Abstract
The impermeability of the outer membrane in combination with drug efflux are major determinants of the natural drug resistance of mycobacteria. β-Lactams are the most widely used antibiotics for treatment of bacterial infections. However, it is unknown how β-lactams enter Mycobacterium tuberculosis and whether efflux pumps exist that can export these drugs out of the cell. To identify the molecular mechanisms of M. tuberculosis resistance to β-lactams, a library of 7,500 transposon mutants was generated in the model organism Mycobacterium bovis BCG. Thirty-three unique insertion sites were determined that conferred medium or high-level (≥2,000 μg/ml) resistance to ampicillin. Three mutants in sulfolipid synthesis or transport were highly resistant to ampicillin, indicating an indirect effect of the lipid composition on the outer membrane permeability of M. bovis BCG to ampicillin. Mutants with insertions in genes encoding surface molecules such as PPE proteins or lipoarabinomannan were also completely resistant to ampicillin, thus suggesting a lack of transport across the outer membrane. Insertion of the transposon in front of bcg0231 increased transcription of the gene and concomitantly the resistance of M. bovis BCG to ampicillin, streptomycin, and chloramphenicol by 32- to 64-fold. Resistance to vancomycin and tetracycline was increased four- to eightfold. Bcg0231 and Rv0194 are almost identical ATP-binding cassette transporters. Expression of rv0194 significantly reduced accumulation of ethidium bromide and conferred multidrug resistance to Mycobacterium smegmatis. Both effects were abrogated in the presence of the efflux pump inhibitor reserpine. These results demonstrate that Rv0194 is a novel multidrug efflux pump of M. tuberculosis.
Tuberculosis (TB) is caused by Mycobacterium tuberculosis and kills approximately 2 million people each year (22). Due to the intrinsic resistance of M. tuberculosis to many antibiotics, chemotherapy of TB is restricted to a very limited number of drugs, which have to be used in combination for at least 6 months. Infections with multidrug-resistant M. tuberculosis strains require a prolonged treatment with second-line drugs for up to 2 years, which increases the cost of treatment drastically (56). Even more alarming are the extensively drug-resistant strains of M. tuberculosis, which are resistant not only to isoniazid and rifampin but also to at least two second-line anti-TB drugs (38). A better understanding of resistance mechanisms to known antibiotics may be one way to overcome these problems. A major determinant of the intrinsic resistance of M. tuberculosis is its unique outer membrane, which presents an effective permeability barrier to both hydrophobic and hydrophilic solutes (7). However, the mycobacterial outer membrane can only slow down the influx of drugs and is required to act in synergy with alternative mechanisms to effectively decrease the intracellular concentration of drugs. Recent evidence suggests that multidrug resistance of M. tuberculosis is associated with constitutive or inducible expression of efflux systems (39). By using a bioinformatical approach it was predicted that at least 26 complete and 11 incomplete ATP-binding cassette superfamily (ABC) transporters and 16 major facilitator superfamily proteins exist in M. tuberculosis. Involvement of several of them in transport of aminoglycosides, fluoroquinolones, chloramphenicol, isoniazid, rifampin, and tetracyclines has been demonstrated (15).
β-Lactams are the most widely used antibiotics for treatment of bacterial infections (43). However, they are not regarded as useful for treatment of tuberculosis, because M. tuberculosis is considered intrinsically resistant to β-lactams (10). The production of the β-lactamase BlaC was assumed to be the most significant factor for the resistance of M. tuberculosis to β-lactam antibiotics (19). However, other factors, such as permeability of the cell wall and low affinity of binding to penicillin-binding proteins, cannot be dismissed (10). M. tuberculosis does not grow in vitro with ampicillin concentrations of 100 μg/ml or higher, indicating that it is somewhat susceptible to these antibiotics. Indeed, imipenem was used successfully to treat TB patients infected with multidrug-resistant strains (11). The aim of our work was to identify mechanisms that account for the susceptibility of M. tuberculosis to β-lactam antibiotics, in particular to identify proteins involved in uptake.
To identify genes associated with high resistance of M. tuberculosis to β-lactam antibiotics, we performed transposon mutagenesis of the model organism Mycobacterium bovis BCG. Here we describe the identification of 33 transposon mutants with increased resistance to ampicillin. Insertions of the transposon were found in genes involved in cell wall synthesis and assembly, metabolism, and in genes of unknown function. Our investigations revealed a mutant with an insertion of the transposon in front of the gene bcg0231, which leads to a drastically increased resistance of M. bovis BCG to ampicillin and several other unrelated drugs. Our results provide evidence that the almost identical gene rv0194 encodes a novel multidrug efflux pump of M. tuberculosis.
MATERIALS AND METHODS
Chemicals and enzymes.
Hygromycin B was purchased from Calbiochem. All other chemicals were purchased from Merck, Roth, or Sigma at the highest purity available. Enzymes for DNA restriction and modification were from New England Biolabs and Invitrogen. Oligonucleotides were obtained from IDT.
Bacterial strains, media, and growth conditions.
Escherichia coli DH5α was used for cloning experiments and was routinely grown in Luria-Bertani broth at 37°C. Mycobacterium smegmatis strains were grown at 37°C in Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol and 0.05% Tween 80 or on Middlebrook 7H10 plates supplemented with 0.5% glycerol. M. bovis BCG (strain Institut Pasteur) was grown in Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% oleic acid-albumin-dextrose-catalase (OADC; Remel) or on Middlebrook 7H10 plates supplemented with 0.5% glycerol and 10% OADC (Remel). Antibiotics were used when required at the following concentrations: hygromycin, 200 μg/ml for E. coli and 50 μg/ml for mycobacteria; kanamycin, 30 μg/ml.
Construction of plasmids.
