quantified and set to a nominal value of 100%. Other values are the mean of three independent experiments where the amount of adduct obtained after the different treatments is expressed relative to the mock treated sample. Values that differ from the mock treated control with a significance of P < 0.02 are indicated by asterisks.
For effective cross-linking to OST subunits, it is necessary to use artificially trapped nascent polypeptide chains (see above); thus, we next established whether the N-glycosylation of these rather artificial substrates reflected that of the full-length protein. When a 170 residue long ribosome-bound translocation intermediate of PPLNKT was analyzed, the overall efficiency of N-glycosylation was substantially less than that obtained with the full-length protein (compare Fig. 3 A and B). Nevertheless, STT3A and STT3B depletion reduced N-glycosylation to background levels (Fig. 3B, lanes 2, 3, and 5), whereas ribophorin I depletion had no effect (Fig. 3B, lanes 1, 4, and 6). When the cross-linking of the same nascent PPLNKT chains to OST subunits was analyzed, depletion of STT3A led to a huge reduction in adduct formation with this component (Fig. 3C, lanes 2, 4, and 5), whereas the levels of trapped PPLNKT chains were unaffected (Fig. 3B, lanes 1–4, 6, PPL-170). This result confirms the effectiveness of the STT3A knockdown as independently assessed by immunoblotting (10) (Fig. S1 A and D) and directly correlates STT3A function with efficient N-glycosylation. In contrast, the knockdown of ribophorin I had no significant effect on STT3A adduct formation (Fig. 3C, lanes 1, 4, and 5), consistent with the normal levels of PPLNKT N-glycosylation observed after ribophorin I depletion (Fig. 3 A and B, lanes 1, 2, and 6).
Ribophorin I−Dependent Substrates.
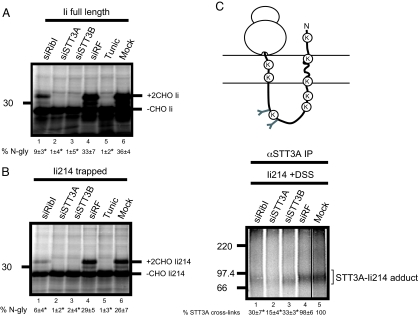
We next performed a cross-linking analysis by using a glycoprotein substrate known to require ribophorin I for efficient modification. The Ii chain has two sites for N-glycosylation, and their use is significantly reduced after ribophorin I depletion and in vitro analysis (10). Furthermore, no detectable N-glycosylation of endogenous Ii occurs after ribophorin I depletion in Mel Juso cells (this study, Fig. 1D). A variety of different length integration intermediates of Ii were analyzed by DSS-dependent cross-linking (cf. ref. 24), and a truncated chain of 214 residues found to form discrete adducts with both the Sec61α and STT3A (Fig. S3). In each case, adducts were lost upon puromycin treatment, consistent with the interactions depending upon the presence of a trapped nascent chain (Fig. S3). When the N-glycosylation of full-length Ii was analyzed by using an in vitro readout, we found that STT3A and STT3B depletions resulted in an almost complete loss of N-linked glycans (Fig. 4A, lanes 2–4, 6) (10). Likewise, the depletion of ribophorin I caused a substantial drop in N-glycosylation, although some modification was still apparent in vitro (Fig. 4A, lanes 1, 4–6) (10). The behavior of the truncated 214-residue long trapped Ii integration-intermediate (Ii214) was almost identical, although N-glycosylation was marginally less efficient (Fig. 4B, compare lanes 1–6).
Fig. 4.
Molecular analysis of ribophorin I dependent substrate. (A) Full-length invariant chain (Ii) was synthesized as above (Fig. 3) to determine the extent of its N-glycosylation. The resulting glycosylated (+2CHOIi) and nonglycosylated (Ii) polypeptides and the proportion of N-glycosylated products obtained after various treatments are indicated as described for Fig. 3. (B) A 214-residue integration intermediate of Ii was synthesized by using a truncated mRNA lacking a stop codon and analyzed as described for A. (C) A proportion of the 214-residue Ii–integration intermediates were treated with DSS and the resulting STT3A adducts recovered by immunoprecipitation. The relative percentage of STT3A adduct formation under the various experimental conditions was calculated as before (compare to Fig. 3). Symbols are as defined in the legend to Fig. 3.
