Abstract
In Klebsiella pneumoniae, the cooccurrence of chromosomal and plasmid-mediated beta-lactamases can hinder their accurate molecular detection. We developed a fast and reliable method that allows the typing of isolates carrying more than one SHV gene. The method is based on pyrosequencing the DNA sequence corresponding to amino acid positions 35, 238, and 240.
Bacteria producing extended-spectrum beta-lactamases (ESBLs) have become a significant clinical problem, and their detection is problematic (4). Many ESBLs have derived from different beta-lactamases by amino acid substitutions that enable the enzymes to hydrolyze various beta-lactam antibiotics. In SHV, the amino acid substitutions leading to an ESBL phenotype occur at a limited number of positions. Especially, the amino acid substitutions G238S and E240K, according to the numbering of Ambler (2), are important for producing the ESBL phenotype (4, 10).
Klebsiella pneumoniae isolates usually carry a chromosomal beta-lactamase, most commonly SHV (3, 8), and the plasmid variants have probably derived from the chromosomal SHV genes (5, 9). The chromosomal beta-lactamases may interfere with detection of the ESBL genes usually residing in plasmids; e.g., the PCR-based methods developed for the detection of ESBLs also amplify the chromosomal variants and form a duplex PCR product whose sequence determination by conventional sequencing requires careful analysis (1). The exact SHV type within an isolate harboring more than one SHV gene can be defined by cloning and sequencing.
Pyrosequencing is a PCR-based sequencing-by-synthesis method (18) that is ideal for mutation analysis and the detection of heterogeneous sequences (6, 7, 12). Pyrosequencing has been applied to the detection of mutations in 23S rRNA (6, 19) and to the typing of beta-lactamase genes (13, 14, 16). In the results of our recent study (15), cyclic sequencing of PCR-amplified SHV genes did not always reveal an unambiguous sequence. This could be explained by the presence of more than one SHV gene in one strain. The present study was designed to test whether the pyrosequencing technique could be used to resolve these ambiguous SHV sequences. To detect the most-common and important sequences found in SHV ESBLs, the DNA sequences corresponding to the amino acid positions 35, 238, and 240 were included to the pyrosequencing assays (10, 17).
We included in this study 40 SHV-positive K. pneumoniae isolates whose SHV gene sequence could not be confirmed by cyclic sequencing due to heterogeneous sequences corresponding to the amino acid positions 35, 238, and/or 240 (15). In addition, 66 K. pneumoniae isolates representing different SHV variants and 10 SHV-positive Escherichia coli isolates were included. The primers used for amplification and pyrosequencing are shown in Table 1. The concentrations of the PCR reagents and temperature-cycling conditions, except for annealing at 54°C and all PCR steps for 30 s, were as described earlier (15).
TABLE 1.
Primers used for amplification and pyrosequencing of the SHV beta-lactamase gene
| Primer | Sequence | Positiona | Reference |
|---|---|---|---|
| SHV_no_PCR-1b | ATGCGTTATATTCGCCTGTG | 199-218 | 20 |
| SHV_pyro_35_Rbio2b,c | Bio-CCGCASAGCASRACTTTAc | 419-402 | This study |
| SHV_cod238_1d | CTGGTTTATCGCCGATAAGA | 870-889 | This study |
| SHV_pyro_238_Rbioc,d | Bio-TTGCCAGTGCTCGATCAGc | 1053-1036 | This study |
| SHV_py35F1mode | CGCAGCCGCTTGAGCAAATTA | 266-286 | This study |
| SHV_seq_238_240e | TATCGCCGATAAGACCGGAG | 876-895 | This study |
Positions are numbered according to the sequence of NCBI accession number AF124984.
PCR primer for position 35.
bio, biotin.
PCR primer for positions 238 and 240.
Pyrosequencing primer for positions 35, 238, and 240.
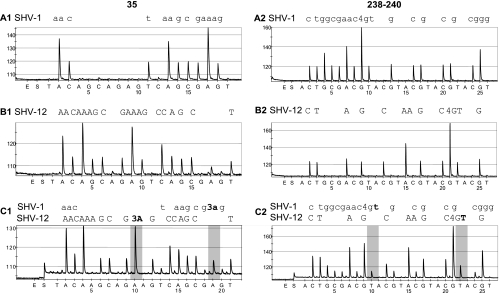
Pyrosequencing was performed by using a PSQ96MA pyrosequencing device and Pyro gold SQA reagents (Biotage AB) according to the manufacturer's instructions. The design of the nucleotide dispensation order for the position 35 assay was TACAGCAGAGTCAGCGAGT and for the assay of positions 238 and 240 was ACTGCG5(ACGT). By combining the results from the two assays, the chromosomal SHV-1 or SHV-11 could be differentiated from plasmid-mediated ESBL SHV genotypes (Table 2). Pyrograms obtained with isolates containing sequences belonging to SHV-1, SHV-12, or both SHV-1 and SHV-12 pyrosequencing groups are presented in Fig. 1.
