Abstract
Our previous studies indicate that hMRE11 plays a role in MMR, and this function of hMRE11 is most likely mediated by the hMLH1-hMRE11 interaction. Here, we explored the functional implications of the hMLH1-hMRE11 interaction in MMR and the effects of hMLH1 mutations on their interaction. Our in vitro MMR assay demonstrated that the dominant-negative hMRE11452–634 mutant peptide (i.e., harboring only the hMLH1-interacting domain) imparted a significant reduction in both 3’ excision and 3’-directed MMR activities. Furthermore, the expression of hMRE11452–634, and to a lesser extent hMRE111–634 (ATLD1), impaired G2/M checkpoint control in response to MNU and cisplatin treatments, rendering cells resistant to killings by these two anticancer drugs. Analysis of 38 hMLH1 missense mutations showed that the majority of mutations caused significant (> 50%) reductions in their interaction with hMRE11, suggesting a potential link between aberrant protein interaction and the pathogenic effects of hMLH1 variants.
Keywords: hMLH1, hMRE11, MMR, cisplatin, MNU, G2/M checkpoint
INTRODUCTION
The DNA mismatch repair (MMR) system is a critical genome surveillance pathway in which mispaired bases, arising during DNA replication and recombination, are corrected [1]. It is evident that the development of Lynch syndrome can be largely attributed to germline mutations in several MMR genes, of which mutations in hMLH1 and hMSH2 account for the majority of cases, with incident rates of 59.2% and 39.5%, respectively [2] (www.insight-group.org). Although many of the identified Lynch syndrome mutations are inactivating sequence alterations that create internal stop codons, and as such may cause complete loss of function, a significant portion of Lynch syndrome mutations result in single amino acid substitutions that are expected to produce functionally hypomorphic full-length proteins. In fact, 25% of the approximately 300 presently known hMLH1 mutations are scattered missense alterations, for which the precise roles of these mutations in the pathogenesis of Lynch syndrome are obscure [2]. Therefore, a thorough understanding of the functions of MMR in the processes of DNA repair and cellular DNA damage response will provide clues in deciphering the molecular basis underlying the pathogenic consequences of hMLH1 variants.
To directly assess the functional effects of hMLH1 missense mutations, S. cerevisiae and human systems have been previously experimented [3, 4]. In human cells, hMLH1 existed as a heterodimeric complex with hPMS2 [5], and the disruption of this heterocomplex by hMLH1 Lynch syndrome mutations, including those reside outside of the PMS2-binding domain, can lead to defective MMR [6]. Studies performed with yeast two-hybrid analysis have revealed that a number of hMLH1 Lynch syndrome-related missense mutations could differentially affect interactions with either hPMS2 or hEXO1 [7], highlighting the functional heterogeneities of disease-associating hMLH1 mutations. It is conceivable that the functional effects of hMLH1 variants are governed by the outcomes of their impacts on protein interaction with different partners. We have previously demonstrated that the RNAi-mediating silencing of hMRE11 in HeLa cells results in microsatellite instability (MSI) and defective 3’-5’ MMR repair [8], and hMRE11 interacted with hMLH1 both in vitro and in vivo [9]. However, the exact functional roles of the hMLH1-hMRE11 interaction in MMR and MMR-related DNA damage response are presently elusive. Our current study provides evidence to support the role of hMLH1-hMRE11 interaction in MMR and DNA damage response.
MATERIALS AND METHODS
Cell lines and transfection
The pPuro-FLAG vector [10] was used to generate mammalian expression constructs pPuro-FLAG/hMRE111–634 and pPuro-FLAG/hMRE11452–634. Full-length hMLH1 cDNA was cloned into pcDNA6 (Invitrogen, Carlsbad, CA) to create the mammalian expression construct pcDNA6/hMLH1. Human embryonic kidney 293T cells were first transfected with either pPuro-FLAG/hMRE111–634 or pPuro-FLAG/hMRE11452–634 by the standard calcium phosphate method. Cells were maintained in DMEM containing 10% FBS (Biomeda) and stable cell lines were selected by using 1 µg/ml puromycin (A.G. Scienfitic, San Diego, CA). The positive clones were then transfected with pcDNA6/hMLH1 and selected with 10 µg/ml Blasticidin (Invitrogen).
In vitro MMR and mismatch-provoked excision assays
Nuclear extracts and the DNA heteroduplex contained an A–C mismatch with a strand break 3’ to the mismatch (3’ A–C) were prepared as described previously [11]. The mismatch-provoked excision assay was adapted from a published procedure [11].