When the association of Ii214 with the STT3A subunit of the OST was analyzed by cross-linking, the depletion of ribophorin I resulted in a significant loss of adduct formation approaching that seen after STT3A depletion (Fig. 4C, lanes 1, 2, 4, and 5). Thus, ribophorin I depletion prevents the efficient access of a ribophorin I−dependent precursor, Ii, to the catalytic STT3A subunit of the OST but has little, if any, effect upon the association of a ribophorin I−independent glycoprotein substrate (PPLNKT). We therefore conclude that ribophorin I acts to present a subset of precursors to the catalytic core of the OST complex in such a way that efficient modification can occur.
Complexity of the Mammalian OST.
To date, we find that simple membrane proteins are most commonly dependent on ribophorin I both in vitro (10) and in vivo (this study). In contrast, other precursors including several different secretory glycoproteins and some other membrane proteins, show no detectable dependency on ribophorin I activity. In this case, such precursors most likely either have direct access to the catalytic core of the OST, or their presentation is dependent on one of the other ‘noncatalytic’ OST subunits. One possibility for ribophorin I function is that this subunit binds newly synthesized membrane proteins and simply delays these precursors in the vicinity of the OST complex to stochastically increase their chances of N-glycosylation (10, 19). On the basis of our current study this possibility seems unlikely. Hence, when an Ii integration-intermediate is artificially trapped at the ER translocon and is close enough to the STT3A subunit to be cross-linked to it (Fig. 4C), the efficiency of N-glycosylation of the trapped nascent chain after ribophorin I depletion is no better than that seen for the same protein without any such trapping (compare Fig. 4 A and B). Thus, artificially retaining the Ii nascent chain in proximity of the OST complex cannot rescue the defect in N-glycosylation caused by ribophorin I depletion, arguing against a simple stochastic effect.
Surprisingly, STT3B depletion has a marked effect on the cross-linking of both PPLNKT and Ii-trapped intermediates to the STT3A subunit, causing a substantial reduction in both cases (Figs. 3C and 4C). Thus, although STT3A levels appear unaffected by STT3B depletion (10) (Fig. S1), it seems that in the absence of STT3B the access of substrates to STT3A is inhibited. The simplest explanation for this observation is that both STT3A and STT3B may contribute to a functional dimeric OST complex (6, 25, 26). However, there is clear evidence that STT3A and STT3B are present in distinct OST complexes (4) and that the roles of the two isoforms are not equivalent (10), suggesting such a model is an oversimplification.
Interestingly, other co- and posttranslational modifications of proteins synthesized at the ER are also mediated by oligomeric complexes where a minority of the subunits has catalytic activity. The glycosylphosphatidylinositol (GPI) transamidase complex is responsible for the addition of lipid anchors to cell surface proteins synthesized at the ER. The mammalian and S. cerevisiae transamidases each consist of five subunits, yet it is the GPI8/Gpi8p subunit alone that appears to be catalytic (27). The precise function of the other subunits is less clear, and a role for one or more of these components in regulating substrate access to the catalytic Center of the transamidase has been suggested (27), akin to the proposed function of ribophorin I. The mammalian signal peptidase complex is responsible for the proteolytic removal of ER targeting signals consists of five subunits, yet only two of these appear to be catalytic (28).
In summary, the elaborate OST complexes found in many eukaryotes most likely reflect their ability to modify a wide range of potential glycoprotein substrates (12) and to refine the selection of the dolichol-linked oligosaccharide donor used for the modification (29). In the case of ribophorin I, we propose that this subunit acts to directly present a subset of potential substrates such as simple membrane proteins, to the catalytic core of the mammalian OST. This model is further supported by a recent structural study of the S. cerevisiae OST that places its ribophorin I homolog, Ost1p, close to the STT3 catalytic Center (6). It should be noted that whereas cultured mammalian cells appear to be able to tolerate ribophorin I depletion for a limited period (ref. 10, this study), studies in C. elegans show that both STT3 and ribophorin I are essential for viability (30) (www.wormbase.org). This indicates that ribophorin I facilitates the biosynthesis and N-glycosylation of developmentally crucial glycoproteins in more complex multicellular organisms.
Materials and Methods
Reagents and Antibodies.