Our previous expression vectors were based on transcriptional fusions in which the Shine-Dalgarno sequence had to be included in the forward primer (26). To provide an alternative cloning strategy with much shorter forward primers, a PacI restriction site which is not present in the M. tuberculosis genome was used, making cloning with this enzyme very convenient, and was engineered between the gene and the Shine-Dalgarno sequence in this vector backbone. To this end, the mspA gene was amplified by PCR using pMN006 (52) as a template with the oligonucleotides pMS-Seq1 and MspA_SD (see Table S1 in the supplemental material) that introduced SphI and PacI restriction sites and a synthetic Shine-Dalgarno sequence which efficiently initiates translation of gfp (unpublished results). The PCR fragment was digested with SphI and HindIII and cloned into the plasmid pMN013 (26) digested with the same restriction endonucleases. This cloning step yielded the vector pML653 in which the Shine-Dalgarno sequence was separated from the translation start site by a PacI restriction site. In this expression vector, genes can be cloned as translational fusions using the restriction sites PacI/HindIII. Promoters can be exchanged using the SpeI and SphI sites.
Then, the rv0194 gene was amplified from genomic DNA of M. tuberculosis H37Rv (obtained from Colorado State University as part of National Institutes of Health, [NIAID] contract HHSN266200400091C entitled “Tuberculosis Vaccine Testing and Research Materials”) using the oligonucleotides rv0194_F4 and rv0194_Hind2, which introduced the HindIII restriction site at the 3′ end (see Table S1 in the supplemental material). The rv0194 PCR fragment was digested with HindIII and cloned into pML653 digested with PacI. The 5′ overhanging ends of the PacI sites were removed by T4 DNA polymerase following HindIII restriction digestion to obtain a 5-bp distance between the Shine-Dalgarno sequence and the rv0194 translation start site in the overexpression vector pML655. In addition the rv0194 expression cassette, pML655, features the pAL5000 origin for replication in mycobacteria, the ColE1 origin for replication in E. coli, and a hyg resistance gene.
Construction and analysis of a transposon library of M. bovis BCG.
The suicide plasmid vector pPR32, containing IS1096::Km, was used to generate a transposon insertion mutant library of M. bovis BCG as described previously (40). The vector pPR32 was electroporated into M. bovis BCG. After recovery at 32°C, the bacteria were plated on 7H10 agar containing kanamycin and incubated at 32°C for 5 to 7 weeks. The colonies were streaked on plates containing kanamycin and gentamicin to prevent the premature loss of the plasmid. Clones were picked from five Kmr/Gmr candidates and transferred into 7H9 liquid medium supplemented with kanamycin and gentamicin. Incubation was at 32°C for 3 to 4 weeks. The cultures were filtered through a filter with a pore size of 5 μm (Sartorius) to remove cell clumps. The filtrate was grown for two further weeks until an optical density at 600 nm (OD600) of 0.4 to 0.5 was achieved. Approximately 107 cells were plated on 7H10 agar containing kanamycin and 2% sucrose. Incubation of the plates occurred at 39°C for 3 weeks. Counterselection against the vector pPR32 and selection with kanamycin resulted in approximately 7,500 transposon mutants. This corresponds to a transposon efficiency of 1.5 × 10−3. Ten colonies were arbitrarily picked, and chromosomal DNA was prepared and analyzed by Southern blotting as described elsewhere (52) to examine the randomness of the IS1096::Km insertions. To select for ampicillin-resistant mutants, the library was washed from the plates, passed through a 5-μm filter, and plated on 7H10 plates supplemented with 100 μg/ml of ampicillin. To identify the insertion sites of the transposon, ligation-mediated PCR was employed as described previously (44). Chromosomal DNA was prepared and used as a template for PCR with the primers Salgd and Tn_mut_seq2 or Tn_mut_seq4 and IS2; primers Salgd and IS2 were used for sequencing (see Table S1 in the supplemental material). The resulting sequences were compared with the M. bovis BCG genome sequence using Blast analysis (http://genolist.pasteur.fr/BCGList/).
RNA preparation.
Total RNA of M. bovis BCG was isolated by the Trizol method as recommended by the manufacturer (Invitrogen). Briefly, cultures were grown in 30 to 60 ml of corresponding medium until late log phase. A 35-ml volume of GTC buffer (5 M guanidium thiocyanate, 0.5% sarcosyl, 0.5% Tween 80, 1% β-mercaptoethanol) was added and centrifuged at 10,000 × g for 10 min at 4°C. The pellet was resuspended in 1.5 ml Trizol and lysed by agitation with glass beads (FastRNA Tubes-Blue) in a FastPrep FP120 bead beater apparatus (Bio-101) three times for 45 seconds at level 6.5. Suspensions were cooled on ice for 5 min between agitation steps. A 500-μl volume of chloroform was added, and centrifugation was done for 5 min at 14,000 × g. The upper phase was transferred to a new tube containing an equal volume of isopropanol. Tubes were incubated for 20 min at −80°C and centrifuged at 14,000 × g for 20 min at 4°C. The pellet was washed with 70% ethanol, dried, and resuspended in 100 μl distilled water. Further purification of samples was performed using a Nucleospin RNAII kit (Macherey-Nagel) following the instructions of the manufacturer.
Dot blot analysis.