Cell cycle and viability analyses
Cell cycle analysis was performed either 24 or 48 hr following treatments with the relevant DNA damaging agents, which included 2 mM N-methyl-N-nitrosourea [MNU] or 20 µM cisplatin [CDDP, cis-diamminedichloroplatinum(II)] (Sigma). DNA content was determined using a fluorescence-activated cell sorter (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ). To assess cell viability in response to various concentrations of MNU and cisplatin, a standard 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was performed in 96-well format in which approximately 2000 cells per well were analyzed. The amounts of soluble formazan were determined by the use of an ELX 808 Ultra Microplate Reader at 570 nm (BioTEK Instruments INC, Winooski, UT).
Analysis of protein interactions
Yeast two-hybrid and far-Western analyses were performed as previously described [8]. The cDNAs encoding various hMLH1 mutants were either generated previously [7] or engineered in this study using a standard site-directed mutagenesis approach with overlapping PCR primers. The resulting PCR products were cloned into pET-28a and pBTMd vectors, and were validated by restriction and DNA sequencing analyses.
Western blot analysis and antibodies
Proteins separated on 7.5–20% SDS-PAGE were transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, NH), and then followed by Western blot analysis using the ECL Western detection system (Amersham Pharmacia Biotech, Piscataway, NJ). The primary antibodies used in this study included: α-hMLH1 monoclonal (1:100 dilution, EMD Biosciences, Inc., San Diego, CA), α-hMLH1 polyclonal (1:500 dilution, Santa Cruz Biotechnology Inc., Santa Cruz, CA), α-FLAG monoclonal (1:2000 dilution, Sigma-Aldrich Co., St. Louis, MO), α-hMRE11 polyclonal (1:1000 dilution, Novus Biologicals Inc., Littleton, CO), and α-T7 monoclonal (1:10000 dilution, Novagen, Madison, WI).
RESULTS
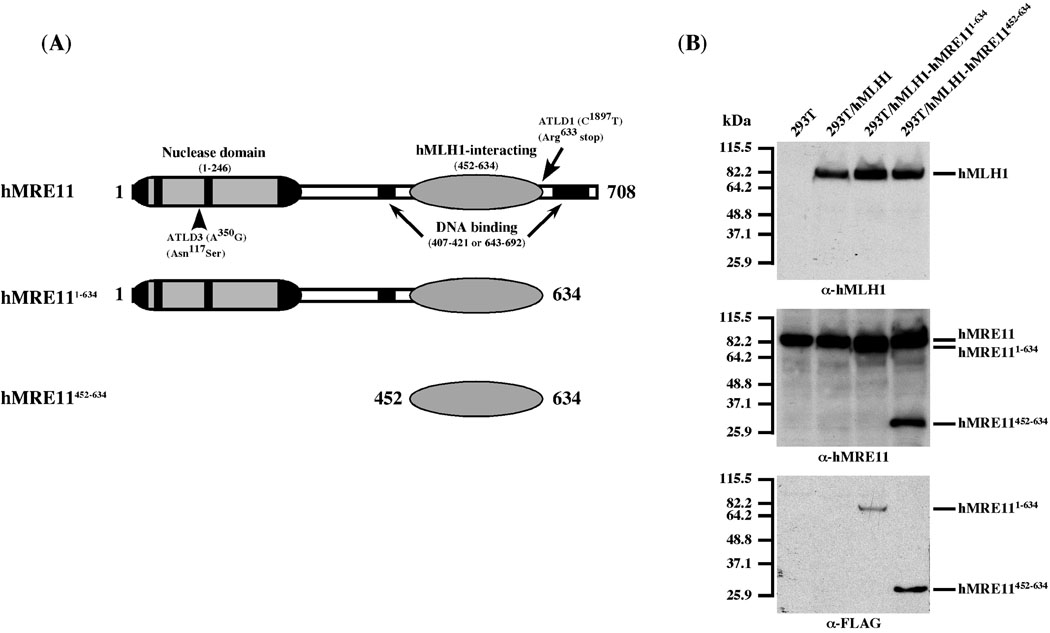
To investigate the role of the hMLH1-hMRE11 interaction in MMR, we generated hMLH1-complemented 293T stable transfectants that are capable of expressing hMRE11452–634 or hMRE111–634 mutant proteins (Fig. 1A). The hMRE11452–634 fragment comprises the minimal hMLH1-interacting domain, while hMRE111–634 (i.e., hMRE11 ATLD1) lacks the last 122 amino acid residues that include a DNA-binding motif [8]. Since 293T cells lack hMLH1 expression [4], the resulting positive clones and the parental cells were stably transfected to express hMLH1. The expression of desired proteins was validated by immunoblots (Fig. 1B). It is evident that over-expression of the hMRE11 fragments has no detectable effects on the levels of the endogenous hMRE11 protein, inferring that the integrity of the hMRE11 complex has not been compromised.