Cell culture reagents were purchased from GIBCO, BRL, Cambrex, or Lonza. RNA polymerase and rabbit reticulocyte lysate were purchased from Promega and EasyTag l-[35S]methionine was purchased from PerkinElmer (U.K.). All other chemicals were purchased from Sigma or BDH/Merck. Rabbit polyclonal antisera recognizing ribophorin I, Sec61α, TRAM, and STT3A were made to order by Invitrogen. Antisera recognizing transferrin and the invariant chain were kindly provided by P. Woodman (University of Manchester, U.K.) and B. Dobberstein (Universität Heidelberg, Germany) respectively. Antiserum specific for α1-antitrypsin was purchased from Abcam. Antiserum recognizing tyrosinase from Santa Cruz was kindly provided by N. Bulleid (University of Manchester, U.K.). 21-nulceotide duplexes corresponding to human ribophorin I (siRibI, aagcgcacagtggacctaagc), STT3A (STT3A, gcagtaggatcatatttgatt), STT3B (STT3B, tatcaacgatgaaagagtatt), scrambled ribophorin I (siRibI scram, aggaatgcgcacccggactaa), and the Risc-free siCONTROL were purchased from Dharmacon Research. Mel Juso cells were kindly provided by J. Trowsdale (University of Cambridge, U.K.).
cDNA Constructs.
Bovine pre-prolactin with arginine replacing the lysines at residues 4 and 9 of the wild type sequence (cf. ref. 23) was used as a template for site-directed mutagenesis, and additional lysines located at residues 72, 78, 99, 136, and 154 were changed to arginine. A consensus N-glycosylation site was then introduced by replacing the sequence YAQ at amino acids 74–76 of the wild type coding region with NKT (cf. ref. 8), the resulting coding region was inserted into the pGEM4 vector and the construct named PPLNKT. The invariant chain cDNA is in the pSPUTK vector as described (10).
RNA Interference.
HepG2, Mel Juso, and HeLa cells grown in 10-cm2 dishes, seeded 24 h before treatment, were transfected with 60 μl of 20 μM siRNA duplex by using Oligofectamine (Invitrogen, Paisley, U.K.) as described (31). HepG2 cells were transfected for a second time after 24 h to achieve the most efficient knockdown. RNAi-treated cells were incubated for 48 h, whereas tunicamycin (2 μg/ml) was added 12 h before the preparation of semipermeabilized cells (32) or metabolic labeling.
Metabolic Labeling of Cells.
Before starvation, Mel Juso cells were either pretreated for 4 h with 150 μg/ml leupeptin (Sigma) for Ii pulse chase analysis or pretreated for 1 h with 10 μM proteasome inhibitor II (Calbiochem) for tyrosinase pulse chase analysis. Cells were starved for 20 min in methionine/cysteine-free MEM (Sigma) with 2 mM glutamine, and then metabolically labeled with the same medium containing 20 μCi/ml of [35S]methionine/cysteine (1,175 Ci/mmol, 11.9 mCi/ml) protein labeling mix for 30–45 min. Cells were rinsed twice with PBS and solubilized in Triton X-100 IP buffer (33) containing 1 mM PMSF and 1 mM cold methionine/cysteine. Samples were pre-cleared and immunoprecipitated with antisera specific for α1-antitrypsin, transferrin, tyrosinase or invariant chain before SDS/PAGE and phosphorimaging.
In Vitro Transcription, Translation, and Cross Linking.
DNA templates for the in vitro transcription of mRNAs, with or without a stop codon as required, were generated by PCR using appropriate primers and transcribed as described (10). Semipermeabilized cells were prepared from siRNA transfected HeLa cells as outlined above, and a rabbit reticulocyte lysate system (Promega) used to translate either truncated mRNAs lacking a stop codon for 35 min at 30°C, or full-length mRNAs for 60 min at 30°C, in the presence of 0.75 mCi/ml [35S]methionine/cysteine and RNAi treated semipermeabilized HeLa cells. Aurintricarboxylic acid (ATCA) was then added to a final concentration of 100 μM to inhibit further initiation, and the samples were incubated at 30°C for 10 min. Membrane-associated products were isolated by centrifugation for 2 min at 16,000 × g and washed by resuspension in KHM buffer (20 mM HEPES pH 7.2, 110 mM potassium acetate, 2 mM magnesium acetate). For cross-linking, samples were incubated at 30°C for 10 min either in the presence of the amine-reactive cross-linking reagent DSS at 1 mM or an equivalent dimethyl sulfoxide (Me2SO) solvent control. The reaction was quenched by the addition of 40 mM glycine for 10 min on ice, and samples were either analyzed directly or after immunoprecipitation in the presence of 0.2% SDS as described (23). All samples were treated with RNase A (7 μg/translation reaction) to remove any tRNA that remained attached to stalled polypeptide chains and then incubated with 25 μl of sample buffer for 10 min at 70°C and analyzed on 10, 12 or 14% SDS-polyacrylamide Tris-glycine gels. The resulting gels were dried and then visualized by using a Fuji BAS 3000 PhosphorImager system (Fuji Photo Film, Tokyo, Japan).
