Abstract
The urokinase receptor (uPAR) can recognize several ligands. The structural basis for this multiple ligand recognition by uPAR is unknown. This study reports the crystal structures of uPAR in complex with both urokinase (uPA) and vitronectin and reveal that uPA occupies the central cavity of the receptor, whereas vitronectin binds at the outer side of the receptor. These results provide a structural understanding of one receptor binding to two ligands.
Many cell-surface receptors can recognize several ligands. uPAR is one such receptor. uPAR binds to uPA at high affinity and is also capable of interacting with other ligands, including the matrix vitronectin, integrins and G protein–coupled receptor1. This ability to interact with multiple ligands localizes plasminogen activation to the pericellular region, confers an adhesive property to the vitronectin matrix and facilitates uPAR's role in signal transduction; subsequently, uPAR has been implicated in many cellular functions and diverse pathophysiological processes1.
Structures of complexes between soluble uPAR (residues 1−277; suPAR), which lacks the glycophosphatidylinositol moiety, and uPAR antagonists2 or the N-terminal fragment of uPA (residues 1−143; ATF)3 have been determined, showing that three domains in uPAR are packed closely and form a unique central cavity that recognizes the growth factor domain of uPA (residues 1−44; GFD). uPAR binds simultaneously to uPA and the somatomedin B domain (residues 1−44; SMB) of vitronectin4,5. However, the structural basis of multiple ligand recognition by uPAR is unknown.
We determined the crystal structures of the ternary complex of human suPAR–ATF–SMB at 2.8Å and the crystal structure of suPAR–ATF–SMB bound to an anti-uPAR antibody at 2.5Å (Fig. 1, Supplementary Fig. 1, Supplementary Table 1 and Supplementary Methods online). These structures show that the GFD of uPA occupies the central cavity of the receptor, whereas SMB binds to the outer side (the D1 domain and the D1-D2 linker region). SMB and GFD are located on opposite sides of the D1 β-sheet of the receptor, and there is no direct contact between SMB and ATF.
Figure 1.
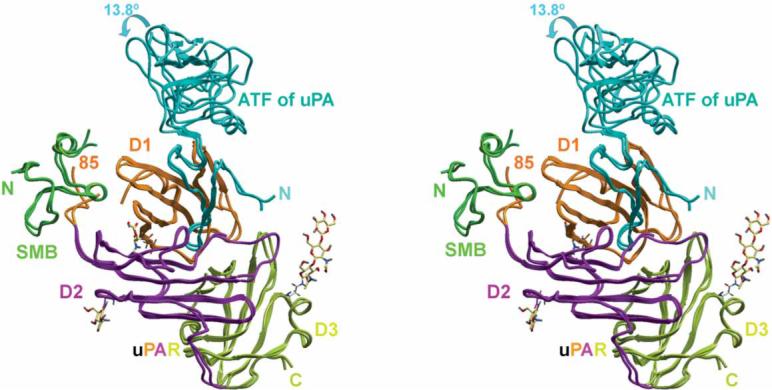
Recognition of both the uPA N-terminal fragment (ATF) and the vitronectin SMB domain by uPAR. Stereoview of superimposed crystal structures of the uPAR–ATF–SMB complex and the uPAR–ATF–SMB-antibody complex are shown (the antibody is omitted for clarity). The carbohydrate moieties of uPAR are shown in sticks. The three domains of uPAR are colored differently.
The superimposition of these two suPAR–ATF–SMB structures shows that the kringle domain of uPA (residues 45−143) undergoes a 13.81 rotation. The remainder of the suPAR–ATF–SMB structures are similar to each other and to our previous suPAR–ATF structure3, suggesting that SMB binding does not perturb the structure of the suPAR–uPA interface.
On the other hand, receptor occupancy by uPA does affect the binding of SMB to uPAR. In the absence of uPA, the suPAR–vitronectin binding affinity was reduced by four-fold6 or more7. uPAR oligomerization was proposed to explain this uPA effect on vitronectin binding to uPAR7. However, it is possible that uPA has a role in stabilizing the active conformation of uPAR, and this may explain the effect of uPA on SMB binding. In the absence of uPA, uPAR may adopt a different conformation8 and thus affect the binding of SMB to uPAR. Determination of the ligand-free structure of uPAR will allow further evaluation of the structural basis of this ligand interplay.