Figure 1.
293T stable cell lines expressing full-length hMLH1 and hMRE11 as well as truncated hMRE11 proteins. (A) Schematic depiction of hMRE11 functional domains and the two truncated hMRE11 polypeptides, hMRE111–634 and hMRE11452–634. (B) Western blot analysis of protein expression in relevant cell lines. The hMLH1 expression was validated by α-hMLH1 immunoblots, and the expression of full-length and truncated hMRE11 proteins was examined by the use of α-hMRE11 and α-FLAG antibodies. kDa, molecular weight (Mr) in thousands.
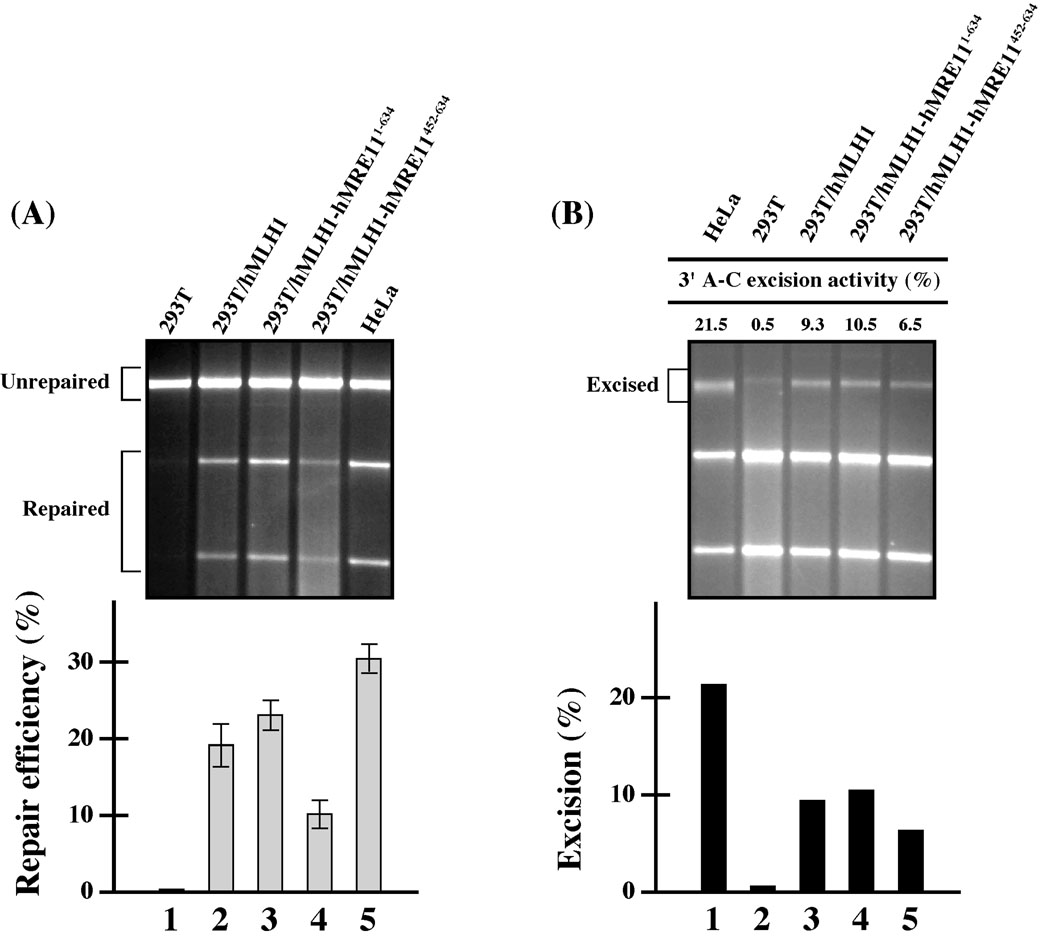
Next, we analyzed whether the expression of hMRE11452–634 and hMRE111–634 fragments in cells would negatively affect MMR activity. Nuclear extracts of 293T derived stable transfectants were used to perform an in vitro MMR assay, in which HeLa nuclear extracts were included as a MMR proficient control. Since it was previously observed that hMRE11 RNAi preferentially affected the 3’ MMR [8], the in vitro MMR assay was conducted with a heteroduplex MMR substrate containing a single A–C mismatch and a strand break 3’ to the mismatch. Consistent with previous observations, HeLa cells were MMR proficient with repair efficiency of 31% for the 3’ A–C substrate (Lane 5, Fig. 2A), while 293T cells were MMR deficient (Lane 1, Fig. 2A) due to methylation-mediated silencing of the hMLH1 gene through hMLH1 promoter suppression [4]. Stable expression of hMLH1 in 293T cells restores 3’ MMR activity to about two-thirds that of HeLa cells (Fig. 2A), and this 3’ repair activity was slightly enhanced when the hMRE111–634 fragment was concomitantly expressed (Lanes 2–3, Fig. 2A). Since hMRE111–634 (i.e., ATLD1) harbors the entire nuclease motif as well as the hMLH1-binding domain (Fig. 1), the above observation is compatible with the view that the hypomorphic hMRE111–634 would be functioning in MMR. Conversely, the repair activity for the 3’ A–C substrate in hMLH1-complemented 293T cells was significantly reduced when the hMRE11452–634 fragment (i.e., the minimal hMLH1-interacting domain) was co-expressed (Lane 4, Fig. 2A). These results are consistent with the idea that hMRE11452–634 could interfere with the MMR process.