Competitive Tripeptide Block.
In vitro translations of truncated, 170-residue long, PPLNKT chains were performed for 25 min in the presence of 1 mM tripeptide inhibitor (N-benzoyl-Asn-Leu-Thr-methylamine), 1 mM control peptide (Asp-Asn-Arg-Glu-Arg-Gln-Glu-His-Asn-Asp-Arg-Ser) or without any additions.
Statistical Analysis.
All N-glycosylated and cross-linking assays were carried out in at least three independent experiments and subsequent statistical analysis (two-sample t test) performed by using SPSS 10.1 software.
Supplementary Material
Acknowledgments.
We are eternally grateful to all our colleagues who have contributed their time and materials toward this paper. We thank Neil Bulleid, Ben Cross, Martin Pool and Blanche Schwappach for their comments during the preparation of the manuscript. This work was supported by a project grant from the Biotechnology and Biological Sciences Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711846105/DCSupplemental.
References
- 1.Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- 2.Freeze HH, Aebi M. Altered glycan structures: The molecular basis of congenital disorders of glycosylation. Curr Opin Struct Biol. 2005;15:490–498. doi: 10.1016/j.sbi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Kelleher DJ, Gilmore R. DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc Natl Acad Sci USA. 1997;94:4994–4999. doi: 10.1073/pnas.94.10.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelleher DJ, Karaoglu D, Mandon EC, Gilmore R. Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol Cell. 2003;12:101–111. doi: 10.1016/s1097-2765(03)00243-0. [DOI] [PubMed] [Google Scholar]
- 5.Shibatani T, David LL, McCormack AL, Frueh K, Skach WR. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry. 2005;44:5982–5992. doi: 10.1021/bi047328f. [DOI] [PubMed] [Google Scholar]
- 6.Hua L, Chavan M, Schindelin H, Lennarz WJ, Li H. Structure of the oligosaccharyl transferase complex at 12 Å resolution. Structure. 2008;16:432–440. doi: 10.1016/j.str.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Igura M, et al. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 2008;27:234–243. doi: 10.1038/sj.emboj.7601940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson I, et al. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol. 2003;161:715–725. doi: 10.1083/jcb.200301043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Q, Lennarz WJ. Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J Biol Chem. 2002;277:47692–47700. doi: 10.1074/jbc.M208136200. [DOI] [PubMed] [Google Scholar]
- 10.Wilson CM, High S. Ribophorin I acts as a substrate-specific facilitator of N-glycosylation. J Cell Sci. 2007;120:648–657. doi: 10.1242/jcs.000729. [DOI] [PubMed] [Google Scholar]
- 11.Igura M, et al. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 2008;27:234–243. doi: 10.1038/sj.emboj.7601940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowarik M, et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wacker M, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 14.Kreibich G, et al. Characterization of the ribosomal binding site in rat liver rough microsomes: Ribophorins I and II, two integral membrane proteins related to ribosome binding. J Supramol Struct Cell Biochem. 1978;8:279–302. doi: 10.1002/jss.400080307. [DOI] [PubMed] [Google Scholar]
- 15.Marcantonio EE, Amar-Costesec A, Kreibich G. Segregation of the polypeptide translocation apparatus to regions of the endoplasmic reticulum containing ribophorins and ribosomes. II. Rat liver microsomal subfractions contain equimolar amounts of ribophorins and ribosomes. J Cell Biol. 1984;99:2254–2259. doi: 10.1083/jcb.99.6.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelleher DJ, Kreibich G, Gilmore R. Oligosaccharyltransferase activity is associated with a protein complex composed of ribophorins I and II and a 48 kd protein. Cell. 1992;69:55–65. doi: 10.1016/0092-8674(92)90118-v. [DOI] [PubMed] [Google Scholar]