The probe for the rv0194 gene was amplified from pML655 by PCR using the primers Rv0194_F1 and Rv0194_rev_T7 (see Table S1 in the supplemental material). The probe for the 16S rRNA gene was amplified from chromosomal DNA of M. bovis BCG using the primers 16SNbfw and 16SrevT7Prom (see Table S1 in the supplemental material). A recognition site for T7 RNA polymerase was added to the 5′ ends of the reverse primers (see Table S1 in the supplemental material). The probes were labeled with digoxigenin by in vitro transcription. The dot blot experiments were carried out as described previously (24). The amount of RNA was quantified photometrically. A 7.2-μg aliquot of RNA was spotted in triplicate onto the blot for each sample. To obtain a visible signal for the bcg0231 mRNA in comparison to the standard 16S rRNA, the exposure time of the blot was increased to 700 s. The LabWorks 4.6 software (UVP) was used for image analysis of the dot blot. The lane profile of the dots was analyzed to examine saturation of the signals. The amount of RNA in the dots was quantified using integrated optical density analysis. The signals for the bcg0231 transcripts were normalized to those of 16S rRNA in the same sample.
Determination of antibiotic susceptibility.
To determine MICs of M. smegmatis and M. bovis BCG strains, a microplate Alamar blue assay (MABA) was used as described previously (20) with some modifications (O. Danilchanka, M. Pavlenol, and M. Niederweis, submitted for publication). Final drug concentrations for M. smegmatis were as follows: ampicillin, 62.5 to 2,000 μg/ml; erythromycin and vancomycin, 0.3125 to 10 μg/ml; chloramphenicol and novobiocin, 2 to 64 μg/ml; tetracycline, 0.01875 to 0.6 μg/ml; kanamycin, 0.156 to 5 μg/ml; ciprofloxacin, ofloxacin, and levofloxacin, 0.8 to 25.6 μg/ml. Final drug concentrations for M. bovis BCG were as follows: ampicillin, 62.5 to 2,000 μg/ml; vancomycin, 1.25 to 40 μg/ml; streptomycin, 0.25 to 8 μg/ml; chloramphenicol, 4 to 128 μg/ml; tetracycline, 0.5 to 16 μg/ml. The MICs were defined as the lowest concentration of antibiotic which reduced the viability of the culture by at least 90% as determined by fluorescence measurements at room temperature in top-reading mode at an excitation wavelength of 530 nm and an emission wavelength of 590 nm using a Synergy HT reader (Bio-Tek).
β-Lactamase activity assay.
The β-lactamase activity of M. bovis BCG was determined by measuring the hydrolysis of nitrocefin by whole cells as described elsewhere (Danilchanka et al., submitted). Briefly, cells of M. bovis BCG strains were grown to saturation (OD600, 2.0 to 4.0). Culture supernatants were filtered through 0.2-μm filters (Pall Corporation) twice to obtain cell-free culture filtrates. To obtain lysates, cells were pelleted and washed in ice-cold 1× phosphate-buffered saline (PBS) buffer (pH 7.4). The cell pellets were resuspended in a 1/30 volume of 1× PBS containing corresponding amounts of protease inhibitor cocktail (Sigma) and DNase I (New England Biolabs). Cells were disrupted by agitation with glass beads (FastRNA Tubes-Blue) in a FastPrep FP120 bead beater apparatus (Bio-101) twice for 30 seconds at level 6.0 with 5 min of rest on ice between agitations. Cell debris was removed by centrifugation and filtered twice through 0.2-μm filters. Protein concentrations were determined using a bicinchoninnic acid protein assay kit (Pierce). Nitrocefin was added to a final concentration of 200 μM in 1× PBS (pH 7.4), and hydrolysis was monitored as a change in absorbance at 490 nm using a microplate reader (Synergy HT; Bio-Tek). The activities of β-lactamases for each strain were determined as the A490 min−1 mg of total protein−1.
Accumulation of ethidium bromide by mycobacteria.
The accumulation of ethidium bromide by mycobacteria was measured as described previously with some modifications (33). M. smegmatis was grown to early exponential phase (OD600, 0.6 to 1.0). The cells were pelleted by centrifugation at room temperature, resuspended in uptake buffer (50 mM KH2PO4 [pH 7.0], 5 mM MgSO4), diluted to an OD600 of 0.5, and preenergized with 25 mM glucose for 5 min. One hundred microliters of cells was added per well of black, clear-bottomed 96-well microplates (Greiner Bio-One). Ethidium bromide was added to a final concentration of 20 μM, and its entry was measured at room temperature in top-reading mode at an excitation wavelength of 530 nm and an emission wavelength of 590 nm using a Synergy HT reader (Bio-Tek). When required, reserpine was added after 8 min of incubation with ethidium bromide at a final concentration of 0.1 mM.
Susceptibility of M. smegmatis to ethidium bromide.
The susceptibility of M. smegmatis to ethidium bromide was tested as described previously (18). Shortly, M. smegmatis was grown overnight in Middlebrook 7H9 medium supplemented with 0.05% Tween 80 and 50 μg/ml hygromycin and filtered through a 5-μm filter (Sartorius) to remove cell clumps. Cells were diluted in the same medium to an approximate OD600 of 0.04. Bacterial growth was monitored by measuring the optical density of the cultures at 600 nm. Ethidium bromide was added to the cultures at final concentrations from 1.56 μM to 12.5 μM. When required reserpine was added at a final concentration of 8 mM.
RESULTS
Isolation of M. bovis BCG mutants resistant to ampicillin.