Figure 2.
Expression of hMRE11 fragments (hMRE111–634 and hMRE11452–634) in cells affects MMR efficiency. (A) In vitro MMR analysis. The average repair efficiencies and standard deviations (error bars) were determined from three independent data points (bar graph, lower panel). (B) In vitro mismatch-provoked excision analysis. Relative excision activities were determined from two independent experiments.
To validate the role of hMRE11 in 3’ MMR, the nuclear extracts of 293T, 293T/hMLH1, 293T/hMLH1-hMRE11452–634, and 293T/hMLH1-hMRE111–634 cells were analyzed in an in vitro excision assay. Specifically, a HindIII restriction site located immediately 5’ to the A–C mismatch was used to assess 3’ excision activities. The results of the excision analysis were very compatible with that of the MMR analysis (Fig. 2B). It is evident that the expression of hMLH1 in 293T cells can partially restore 3’ excision activity (Fig. 2B). Noticeably, this functional restoration can be compromised by the expression of hMRE11452–634 but not the larger hMRE111–634 fragment (Fig. 2B). The differential effects of these two fragments in MMR underscores the potential involvement of the the nuclease domain of hMRE11 in MMR.
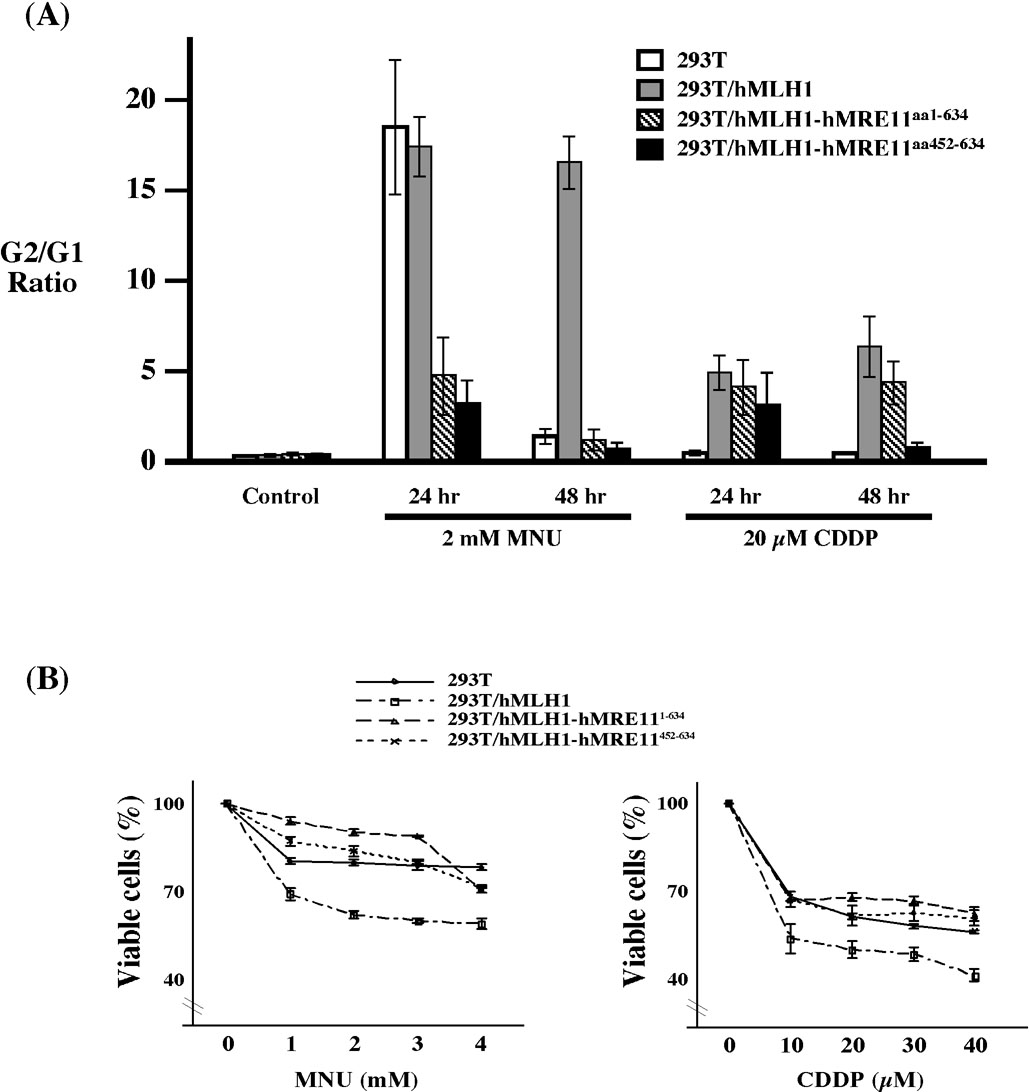
The integrity of the MMR pathway is also known to play critical roles in mediating cellular responses to DNA damaging agents, and MMR deficiency is commonly associated with increased resistance to alkylating agents and platinum compounds [12]. Thus, we investigated cellular responses to MNU and cisplatin to delineate the role of hMLH1-hMRE11 interaction in cell cycle modulation and damage response. In response to MNU, 293T, 293T/hMLH1, 293T/hMLH1-hMRE11452–634, and 293T/hMLH1-hMRE111–634 cells displayed different G2 arrest profiles (Fig. 3A). Cell cycle analysis at 24 hr and 48 hr post treatment suggested