- 17.Bause E, Wesemann M, Bartoschek A, Breuer W. Epoxyethylglycyl peptides as inhibitors of oligosaccharyltransferase: Double-labelling of the active site. Biochem J. 1997;322(Pt 1):95–102. doi: 10.1042/bj3220095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Q, Prestwich GD, Lennarz WJ. The Ost1p subunit of yeast oligosaccharyl transferase recognizes the peptide glycosylation site sequence, -Asn-X-Ser/Thr. J Biol Chem. 1999;274:5021–5025. doi: 10.1074/jbc.274.8.5021. [DOI] [PubMed] [Google Scholar]
- 19.Wilson CM, et al. Ribophorin I associates with a subset of membrane proteins after their integration at the sec61 translocon. J Biol Chem. 2005;280:4195–4206. doi: 10.1074/jbc.M410329200. [DOI] [PubMed] [Google Scholar]
- 20.Ou WJ, Cameron PH, Thomas DY, Bergeron JJ. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 21.Zachgo S, Dobberstein B, Griffiths G. A block in degradation of MHC class II-associated invariant chain correlates with a reduction in transport from endosome carrier vesicles to the prelysosome compartment. J Cell Sci. 1992;103(Pt 3):811–822. doi: 10.1242/jcs.103.3.811. [DOI] [PubMed] [Google Scholar]
- 22.Ujvari A, et al. Translation rate of human tyrosinase determines its N-linked glycosylation level. J Biol Chem. 2001;276:5924–5931. doi: 10.1074/jbc.M009203200. [DOI] [PubMed] [Google Scholar]
- 23.High S, et al. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J Biol Chem. 1993;268:26745–26751. [PubMed] [Google Scholar]
- 24.High S, et al. Sec61p is adjacent to nascent type I and type II signal-anchor proteins during their membrane insertion. J Cell Biol. 1993;121:743–750. doi: 10.1083/jcb.121.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chavan M, et al. Dimeric organization of the yeast oligosaccharyl transferase complex. Proc Natl Acad Sci USA. 2006;103:8947–8952. doi: 10.1073/pnas.0603262103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Dobberstein B. Oligomeric complexes involved in translocation of proteins across the membrane of the endoplasmic reticulum. FEBS Lett. 1999;457:316–322. doi: 10.1016/s0014-5793(99)01075-3. [DOI] [PubMed] [Google Scholar]
- 27.Orlean P, Menon AK. Thematic review series: Lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Liang H, et al. Genetic complementation in yeast reveals functional similarities between the catalytic subunits of mammalian signal peptidase complex. J Biol Chem. 2003;278:50932–50939. doi: 10.1074/jbc.M307542200. [DOI] [PubMed] [Google Scholar]
- 29.Kelleher DJ, Banerjee S, Cura AJ, Samuelson J, Gilmore R. Dolichol-linked oligosaccharide selection by the oligosaccharyltransferase in protist and fungal organisms. J Cell Biol. 2007;177:29–37. doi: 10.1083/jcb.200611079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 31.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 32.Wilson R, et al. The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem J. 1995;307(Pt 3):679–687. doi: 10.1042/bj3070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson CM, Farmery MR, Bulleid NJ. Pivotal role of calnexin and mannose trimming in regulating the endoplasmic reticulum-associated degradation of major histocompatibility complex class I heavy chain. J Biol Chem. 2000;275:21224–21232. doi: 10.1074/jbc.M000567200. [DOI] [PubMed] [Google Scholar]
- 34.Hosokawa N, Wada I, Natsuka Y, Nagata K. EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-antitrypsin. Genes Cells. 2006;11:465–476. doi: 10.1111/j.1365-2443.2006.00957.x. [DOI] [PubMed] [Google Scholar]
- 35.Kelsall SR, Le Fur N, Mintz B. Qualitative and quantitative catalog of tyrosinase alternative transcripts in normal murine skin melanocytes as a basis for detecting melanoma-specific changes. Biochem Biophys Res Commun. 1997;236:173–177. doi: 10.1006/bbrc.1997.6925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.