TABLE 2.
Detection of the SHV substitutions at Ambler positions 35, 238, and 240 by pyrosequencing
| Speciesa | No. of strains with results | SHV gene identified by cyclic sequencing | SHV gene and no. of variants identified by pyrosequencingb
|
Nucleotide sequencec of position:
|
Amino acid(s)d of position:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of variants | SHV(A) | SHV(B) | 35(A) | 35(B) | 238(A) | 238(B) | 240(A) | 240(B) | 35 | 238 | 240 | |||
| K. pneumoniae | 14 | SHV-1 | 1 | SHV-1 | CTA | GGC | GAA | L | G | E | ||||
| K. pneumoniae | 2 | SHV-1 | 1 | SHV-1 | CTA | GGC | GAG | L | G | E | ||||
| K. pneumoniae | 8 | SHV-11 | 1 | SHV-11 | CAA | GGC | GAA | Q | G | E | ||||
| K. pneumoniae | 6 | SHV-11 | 1 | SHV-11 | CAA | GGC | GAG | Q | G | E | ||||
| K. pneumoniae | 1 | SHV-2 | 1 | SHV-2 | CTA | AGC | GAG | L | S | E | ||||
| K. pneumoniae | 3 | SHV-12 | 1 | SHV-12 | CAA | AGC | AAG | Q | S | K | ||||
| K. pneumoniae | 3 | SHV-1 | 2 | SHV-1 | SHV-1 | CTA | CTA | GGC | GGC | GAG | GAA | L | G | E |
| K. pneumoniae | 1 | SHV-1 | 2 | SHV-11 | SHV-1 | CAA | CTA | GGC | GGC | GAG | GAA | L, Q | G | E |
| K. pneumoniae | 1 | SHV-2 | 2 | SHV-2 | SHV-1 | CTA | CTA | AGC | GGC | GAG | GAA | L | G, S | E |
| K. pneumoniae | 1 | SHV-2 | 2 | SHV-11 | SHV-5 | CAA | CTA | GGC | AGC | GAA | AAG | L, Q | G, S | E, K |
| K. pneumoniae | 2 | SHV-2A | 2 | SHV-2A | SHV-11 | CAA | CAA | AGC | GGC | GAG | GAA | Q | G, S | E |
| K. pneumoniae | 3 | SHV-5 | 2 | SHV-5 | SHV-1 | CTA | CTA | AGC | GGC | AAG | GAA | L | G, S | E, K |
| K. pneumoniae | 2 | SHV-5 | 2 | SHV-5 | SHV-11 | CTA | CAA | AGC | GGC | AAG | GAA | L, Q | G, S | E, K |
| K. pneumoniae | 1 | SHV-5 | 2 | SHV-5 | SHV-12 | CTA | CAA | AGC | AGC | AAG | AAG | L, Q | S | K |
| K. pneumoniae | 2 | SHV-5 | 2 | SHV-12 | SHV-1 | CAA | CTA | AGC | GGC | AAG | GAA | L, Q | G, S | E, K |
| K. pneumoniae | 1 | SHV-5 | 2 | SHV-12 | SHV-11 | CAA | CAA | AGC | GGC | AAG | GAA | Q | G, S | E, K |
| K. pneumoniae | 1 | SHV-12/SHV-1 | 2 | SHV-11 | SHV-12 | CAA | CAA | GGC | AGC | GAA | AAG | Q | G, S | E, K |
| K. pneumoniae | 1 | SHV-12 | 2 | SHV-12 | SHV-1 | CAA | CTA | AGC | GGC | AAG | GAA | L, Q | G, S | E, K |
| K. pneumoniae | 12 | SHV-12 | 2 | SHV-12 | SHV-11 | CAA | CAA | AGC | GGC | AAG | GAA | Q | G, S | E, K |
| K. pneumoniae | 1 | SHV-X | 2 | SHV-1 | SHV-11 | CTA | CAA | GGC | GGC | GAG | GAG | L, Q | G | E |
| K. pneumoniae | 1 | SHV-X | 2 | SHV-2 | SHV-1 | CTA | CTA | AGC | GGC | GAG | GAG | L | G, S | E |
| K. pneumoniae | 2 | SHV-X | 2 | SHV-2 | SHV-1 | CTA | CTA | AGC | GGC | GAG | GAA | L | G, S | E |