To identify molecular mechanisms of resistance of slowly growing mycobacteria such as M. tuberculosis to β-lactam antibiotics, we used M. bovis BCG as a model organism and generated a transposon library. The transposon IS1096::Km was chosen, which inserts randomly with a high frequency into mycobacterial genomes (36). A culture of a clone from M. bovis BCG containing pPR32 with the transposon IS1096::Km (40) was plated under conditions nonpermissive for replication of the vector, and this yielded approximately 7,500 kanamycin-resistant clones. To examine the uniqueness of the insertions, chromosomal DNA was prepared from 10 randomly selected clones from the library. Southern blot analysis showed the presence of the transposon in all clones at different positions in the chromosome (not shown). This indicated a random transposition of IS1096::Km into the chromosome of M. bovis BCG. To select mutants with a high resistance to β-lactam antibiotics, the library was washed from the plates and filtered to remove cell clumps. Serial dilutions were plated on 7H10 agar with 100 μg/ml ampicillin, on which wild-type (wt) M. bovis BCG did not grow. Seventy-eight ampicillin-resistant mutants were obtained. MICs of ampicillin for all transposon mutants were higher than 62.5 μg/ml for wt M. bovis BCG as determined in a MABA. Twenty-one mutants were completely resistant to ampicillin (≥2,000 μg/ml), while 11 mutants showed a moderate resistance with MICs of 250 to 500 μg/ml (Table 1). Twenty mutants with MICs lower than 250 μg/ml were excluded from further analysis.
TABLE 1.
Bioinformatic analysis of insertion sites of the mutants resistant to ampicillina
| Group and strain no. | M. bovis BCG gene | M. tuberculosis gene | Position (bp) of insertion | MIC of ampicillin (μg/ml) | Gene function |
|---|---|---|---|---|---|
| Group A | |||||
| ML1075 | gca | rv0112 | +899 | 2,000 | Putative GDP-mannose 4,6-dehydratase; LAM synthesis (51) |
| ML1010 | ppe12 | rv0755c | +43 | 500 | PPE family protein |
| ML1041 | ppe24* | rv1753c | +1707 | >2,000 | PPE family protein; required for growth of M. marinum in macrophages (37) |
| ML1061 | lppA | rv2543 | +19 | >2,000 | Putative lipoprotein |
| ML1058 | lprR* | rv2203c | +320 | 2,000 | Putative lipoprotein |
| ML1064 | lppB | rv2544 | +569 | 2,000 | Putative lipoprotein |
| ML1036 | agpS | rv3107c | +277 | 250 | Putative alkyldihydroxyacetonephosphate synthase (lipid biosynthesis) |
| ML1007 | ppe53* | rv3159c | +988 | 2,000 | PPE family protein; virulence factor of M. marinum (49); required for growth of M. marinum in macrophages (37) |
| ML1040 | papA2 | rv3820c | +1390 | 2,000 | Putative polyketide synthase-associated protein; sulfolipid synthesis (5) |
| ML1047 | fadD23 | rv3826 | +544 | 2,000 | Putative fatty-acid-coenzyme A ligase; sulfolipid synthesis (31) |
| Group B | |||||
| ML1069 | gmhA | rv0113 | +205 | 2,000 | Putative phosphoheptose isomerase |
| ML1013 | cpsY* | rv0806c | +923 | 2,000 | Putative UDP-glucose-4-epimerase |
| ML1009 | cyp121* | rv2276 | +368 | 2,000 | Cytochrome P450 |
| ML1006 | bcg3787* | rv3727 | +720 | 2,000 | Putative oxidoreductase |
| Group C | |||||
| ML1025 | bcg3145 | rv3124 | +435 | 250 | Putative transcriptional regulator |
| Group D | |||||
| ML1065 | bcg0061 | rv0030 | +96 | >2,000 | Unknown |
| ML1048 | bcg1567c | rv1503c | +519 | 250 | Unknown; survival in macrophages (47) |
| ML1030 | bcg1988c | rv1949c | +849 | 2,000 | unknown; LAM synthesis (51) |
| ML1053 | bcg2326c* | rv2307B | +271 | 2,000 | Unknown |
| ML1029 | bcg2734c* | rv2721c | +524 | >2,000 | Putative conserved transmembrane Ala/Gly-rich protein |
| ML1038 | bcg2735 | rv2722 | +78 | 2,000 | Unknown |
| ML1046 | bcg2743 | rv2730 | +33 | 500 | Unknown |
| ML1052 | bcg2824 | rv2806 | +78 | >500 | Unknown |
| ML1050 | bcg2827 | rv2809 | +41 | 2,000 | Unknown |
| ML1037 | bcg3693 | rv3635 | +1765 | 250 | Unknown |
| ML1012 | bcg3960c* | rv3903c | +1255 | >2,000 | Unknown |
| Group E | |||||
| ML1034 | bcg0231 | rv0194 | −54 | 2,000 | ABC transporter |
| ML1060 | fadD25/mmpL12 | rv1521/rv1522c | −206 | 250 | Fatty acid-coenzyme A ligase/conserved transmembrane protein |
| ML1062 | pks11 | rv1665 | −18 | >2,000 | Chalcone synthase |
| ML1044 | ppe33b | rv1810 | −178 | 250 | PPE family protein |
| ML1042 | bcg2123c/pe22 | rv2104c/rv2107 | −509 | 500 | Putative transposase/PE family protein |
| ML1051 | bcg2965 | rv2943 | −420 | >500 | Probable transposase for insertion sequence IS1533 |
| ML1005 | mmpL8 | rv3823c | −76 | ND | Conserved transmembrane protein; sulfolipid transport (13) |
MICs were determined by MABA. The MIC of ampicillin for wt M. bovis BCG was 62.5 μg/ml. Transposon mutants that were resistant to 125 μg/ml are not listed. The open reading frame numbers and annotations of transposon mutants that were resistant to at least 250 μg/ml were taken from the genome websites for M. bovis BCG Pasteur 1173P2 (bcg) and M. tuberculosis H37Rv (rv), respectively, at http://genolist.pasteur.fr. Group A contains proteins associated with cell wall synthesis and assembly, group B contains enzymes involved in metabolic functions, group C contains regulators, group D contains proteins with unknown functions, and group E contains proteins with unknown effects of mutations (intergenic insertions). The mutants are ordered according to their gene tag within the functional groups. The position of insertion was determined relative to the predicted gene start in M. bovis BCG. Negative values indicate insertion of the transposon upstream of the gene. Two or more transposon mutants with identical insertion sites were obtained for genes marked with an asterisk. ND, not determined.