that hMLH1 complementation was required for the maintenance of MNU triggered G2 arrest; whereas in the absence of hMLH1, G2 arrest could briefly occur at 24 hr post MNU treatment (Fig. 3A). Remarkably, the MNU-triggered G2 arrest was almost completely blocked after 48 hr, and to a lesser extent at 24 hr, by the presence of hMRE111–634 or hMRE11452–634 fragments (Fig. 3A), suggesting that the hMLH1-hMRE11 complex plays a crucial role in mediating MNU-triggered cellular response. Expression of hMLH1 was also necessary for execution of G2 arrest in response to cisplatin challenge, while cells concomitantly expressing hMRE11452–634 failed to maintain G2 arrest at 48 hr (Fig. 3A). Coherent with the above observation were the results of cell viability analysis revealing that, in contrast to MMR deficient 293T cells, hMLH1 expression sensitized cells to MNU or cisplatin exposure. In contrast, expression of the hMLH1-binding domain containing hMRE111–634 or hMRE11452–634 polypeptides rendered hMLH1-complemented cells resistant to these two drugs (Fig. 3B). Together, this series of experiments clearly implicates the involvement of the hMLH1-hMRE11 complex in the processes of MMR and DNA damage response.
Figure 3.
Expression of hMRE111–634 and hMRE11452–634 abolished hMLH1-dependent G2 arrest triggered by MNU and cisplatin treatments, and rendered cells sensitive to these two drugs. (A) G2/G1 ratio of MNU and cisplatin treated cells. Average G2/G1 ratio and standard deviations (error bars) were determined from three independent experiments. Control, cells that were not treated with either MNU or cisplatin. (B) MTT analysis of cell viability in response to MNU and cisplain treatments. Percentage of cell viability was determined in reference to that of untreated controls, and error bars represent standard deviations of three independent measurements.