| K. pneumoniae | 1 | SHV-X | 2 | SHV-2A | SHV-1 | CAA | CTA | AGC | GGC | GAG | GAA | L, Q | G, S | E |
| K. pneumoniae | 4 | SHV-X | 2 | SHV-5 | SHV-1 | CTA | CTA | AGC | GGC | AAG | GAA | L | G, S | E, K |
| K. pneumoniae | 1 | SHV-X | 2 | SHV-11 | SHV-1 | CAA | CTA | GGC | GGC | GAG | GAA | L, Q | G | E |
| K. pneumoniae | 3 | SHV-X | 2 | SHV-11 | SHV-12 | CAA | CAA | GGC | AGC | GAA | AAG | Q | G, S | E, K |
| K. pneumoniae | 9 | SHV-X | 2 | SHV-12 | SHV-1 | CAA | CTA | AGC | GGC | AAG | GAA | L, Q | G, S | E, K |
| K. pneumoniae | 18 | SHV-X | 2 | SHV-12 | SHV-11 | CAA | CAA | AGC | GGC | AAG | GAA | Q | G, S | E, K |
| E. coli | 1 | SHV-11 | 1 | SHV-11 | CAA | GGC | GAG | Q | G | E | ||||
| E. coli | 2 | SHV-2 | 1 | SHV-2 | CTA | AGC | GAG | L | S | E | ||||
| E. coli | 1 | SHV-2A | 1 | SHV-2A | CAA | AGC | GAG | Q | S | E | ||||
| E. coli | 2 | SHV-5 | 1 | SHV-5 | CTA | AGC | AAG | L | S | K | ||||
| E. coli | 4 | SHV-12 | 1 | SHV-12 | CAA | AGC | AAG | Q | S | K | ||||
E. coli strains were used as controls.
“A” sequences are the more-prevalent SHV type determined from the pyrosequencing results for positions 35, 238, and 240. As not all positions are determined, the result is to be considered as a sequence group. “B” sequences are another SHV type detected by pyrosequencing.
Nucleotide sequences determined by pyrosequencing. “A” sequences are from the more-prevalent gene copy. Bold letters indicate nucleotides that differ from SHV-1.
Amino acids were translated from the pyrosequencing results.
FIG. 1.
Sample pyrograms of the SHV 35 (A1, B1, and C1) and 238 and 240 (A2, B2, and C2) assays. The sequences obtained from the traces are indicated above the pyrograms. Pyrograms A1 and A2 are from an isolate harboring only an SHV-1 gene, B1 and B2 are from an isolate with an SHV-12 gene, and C1 and C2 are from an isolate containing sequences belonging to both SHV-1 and SHV-12 pyrosequencing groups. In pyrogram A1, the sequence of position 35 is CTA (Leu), and the sequence of positions 238 and 240 from A2 is GGC GAA, the codons for gly and glu. In pyrogram B1, the mutant codon CAA (gly) is detected, and in B2, codons 238 and 240 are mutated; the sequence is AGC AAG (gly → ser and glu → lys). In pyrogram C1, the proportion of the wild type is lower than that of the mutant since, for example, the highlighted A peak at dispensation 10 deriving from SHV-12 is higher than the peak of dispensation 19 from the SHV-1 copy. Similarly, in pyrogram C2, the proportion of the wild type is lower than that of the mutant since the highlighted peak deriving from SHV-1 is lower than the peak deriving from the mutated copy.