Sequence analysis and functional classification of ampicillin-resistant transposon mutants.
To determine the insertion sites of the transposon, chromosomal DNA was prepared from all mutants and was analyzed by ligation-mediated PCR (44). Thirty-three unique insertion sites in M. bovis BCG were determined that conferred a medium or high level of resistance to ampicillin (Table 1). The mutants were grouped into four functional classes based on the predicted or known functions of the disrupted genes (Table 1). The vast majority of the disrupted genes (11/31) encode proteins involved in cell wall biosynthesis or assembly (Table 1, group A). This was not unexpected, considering the periplasmic location of the transpeptidases and the necessity of ampicillin to cross the permeability barrier established by the outer membrane to reach these targets. Other mutant classes included genes involved in general metabolism and genes of unknown function. Six of the sequenced mutants had insertions in intergenic regions (Table 1).
The mutant ML1034 is highly resistant to multiple drugs.
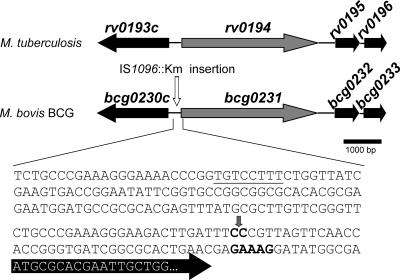
In the ML1034 mutant, the transposon had inserted 54 bp in front of the predicted start codon of the open reading frame bcg0231 in M. bovis BCG, which is almost identical to rv0194 from M. tuberculosis (Table 1; Fig. 1). Blast analysis revealed that rv0194 encodes a putative ATP-binding cassette (ABC) transporter. The Rv0194 protein consists of two membrane-spanning domains, each consisting of six predicted transmembrane helices and two cytoplasmic nucleotide-binding domains fused together. Hence, Rv0194 constitutes a complete multidrug efflux pump (6). However, the function of this protein has not been demonstrated experimentally. The bcg0231 and rv0194 genes differ only by one base pair which causes a proline-to-leucine exchange at position 328. This amino acid is located in one of the cytoplasmic loops, is not part of known functional domains of ABC transporters, and should, therefore, not cause any functional difference. The mutant ML1034 was extremely resistant to ampicillin, with its MIC increased by 32-fold from 62.5 μg/ml to 2,000 μg/ml (see Fig. S1 in the supplemental material).
FIG. 1.
Genomic region of the mutant M. bovis BCG ML1034 and its corresponding region in M. tuberculosis H37Rv. The bcg0231 gene and its flanking genes are depicted. Block arrows represent open reading frames. A vertical arrow depicts the insertion of the transposon IS1096::Km. The sequence of the DNA −200 to +18 relative to the rv0194/bcg0231 start codon is shown. This sequence is identical in M. tuberculosis H37Rv and M. bovis BCG. The black arrow depicts the start of the rv0194 gene with the potential start codon ATG. Putative Shine-Dalgarno and extended −10 promoter sequences (4) are shown in bold and underlined, respectively. The annotated functions of the encoded proteins are as follows: Bcg0230c, hypothetical protein; Bcg0231, probable drug transport transmembrane ATP-binding protein cassette transporter; Bcg0232, possible two-component transcriptional regulatory protein; Bcg0233, possible transcriptional regulatory protein.
To examine whether the resistance of this mutant was specific for ampicillin, its sensitivity to several structurally unrelated antibiotics was determined in a MABA (20) (Table 2). The resistance to the small antibiotics chloramphenicol and tetracycline was 64- and more than 8-fold increased, respectively, compared to wt M. bovis BCG. Resistance to streptomycin, one of the first-line drugs used for the treatment of tuberculosis, was increased by 32-fold compared to wt M. bovis BCG. A fourfold increase in the resistance to the large and hydrophilic antibiotic vancomycin was also observed for the ML1034 mutant (Table 2), therefore suggesting that this mutant is highly resistant to multiple antibiotics.
TABLE 2.
Susceptibility of wild-type M. bovis BCG and the ML1034 mutanta
| Antibiotic | MIC (μg/ml) for M. bovis BCG
|
Resistance factor | |
|---|---|---|---|
| Wild type | ML1034 | ||
| Ampicillin | 62.5 | 2,000 | 32 |
| Chloramphenicol | 8 | 512 | 64 |
| Streptomycin | 0.5 | 16 | 32 |
| Tetracyclin | 2 | >16 | >8 |
| Vancomycin | 5 | 20 | 4 |
The ML1034 mutant carries a transposon in front of the rv0194 gene of M. tuberculosis. The MICs were determined in a microplate Alamar Blue assay. The resistance factor is defined as the MICML1034/MICwt ratio.
Insertion of the transposon increases transcription of bcg0231 in the ML1034 mutant.
In case of insertion of the transposon in front of a gene, two possibilities exist: the transposon can inactivate the gene by inactivating the promoter or other signals required for transcription, or alternatively, gene expression can be upregulated or the gene can be expressed constitutively from a promoter inside the transposon. To distinguish between these two possibilities, total RNA was prepared from wt M. bovis BCG and the ML1034 mutant grown to late logarithmic phase. Dot blot experiments were used to quantify the relative amount of bcg0231 mRNA in both strains. While bcg0231 mRNA was barely detectable in wt M. bovis BCG, it was clearly more visible in the ML1034 mutant (Fig. 2A). Quantitative image analysis showed a threefold increase in the amount of bcg0231 mRNA relative to the 16S rRNA in the ML1034 mutant (Fig. 2B). The lane profile across each dot revealed that the signal for 16S rRNA was not saturated in samples 3 and 6 (Fig. 2A). Quantification of these signals revealed an 8.5-fold-increased amount of bcg0231 mRNA in the ML1034 mutant. In conclusion, these results demonstrated that insertion of the transposon increased transcription of the bcg0231 gene in the ML1034 mutant by at least threefold. This result suggested that the ABC transporter Bcg0231 constitutes an efflux pump and its increased expression caused the multidrug resistance of the ML1034 mutant. A similar activation of gene expression by insertions in front of genes due to promoters inside the transposon has been observed previously, for example, for the mpr gene of M. smegmatis (48).