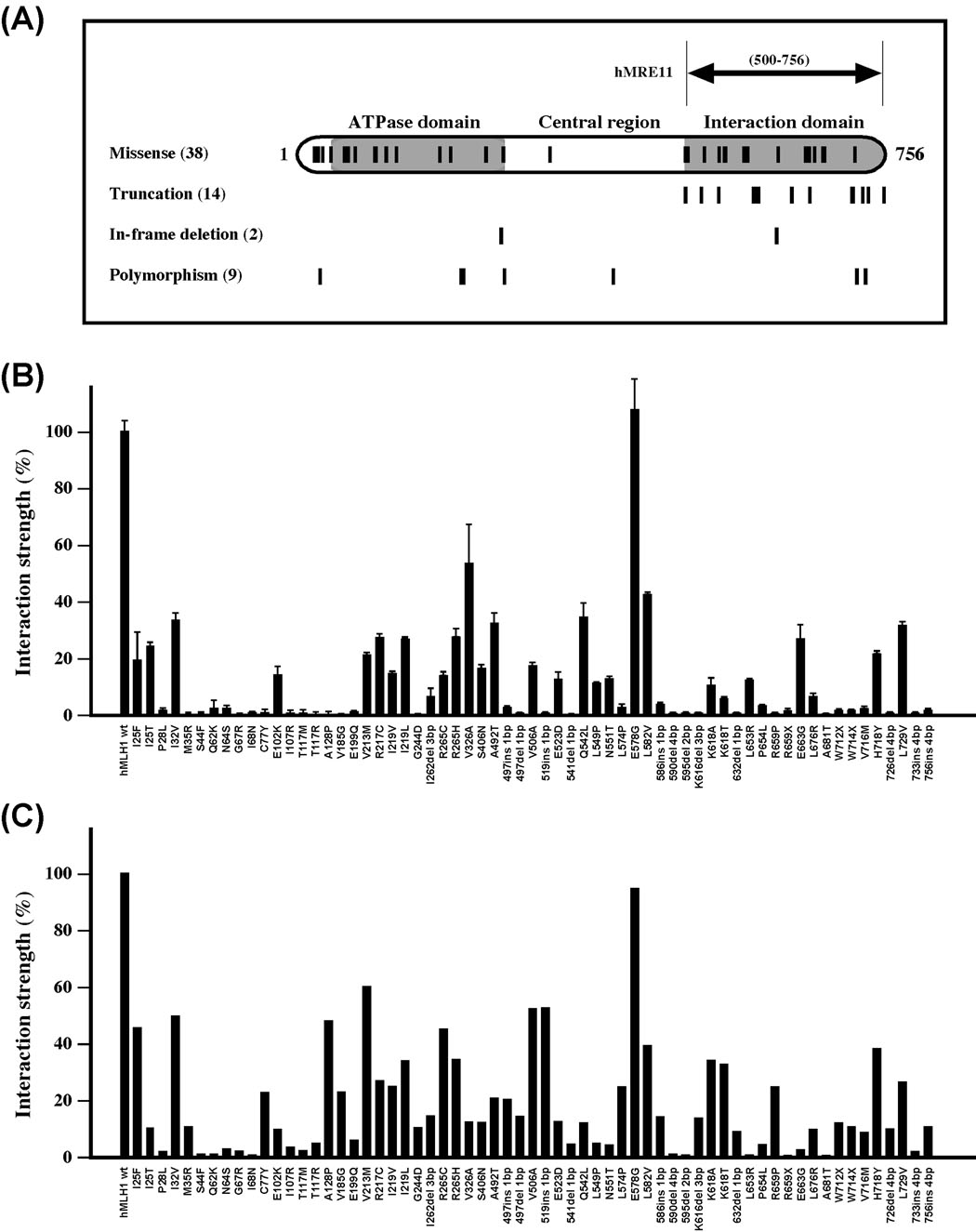
Given that mutations in the hMLH1 gene alone account for over 50% of Lynch syndrome cases (www.insight-group.org) and approximately 25% of identified mutations are scattered single amino acid alterations, a full appreciation of their functional impacts is essential for a better understanding of their roles in the pathogenesis of Lynch syndrome. To this end, we investigated the effects of 63 hMLH1 mutations on hMLH1-hMRE11 interaction by the use of yeast two-hybrid and far-Western analyses (Fig. 4 and Table S1). Specifically, these scattered hMLH1 mutations included 38 missense mutations, 3 nonsense mutations, 11 frame-shift mutations, and 2 in-frame deletion mutants (Fig. 4A and Table S1). In addition, 9 polymorphisms causing single amino acid substitutions (I32V, V213M, R217C, I219V, I219L, R265H, S406N, H718Y, and L729V) were also included in the analysis for comparison purposes (Fig. 4A and Table S1). As shown in Fig. 4B, the results of yeast two-hybrid analysis indicated that the interaction of hMRE11 with hMLH1 variants containing nonsense or frameshift mutations were significantly compromised in comparison with that of the wildtype hMLH1. Twenty-six out of 38 (68.4%) missense mutations and 3 out of 9 (33.3%) polymorphisms also severely impaired the ability of hMLH1 to interact with hMRE11 (Fig. 4B). Importantly, many single amino acid mutations located outside of the hMRE11-interacting domain also significantly disrupted protein interaction (Fig. 4B). The effects of hMLH1 mutations on hMRE11-hMLH1 interaction revealed by two-hybrid assay were largely consistent with the results of far-Western analysis (Fig. 4C, Fig. S1). Since various hMLH1 mutations also significantly affected the levels of protein expression (Fig. S2), the interaction strength measured through in vivo analysis most likely represents a combined effect of both the level of protein expression and the structural alteration of the mutant protein. Thus, under a saturated condition, the far-Western approach could provide a complementary measure of the effects of hMLH1 mutations on protein interaction through minimizing the influence by levels of protein expression. Interestingly, 76% of hMLH1 variants displayed positive concordance between hMLH1-hMRE11 interaction and MMR status (Table S1). Of the hMLH1 variants analyzed in this study, 31 variants were previously analyzed for their impact on MMR as well as for interactions with hPMS2 and hEXO1 by a yeast two-hybrid system [7, 13]. Among this subgroup, 14 hMLH1 mutants (45%) were defective in mediating protein interactions with hPMS2, hEXO1, and hMRE11, and all 14 mutations were MMR defective as well (Table S1). Thirteen mutations displayed differential effects on MMR impairment and protein-protein interactions, suggesting these mutations could cause subtle changes in protein function (Table S1). Four hMLH1 variants (I219L, R265H, L582V, L729V) displayed no apparent defects with regard to protein interaction and MMR, and 5 hMLH1 variants (R217C, V326A, A492T, Q542L, E578G), scattered along the N-terminal ATPase, central, and C-terminal protein interaction domains, showed no apparent defects on protein-protein interaction and yet displayed MMR deficiency (Table S1) – indicative of the existence of yet-to-be-identified molecular mechanisms underlying the functional defects of these mutations. Among the variants of which the status of protein interaction are known, 9 out of 33 missense mutations showed preferential disruption of hMLH1-hMRE11 interaction (Table S1).