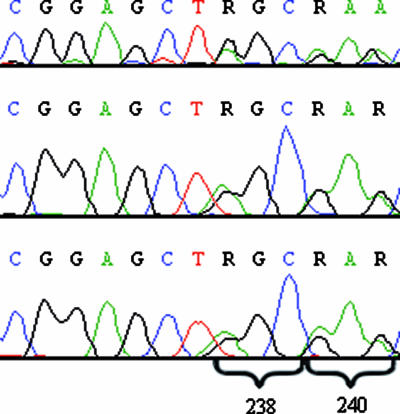
In the 10 E. coli control strains, SHV-2, SHV-2A, SHV-5, SHV-11, and SHV-12 genes were found by cyclic sequencing (Table 2), and the results were confirmed by pyrosequencing. All the 40 K. pneumoniae isolates that could not be typed by cyclic sequencing (Fig. 2) were found by pyrosequencing to have divergent sequences at the nucleotides corresponding to positions 35, 238, and/or 240 (Table 2). The exact sequence combinations of the isolates containing two SHV sequences could be resolved by pyrosequencing, because the most-probable sequences present at the analyzed positions are known and because the pyrosequencing peaks are quantitative (Fig. 1; Table 2). Only 34 of the 66 K. pneumoniae isolates whose SHV type could be determined by cyclic sequencing were found by pyrosequencing to contain only one SHV sequence, SHV-1 (n = 16), SHV-11 (n = 14), SHV-12 (n = 3), or SHV-2 (n = 1), and these pyrosequencing results were in agreement with the results of the cyclic sequencing. Thus, 32 strains found by pyrosequencing to carry more than one SHV gene had been erroneously typed by traditional sequencing to carry one SHV gene (Table 2). Consequently, 72 (67.9%) of the 106 K. pneumoniae isolates were found by pyrosequencing to contain more than one SHV gene (Table 2), indicating the presence of both a plasmid-mediated and a chromosomal copy. The most-common SHV combinations in our material were the SHV-12 pyrosequencing type with the SHV-11 (n = 35) or SHV-1 (n = 12) pyrosequencing type. Contrary to the results of the study of Lee et al. (11), no great difference in the carriage of ESBL genes by K. pneumoniae isolates with SHV-1 or SHV-11 was detected. However, similar to the results of the Korean study, the cocarriage of SHV-12 and SHV-11 was more common than cocarriage of SHV-12 and SHV-1 (11). In addition to the isolates listed in Table 2, one K. pneumoniae isolate was found to apparently carry three SHV genes: two different SHV-1 copies and one SHV-2 copy. The SHV type of this isolate was determined as SHV-2 by cyclic sequencing.
FIG. 2.
Electropherograms of the DNA sequence demonstrating the ambiguous sequences at the positions corresponding to amino acid positions 238 and 240 obtained with three different sequencing primers. The first position of codon 238 and the first and third positions of codon 240 are R, and the exact sequence combinations of the codons cannot be determined from these electropherograms.
The 30 K. pneumoniae isolates carrying only an SHV-1 or SHV-11 copy were considered to contain only a chromosomal SHV copy. In addition, 71 of the 72 isolates carrying more than one SHV copy contained either SHV-1 or SHV-11. Consequently, 101 (95.3%) of the K. pneumoniae strains studied were probable chromosomal SHV carriers.
The quantitative pyrosequencing peaks have been shown to indicate the proportion of mutant copies in 23S rRNA (6, 19). In this study, the most-prevalent SHV copy in the PCR product may be determined by comparing the peak heights deriving from the same, invariable nucleotide positions of different SHV variants. For example, the peak heights in pyrograms C1 and C2 of Fig. 1 indicate that the isolate contains SHV-12 and SHV-1 gene copies, of which SHV-12 is prevalent. In this study, however, the prevalence in the PCR product is not necessarily the same as the prevalence in the organism, as the plasmid and chromosomal copies may be amplified with different efficiencies. Nevertheless, the results of this study agree with the hypothesis that most ESBL-type SHVs are plasmid mediated and are present in higher numbers in bacteria, whereas SHV-1 and SHV-11 are usually chromosomally encoded and present only in low copy numbers (5, 11).
In this study, a pyrosequencing method for the identification of SHV ESBL genes in the presence of a chromosomal beta-lactamase is described. The method is also useful in epidemiological studies where the exact identification of SHV type is a prerequisite for analyzing the spread of certain SHV types. However, the exact SHV type has to be determined by using a method that also covers other amino acid positions.
Acknowledgments
We thank the FiRe group for the isolates. Monica Österblad is thanked for language revision.
This work was supported by grants from the Finnish Ministry of Social Affairs and Health, the Valto Takala Foundation, the Paulo Foundation, and the Maud Kuistila Memorial Foundation.
Footnotes
Published ahead of print on 5 May 2008.