FIG. 2.
bcg0231 mRNA levels are increased in the ML1034 mutant of M. bovis BCG. A. Total RNA was prepared from M. bovis BCG cultures in late logarithmic phase. A 7.2-μg sample of RNA was spotted onto duplicate membranes in triplicate. The bcg0231 mRNA and the 16S rRNA were detected using digoxigenin-labeled probes which were visualized with an antidigoxigenin antibody-alkaline phosphatase conjugate and a chemiluminescent substrate. B. Quantification of the dot blot results. The chemiluminescence of the dots was quantified using integrative optical analysis. The lane profile of the dots was analyzed to examine saturation of the signals. The amount of bcg0231 transcripts was normalized to that of 16S rRNA in the same sample. The bcg0231 amounts detected for the ML1034 mutant were set as 100%.
The β-lactamase activities of wt M. bovis BCG and the mutant ML1034 are identical.
To examine whether altered expression of β-lactamases contributed to the high resistance of the ML1034 mutant to ampicillin, we measured the β-lactamase activity of M. bovis BCG using the nitrocefin hydrolysis assay. The vast majority of the β-lactamase activity of wt M. bovis BCG was cell associated and was approximately 10-fold higher than the activity of the culture filtrate (see Fig. S2 in the supplemental material). Importantly, the β-lactamase activity of the ML1034 mutant was not higher than in the wt strain. Therefore, we concluded that the increased resistance of the ML1034 mutant to ampicillin is not caused by faster hydrolysis of the drug. This result is consistent with the hypothesis that the high resistance of ML1034 to ampicillin is a direct result of the overexpression of the Bcg0231 pump that reduces the accumulation of ampicillin inside the cell. It should be noted that the multidrug resistance of the ML1034 strain also strongly argues against secondary mutations as a cause of this phenotype, because such mutations are specific for a single antibiotic in most cases (57).
Rv0194 confers multidrug resistance to M. smegmatis.
To examine whether the multidrug-resistant phenotype of the ML1034 mutant was directly associated with overexpression of the ABC transporter, the rv0194 expression vector pML655 was transformed into M. smegmatis SMR5 and M. bovis BCG. In several attempts, colonies were only obtained for M. smegmatis, and not for M. bovis BCG. Importantly, overexpression of rv0194 increased the MICs of ampicillin, vancomycin, novobiocin, and erythromycin for M. smegmatis (Table 3). Similar resistance factors were obtained when rv0194 was overexpressed in M. smegmatis mc2155 (data not shown). These results confirmed that the multidrug resistance of the ML1034 mutant was directly associated with overexpression of rv0194.
TABLE 3.
Susceptibility of M. smegmatis overexpressing the rv0194 gene of M. tuberculosisa
| Antibiotic | MIC for M. smegmatis (μg/ml)
|
Resistance factor | |
|---|---|---|---|
| Wild type | rv0194 overexpression | ||
| Ampicillin | 125 | 250 | 2 |
| Chloramphenicol | 32 | 32 | 1 |
| Erythromycin | 2.5 | 10 | 4 |
| Novobiocin | 4 | 8 | 2 |
| Tetracyclin | 0.3 | 0.3 | 1 |
| Vancomycin | 1.25 | 2.5 | 2 |
The MICs were determined in a microplate Alamar Blue assay. The resistance factor is defined as the MICrv0194/MICwt ratio. The wt strain was M. smegmatis SMR5.
Rv0194 reduces accumulation of ethidium bromide in M. smegmatis.
An obvious experiment to examine the function of Bcg0231/Rv0194 would be to measure accumulation of antibiotics whose MICs are increased drastically for the ML1034 mutant. However, uptake by slow-growing mycobacteria is very slow (32), and interpretation of the uptake experiments is complicated by the high amount of surface-adsorbed compounds (unpublished data). This problem is even more pronounced for antibiotics which are substrates of drug efflux pumps (15). Therefore, we have chosen ethidium bromide as a model compound whose uptake can be measured continuously. This assay is based on the increased fluorescence of ethidium bromide by binding to nucleic acids after entry into the bacterial cell (33) and does not therefore suffer from surface adsorption as all radiolabeled compounds. Importantly, expression of rv0194 reduced accumulation of ethidium bromide in M. smegmatis compared to the wt strain (Fig. 3A). When reserpine, an inhibitor of multidrug transporters (1), was added to the rv0194-expressing strain, accumulation of ethidium bromide quickly reached levels observed for wt M. smegmatis (Fig. 3A). Then, we determined the influence of Rv0194 on the ability of M. smegmatis to grow in the presence of ethidium bromide in order to examine whether the efflux activity of Rv0194 also increased the resistance of M. smegmatis. Growth of wt M. smegmatis and the rv0194-overexpressing strain in Middlebrook 7H9 medium did not differ (data not shown). By contrast, addition of 1.56 μM ethidium bromide drastically reduced the growth of wt M. smegmatis compared to the rv0194-expressing strain (Fig. 3B). This was caused by an increase of the MIC of ethidium bromide from 1.25 μg/ml for the wt to 2.5 μg/ml for the rv0194-expressing strain. This effect was reversed by addition of 8 mM reserpine, which completely inhibited growth of both strains (Fig. 3B). These results demonstrate that Rv0194 directly extrudes ethidium bromide and that this activity increases the resistance of M. smegmatis to ethidium bromide. This clearly established the link between the efflux activity of Rv0194 and increased drug resistance of M. smegmatis and strongly suggests that the multidrug resistance of M. bovis BCG ML1034 is associated with increased efflux of antibiotics due to expression of Rv0194.