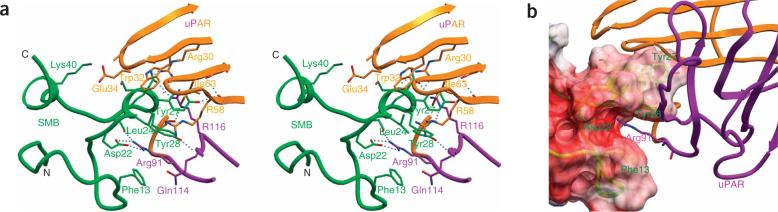
Figure 2a shows the uPAR–SMB interface. uPAR residues Arg91, Trp32, Arg116, Arg30, Ile63, Gln114, Arg58, Ser88, Ser56 and Ser65 (ranked according to their contact areas, Supplementary Table 2 online) contact with SMB residues Tyr27, Tyr28, Leu24, Phe13, Ser26, Asp22, Q29, Glu23, Ser30 and Glu20. This interface consists of an arginine-recognition area and a hydrophobic binding area. The arginine-recognition area includes an open pocket on SMB (residues Phe13, Tyr28 and Asp22) that binds uPAR Arg91 (Fig. 2b). Tyr27 and Tyr28 of SMB insert into a large cavity on uPAR's surface (Fig. 2b). The phenyl ring of SMB Tyr28 contacts with uPAR residues Ile63 and Trp32, mainly through hydrophobic interactions. Taken together, the uPAR–SMB interface shows a high degree of shape and charge complementarity.
Figure 2.
Details of the uPAR–SMB interface. (a) Interaction of the vitronectin SMB domain with uPAR D1 (orange) and D2 domains (magenta) in stereoview. Selected contacting residues in ball-and-stick representation; hydrogen bonds are shown in dashed lines. (b) Residues Phe13, Tyr28 and Asp22 of vitronectin (in ribbon and transparent surface representation) form an open pocket to bind Arg91 of uPAR (in ribbon and stick). Tyr27 and Tyr28 of the SMB domain insert into a large cavity of uPAR, showing shape complementarity of this interface.
The vitronectin binding surface of uPAR was mapped using a single-site alanine-scanning point mutant library of either purified suPAR6 or full-length uPAR expressed on HEK293 cells9. In both studies, uPAR residues Try32, Arg58, Ile63, Arg91 and Tyr92 were identified as key residues for SMB binding in the presence of uPA. On the SMB side, Gly12, Asp22, Leu34, Tyr27, Tyr28, Asp34 and each cysteine residue forming disulfide bonds were identified as key residues for PAI-1 binding. These results are consistent with the current structural model.
Besides uPAR, the SMB of vitronectin also binds to a very different protein, plasminogen activation inhibitor 1 (PAI1)10. Despite the great differences between these two receptors, the SMB uses the same set of residues for receptor recognition. Furthermore, many of these key SMB residues (Phe13, Asp22 and Tyr27, and Leu24) adopt identical conformations, regardless of which receptor they are binding to. The only major differences between these two SMBs are the different side chain conformation of Tyr28 and the use of additional residues (Thr10 and Glu23) for PAI1 binding. Thus, it seems that the SMB of vitronectin uses a structurally conserved set of residues to recognize a diverging pattern on receptors.
Supplementary Material
ACKNOWLEDGMENTS
This research is supported by grants from the US National Institutes of Health (HL08658) and the American Heart Association (0330089N) (to M.H.) and from the British Heart Foundation (to A.Z.). Data for this study were measured at beam lines ID19 and ID24 of Advanced Photon Sources and at beam line X29 of the the National Synchrotron Light Source.
Footnotes
AUTHOR CONTRIBUTIONS Q.H. and L.L. performed the experiments and solved the structures; M.H. refined the structures and wrote the paper; A.Z., A.P.M., G.C.P., J.C., D.E.S., B.F. and B.C.F. provided reagents and/or ideas to this work.
Accession codes. Protein Data Bank: the atomic coordinates of the crystal structures of uPAR in complex with uPA and vitronectin have been deposited with the accession codes 3BT1 and 3BT2, respectively.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Blasi F, Carmeliet P. Nat. Rev. Mol. Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 2.Llinas P, et al. EMBO J. 2005;24:1655–1663. doi: 10.1038/sj.emboj.7600635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huai Q, et al. Science. 2006;311:656–659. doi: 10.1126/science.1121143. [DOI] [PubMed] [Google Scholar]
- 4.Wei Y, et al. J. Biol. Chem. 1994;269:32380–32388. [PubMed] [Google Scholar]
- 5.Deng G, Royle G, Wang S, Crain K, Loskutoff DJ. J. Biol. Chem. 1996;271:12716–12723. doi: 10.1074/jbc.271.22.12716. [DOI] [PubMed] [Google Scholar]
- 6.Gardsvoll H, Ploug M. J. Biol. Chem. 2007;282:13561–13572. doi: 10.1074/jbc.M610184200. [DOI] [PubMed] [Google Scholar]
- 7.Sidenius N, Andolfo A, Fesce R, Blasi F. J. Biol. Chem. 2002;277:27982–27990. doi: 10.1074/jbc.M111736200. [DOI] [PubMed] [Google Scholar]
- 8.Yuan C, Huang M. Cell. Mol. Life Sci. 2007;64:1033–1037. doi: 10.1007/s00018-007-6498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madsen CD, Ferraris GM, Andolfo A, Cunningham O, Sidenius N. J. Cell Biol. 2007;177:927–939. doi: 10.1083/jcb.200612058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou A, Huntington JA, Pannu NS, Carrell RW, Read RJ. Nat. Struct. Biol. 2003;10:541–544. doi: 10.1038/nsb943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.