Figure 4.
Effects of hMLH1 mutations on hMLH1-hMRE11 interaction. (A) Schematic depiction of hMLH1 functional domains and the distribution of hMLH1 mutations analyzed in this study. The short vertical lines mark the locations of affected amino acid residues of 38 hMLH1 HNPCC missense mutations, as well as 14 truncations, 2 in-frame deletions, and 9 polymorphisms. (B) Yeast two-hybrid analysis of protein interactions. Relative interaction strength between hMRE11 and various mutant hMLH1 proteins were determined in reference to that of the wildtype control, and standard deviations (error bars) were calculated with three independent data points. (C) Far-Western blot analysis of the effects of hMLH1 mutations on the hMLH1-hMRE11 interaction. Membranes that were immobilized with purified hMRE11452–634 polypeptide were probed with crude lysate containing wildtype and various hMLH1 mutant proteins. Standard immunoblotting analysis was used to detect the captured hMLH1 proteins. Protein interactions between hMRE11 and various hMLH1 variants were normalized by comparison with that of the wildtype controls, in which the interaction between wildtype proteins was set to 100%.
DISCUSSION
The current study demonstrates that the integrity of hMLH1-hMRE11 complex is essential for 3’ MMR reaction and DNA damage triggered cellular response, and many hMLH1 mutations can affect the hMLH1-hMRE11 interaction. The results presented in this report are consistent with our previous observation that RNAi-mediated hMRE11 silencing in HeLa cells specifically impaired 3’ MMR and mutations affecting the hMRE11-interacting domain could disrupt the hMLH1-hMRE11 interaction [8].
Given the fact that hMRE11 possesses 3’ to 5’ exonuclease activity [14], our data is compatible with the idea that hMRE11 or the hMRE11-associated complex can act as an alternative nuclease in MMR. It is worthy of note that four redundant exonucleases, two 3’-directed (I and X) and two 5’-directed (VII and RecJ), have been identified in E. coli methyl-directed MMR [1], whereas only Exo1 has been experimentally implicated as an MMR-associated 5’-directed exonuclease in eukaryotic cells [15, 16]. In fact, the moderate functional effects of Exo1 deficiency on MMR in both yeast and mice have strongly suggested the existence of multiple functionally redundant exonucleases in these organisms [15, 16]. However, the appreciation of the precise in vivo role of hMRE11 in MMR has been convoluted by both the essentiality and pleiotropy of this gene in human cells. The different effects that hMRE11452–634 and hMRE111–634 fragments posed on the efficiency of 3’ MMR most likely reflect the role of this protein in MMR. Though the expression of either form of truncated hMRE11 would be expected to affect the participation of hMRE11 in MMR, the observation that only hMRE11452–634 could significantly impair 3’ MMR suggests that the hypomorphic hMRE111–634 (i.e., ATLD1), harboring the entire nuclease domain, is functional in MMR.