FIG. 3.
Effects of rv0194 expression in M. smegmatis on accumulation of and killing by ethidium bromide. A. Accumulation of 20 μM ethidium bromide by M. smegmatis SMR5 transformed with control plasmid pMS2 (closed circles) and with the rv0194 expression vector pML655 (open triangles) was measured by fluorescence. A 0.1 mM solution of reserpine was added to half of the culture of SMR5/pML655 after 8 min of incubation with ethidium bromide (closed squares). Fluorescence was measured as relative fluorescence units (RFU) at an excitation wavelength of 530 nm and an emission wavelength of 590 nm. B. Growth of M. smegmatis SMR5 transformed with control plasmid pMS2 (closed circles) and with the rv0194 expression vector pML655 (closed triangles) was measured in the presence of 1.56 mM ethidium bromide. Reserpine at a final concentration of 8 mM was added to cultures of M. smegmatis SMR5 with a control plasmid pMS2 (open circles) or with the rv0194 expression vector pML655 (open triangles) containing 1.56 mM ethidium bromide.
DISCUSSION
Mechanisms of ampicillin resistance in slow-growing mycobacteria.
In this study, we identified 21 transposon mutants that increased the resistance of M. bovis BCG to ampicillin by 32-fold. None of these mutations was in a gene known to be involved in resistance to β-lactam antibiotics. In gram-negative bacteria, resistance to β-lactam antibiotics is mainly mediated by four mechanisms: (i) enzymatic hydrolysis by β-lactamases, (ii) target alterations, i.e., mutations in the penicillin-binding proteins, (iii) reduced uptake across the outer membrane, and (iv) drug efflux. Augmented levels of β-lactamases can drastically increase the resistance to β-lactams, as has been shown for Salmonella enterica (12). However, mutants with insertion of the transposon upstream of any of the β-lactamases of M. bovis BCG or M. tuberculosis were not observed. Mutants with transposon insertions in penicillin-binding protein genes were also not observed, because the activity of these proteins is essential for cell growth (27). However, we expected to obtain mutants with reduced uptake across the outer membrane, as they are found frequently in gram-negative bacteria (23, 45). In these resistant mutants, expression or the activity of porins that provide an uptake pathway across the outer membrane for hydrophilic β-lactams was reduced (36). We showed that the lack of MspA and other Msp porins drastically decreases the permeability of β-lactams across the outer membrane of M. smegmatis and concomitantly increases the resistance of M. smegmatis to β-lactams (52, 54; Danilchanka et al., submitted). Furthermore, expression of MspA significantly increased the susceptibility of M. tuberculosis and M. bovis BCG to these antibiotics (32), indicating that uptake across the outer membrane is not as efficient in these organism as in M. smegmatis. However, endogenous porins of M. tuberculosis and M. bovis BCG that allow diffusion of β-lactams are yet to be identified. OmpA forms pores in vitro (3) and is located in the outer membrane of M. tuberculosis (50), possibly representing a channel accessible for β-lactam antibiotics. However, the OmpA mutant is not more resistant to β-lactams or other antibiotics than wt M. tuberculosis (H. Song et al., unpublished data), indicating that OmpA does not play a critical role in the transport of these drugs. In addition, none of the ampicillin-resistant mutants (Table 1) appears to have an insertion in a gene encoding an outer membrane protein, according to the criteria recently established for mycobacteria (50). One reason why putative porins are missing in the list of ampicillin-resistant mutants might be that a lack of porins can inhibit the growth of the mutant, as recently shown for M. smegmatis (53).
Unexpectedly, our screen revealed mutants with insertions in genes encoding polyketide synthases, such as Pks11 (Rv1665), or polyketide synthase-associated proteins, such as PapA2 (Rv3820c), which conferred high-level resistance to ampicillin (Table 1). Polyketide synthases are involved in the synthesis of complex mycobacterial lipids (21). In particular, synthesis or transport of sulfolipids appears to be important for susceptibility of M. bovis BCG to ampicillin (Table 1). Since ampicillin is a small and hydrophilic antibiotic, direct diffusion through the outer membrane would be very slow. This has been shown for other zwitterions, such as tryptophan and model lipid membranes (9). This indicates an indirect effect of the lipid composition on the outer membrane permeability of M. bovis BCG to ampicillin, possibly through channel proteins such as porins. Indeed, such a mechanism exists in E. coli, which does not synthesize porins in mutants with defective lipopolysaccharides, whereas other outer membrane proteins, such as OmpA, are produced at wt levels (46).
Other genes whose inactivation causes high-level resistance to ampicillin encode lipoproteins and PPE proteins, some of which have been shown to be surface proteins (14). It is possible that these proteins also play a role in the transport of β-lactams. Interestingly, the PPE24 and PPE53 proteins were also identified in a screen for Mycobacterium marinum mutants defective for infection of and growth in macrophages (37). Also, PPE53 was shown to be a virulence factor of M. marinum (49). These results suggest multiples roles of these PPE proteins in resistance to toxic compounds.
The functions of most genes identified in our screen have not been described previously. However, several genes were found previously in other screens. For example, gca and rv1949c (bcg1988c) were shown to be important for normal surface expression of lipoarabinomannan (LAM) (51). LAM is one of the major lipoglycans of M. tuberculosis and is thought to play a role in the cell wall structure and for interaction of M. tuberculosis with host immune cells (8). However, it is unclear how the lack of LAM promotes the resistance of M. bovis BCG to ampicillin.