REFERENCES
- 1.al Naiemi, N., K. Schipper, B. Duim, and A. Bart. 2006. Application of minimal sequence quality values prevents misidentification of the blaSHV type in single bacterial isolates carrying different SHV extended-spectrum β-lactamase genes. J. Clin. Microbiol. 44:1896-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babini, G. S., and D. M. Livermore. 2000. Are SHV β-lactamases universal in Klebsiella pneumoniae? Antimicrob. Agents Chemother. 44:2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaves, J., M. G. Ladona, C. Segura, A. Coira, R. Reig, and C. Ampurdanes. 2001. SHV-1 β-lactamase is mainly a chromosomally encoded species-specific enzyme in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:2856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haanperä, M., P. Huovinen, and J. Jalava. 2005. Detection and quantification of macrolide resistance mutations at positions 2058 and 2059 of the 23S rRNA gene by pyrosequencing. Antimicrob. Agents Chemother. 49:457-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haanperä, M., J. Jalava, P. Huovinen, O. Meurman, and K. Rantakokko-Jalava. 2007. Identification of alpha-hemolytic streptococci by pyrosequencing the 16S rRNA gene and by Vitek 2. J. Clin. Microbiol. 45:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hæggman, S., S. Löfdahl, A. Paauw, J. Verhoef, and S. Brisse. 2004. Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:2400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hæggman, S., S. Löfdahl, and L. G. Burman. 1997. An allelic variant of the chromosomal gene for class A β-lactamase K2, specific for Klebsiella pneumoniae, is the ancestor of SHV-1. Antimicrob. Agents Chemother. 41:2705-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huletsky, A., J. R. Knox, and R. C. Levesque. 1993. Role of Ser-238 and Lys-240 in the hydrolysis of third-generation cephalosporins by SHV-type beta-lactamases probed by site-directed mutagenesis and three-dimensional modeling. J. Biol. Chem. 268:3690-3697. [PubMed] [Google Scholar]
- 11.Lee, Y. H., B. Cho, I. K. Bae, C. L. Chang, and S. H. Jeong. 2006. Klebsiella pneumoniae strains carrying the chromosomal SHV-11 beta-lactamase gene produce the plasmid-mediated SHV-12 extended-spectrum beta-lactamase more frequently than those carrying the chromosomal SHV-1 beta-lactamase gene. J. Antimicrob. Chemother. 57:1259-1261. [DOI] [PubMed] [Google Scholar]
- 12.Lindström, A., J. Odeberg, and J. Albert. 2004. Pyrosequencing for detection of lamivudine-resistant hepatitis B virus. J. Clin. Microbiol. 42:4788-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naas, T., C. Oxacelay, and P. Nordmann. 2007. Identification of CTX-M-type extended-spectrum-β-lactamase genes using real-time PCR and pyrosequencing. Antimicrob. Agents Chemother. 51:223-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naas, T., L. Poirel, and P. Nordmann. 2006. Pyrosequencing for rapid identification of carbapenem-hydrolysing OXA-type β-lactamases in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:1236-1240. [DOI] [PubMed] [Google Scholar]
- 15.Nyberg, S. D., M. Österblad, A. J. Hakanen, P. Huovinen, J. Jalava, and the Finnish Study Group for Antimicrobial Resistance. 2007. Detection and molecular genetics of extended-spectrum beta-lactamases among cefuroxime-resistant Escherichia coli and Klebsiella spp. isolates from Finland, 2002-2004. Scand. J. Infect. Dis. 39:417-424. [DOI] [PubMed] [Google Scholar]
- 16.Poirel, L., T. Naas, and P. Nordmann. 2006. Pyrosequencing as a rapid tool for identification of GES-type extended-spectrum β-lactamases. J. Clin. Microbiol. 44:3008-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Randegger, C. C., A. Keller, M. Irla, A. Wada, and H. Hachler. 2000. Contribution of natural amino acid substitutions in SHV extended-spectrum β-lactamases to resistance against various β-lactams. Antimicrob. Agents Chemother. 44:2759-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronaghi, M., M. Uhlen, and P. Nyren. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363-365. [DOI] [PubMed] [Google Scholar]
- 19.Sinclair, A., C. Arnold, and N. Woodford. 2003. Rapid detection and estimation by pyrosequencing of 23S rRNA genes with a single nucleotide polymorphism conferring linezolid resistance in enterococci. Antimicrob. Agents Chemother. 47:3620-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tofteland, S., B. Haldorsen, K. H. Dahl, G. S. Simonsen, M. Steinbakk, T. R. Walsh, A. Sundsfjord, and the Norwegian ESBL Study Group. 2007. Effects of phenotype and genotype on methods for detection of extended-spectrum-β-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J. Clin. Microbiol. 45:199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]