Our study has also revealed a role for hMLH1-hMRE11 in DNA damage response. Although hMRE11452–634 could act as an effective blockade of both MNU and cisplatin triggered G2 arrest, expression of the hypomorphic hMRE111–634 only affected MNU induced G2 arrest (Fig. 3A), suggesting that the effects of hMRE111–634 in damage response is not directly related to its role in MMR. It has been reported recently that RNAi-mediated hMRE11 silencing phenocopied that of defective MMR, as both led to a comparable reduction of temozolomide-induced G2 arrest as well as an increase in cellular tolerance to the drug [17].
The observation that 68% of scattered hMLH1 Lynch syndrome missense mutations could cause considerable disruption of the interaction between hMLH1 and hMRE11 has raised the possibility that cancer cells harboring hMLH1 missense mutations will likely display aberrant cellular responses to chemotherapeutics such as methylating agents and platinum compounds. Hence, a thorough mechanistic characterization of the multifaceted hMLH1-hMRE11 interaction will be a necessity not only for a better understanding of their functional role in MMR but also for thorough delineation of the interrelationship between hMLH1-hMRE11 and cellular response to chemotherapeutics, which might be useful in predicting drug effectiveness as well as the development of individually tailored therapeutic strategies in cancer treatment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Y.-S. Chen for her technical assistance. This work was supported by NIH grant CA101796 (C. H.), and F. Yuan was supported by GM072756 (G. L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Peltomaki P, Vasen HF. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis. Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimodaira H, Filosi N, Shibata H, Suzuki T, Radice P, Kanamaru R, Friend SH, Kolodner RD, Ishioka C. Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat. Genet. 1998;19:384–389. doi: 10.1038/1277. [DOI] [PubMed] [Google Scholar]
- 4.Trojan J, Zeuzem S, Randolph A, Hemmerle C, Brieger A, Raedle J, Plotz G, Jiricny J, Marra G. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122:211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- 5.Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nystrom-Lahti M, Perrera C, Raschle M, Panyushkina-Seiler E, Marra G, Curci A, Quaresima B, Costanzo F, D‘Urso M, Venuta S, Jiricny J. Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes Cancer. 2002;33:160–167. [PubMed] [Google Scholar]
- 7.Kondo E, Suzuki H, Horii A, Fukushige S. A yeast two-hybrid assay provides a simple way to evaluate the vast majority of hMLH1 germ-line mutations. Cancer Res. 2003;63:3302–3308. [PubMed] [Google Scholar]
- 8.Vo AT, Zhu F, Wu X, Yuan F, Gao Y, Gu L, Li GM, Lee TH, Her C. hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 2005;6:438–444. doi: 10.1038/sj.embor.7400392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Her C, Vo AT, Wu X. Evidence for a direct association of hMRE11 with the human mismatch repair protein hMLH1. DNA Repair (Amst) 2002;1:719–729. doi: 10.1016/s1568-7864(02)00079-4. [DOI] [PubMed] [Google Scholar]
- 10.Yi W, Wu X, Lee TH, Doggett NA, Her C. Two variants of MutS homolog hMSH5: prevalence in humans and effects on protein interaction. Biochem. Biophys. Res. Commun. 2005;332:524–532. doi: 10.1016/j.bbrc.2005.04.154. [DOI] [PubMed] [Google Scholar]
- 11.Guo S, Presnell SR, Yuan F, Zhang Y, Gu L, Li GM. Differential requirement for proliferating cell nuclear antigen in 5‘ and 3’ nick-directed excision in human mismatch repair. J. Biol. Chem. 2004;279:16912–16917. doi: 10.1074/jbc.M313213200. [DOI] [PubMed] [Google Scholar]
- 12.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signaling. DNA Repair (Amst) 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67:4595–4604. doi: 10.1158/0008-5472.CAN-06-3509. [DOI] [PubMed] [Google Scholar]
- 14.Paull TT, Gellert M. The 3‘ to 5’ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 15.Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Jr, Kneitz B, Yang G, Kunkel TA, Kolodner RD, Cohen PE, Edelmann W. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirzoeva OK, Kawaguchi T, Pieper RO. The Mre11/Rad50/Nbs1 complex interacts with the mismatch repair system and contributes to temozolomide-induced G2 arrest and cytotoxicity. Mol. Cancer Ther. 2006;5:2757–2766. doi: 10.1158/1535-7163.MCT-06-0183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.