The ABC transporter Rv0194 is a drug efflux pump of M. tuberculosis.
Our experiments clearly show that overexpression of the ABC transporter Rv0194 leads to increased resistance of both M. smegmatis and M. bovis BCG to multiple drugs and to ethidium bromide. The easiest and most likely explanation of these results is that Rv0194 constitutes a multidrug efflux pump which extrudes ampicillin, chloramphenicol, streptomycin, tetracyclin, vancomycin, erythromycin, and novobiocin (Tables 2 and 3). In fact, multidrug efflux pumps such as AcrAB of E. coli have similar broad transport capacities (41). AcrAB belongs to the resistance-nodulation-division family of efflux pumps, which play a dominant role in multidrug resistance in gram-negative bacteria, in contrast to ABC transporters (30). VcaM of Vibrio cholerae (25) and now Rv0194 appear to be among the very few ABC transporters involved in resistance to several structurally unrelated drugs. It should be noted that export of lipids by Rv0194 cannot be ruled out as an additional explanation for the increased drug resistance of the rv0194-expressing strain. A larger amount of a particular lipid in the outer membrane might reduce uptake of antibiotics and hence indirectly cause resistance in mycobacteria. Such a mechanism has been observed in gram-negative bacteria for the essential ABC transporter MsbA, which transports lipid A, lipopolysaccharides, and hydrophobic drugs across the inner membrane (42). A connection between lipid transport and drug resistance has been also observed for several mmpL mutants of M. tuberculosis (17). In vitro reconstitution experiments are required to identify the substrate specificities of a transporter (33). However, in all of these cases, the multidrug efflux pump transported both lipids and several unrelated drugs. This supports our conclusion that Rv0194 is a multidrug efflux pump.
The Rv0194 efflux pump has unusual properties.
First, no efflux pump is known to be involved in resistance to β-lactams in gram-positive bacteria (34) or in mycobacteria (15). This is a significant discovery, because the transpeptidases as targets of the β-lactam antibiotics are localized in the periplasm (27). A mechanism for how an inner membrane efflux transporter can confer resistance to an antibiotic that targets a periplasmic protein is known only for gram-negative bacteria so far. Here, the bacteria have a tripartite efflux system consisting of an inner membrane efflux pump that connects to an outer membrane channel protein, such as TolC. This complex is stabilized by periplasmic adapter proteins (28). These findings suggest that M. tuberculosis employs a similar drug efflux system.
Second, the resistance factors upon overexpression of a multidrug efflux pump of M. tuberculosis in M. smegmatis or other mycobacteria resulted in most cases in MICs that increased by 2- to 4-fold (2, 16, 55), while MICs were increased 4- to 64-fold upon expression of rv0194. At first sight, it appears surprising that a threefold-increased expression of rv0194 can lead to such high resistance factors in M. bovis BCG. However, this pump does not seem to be expressed in wt bacteria (Fig. 2). Thus, artificial expression of an efficient drug efflux pump might indeed account for the observed drastic increases in MICs for M. bovis BCG ML1034. By contrast, the resistance factors of M. smegmatis expressing rv0194 were low and similar to those observed for heterologous expression of other drug efflux pumps of M. tuberculosis and M. smegmatis. This might be due to the very different background of multidrug transporters and different levels of β-lactamases in both organisms. Indeed, it was only possible to measure the contribution of the efflux pumps LfrA and LfrX to β-lactam resistance of M. smegmatis after deletion of the major β-lactamase gene blaA (blaS) (29).
Conclusions.
We have identified Rv0194 as the first drug efflux pump of M. tuberculosis that is involved in resistance to β-lactam antibiotics. Furthermore, low-level expression of rv0194 drastically increased resistance of M. bovis BCG to multiple antibiotics. These findings suggest a novel molecular mechanism by which M. tuberculosis can easily acquire a multidrug-resistant phenotype.
Supplementary Material
Acknowledgments
We thank Mary Jackson for providing the plasmid pPR32 and for valuable advice regarding the construction and analysis of the transposon library and K. C. Walls and Jason Huff for critically reading of the manuscript.
This work was funded by grant AI63432 from the National Institutes of Health to M.N. and travel grants from the American Society of Microbiology and the University of Alabama at Birmingham to O.D.
Footnotes
Published ahead of print on 5 May 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, A. A. Neyfakh, and S. Schuldiner. 1993. Mutants of the Bacillus subtilis multidrug transporter Bmr with altered sensitivity to the antihypertensive alkaloid reserpine. J. Biol. Chem. 268:11086-11089. [PubMed] [Google Scholar]
- 2.Ainsa, J. A., M. C. Blokpoel, I. Otal, D. B. Young, K. A. De Smet, and C. Martin. 1998. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J. Bacteriol. 180:5836-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alahari, A., N. Saint, S. Campagna, V. Molle, G. Molle, and L. Kremer. 2007. The N-terminal domain of OmpATb is required for membrane translocation and pore-forming activity in mycobacteria. J. Bacteriol. 189:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashyam, M. D., and A. K. Tyagi. 1998. Identification and analysis of “extended −10” promoters from mycobacteria. J. Bacteriol. 180:2568-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatt, A., N. Fujiwara, K. Bhatt, S. S. Gurcha, L. Kremer, B. Chen, J. Chan, S. A. Porcelli, K. Kobayashi, G. S. Besra, and W. R. Jacobs, Jr. 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. USA 104:5157-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braibant, M., P. Gilot, and J. Content. 2000. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24:449-467. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 8.Briken, V., S. A. Porcelli, G. S. Besra, and L. Kremer. 2004. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol. Microbiol. 53:391-403. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti, A. C., and D. W. Deamer. 1992. Permeability of lipid bilayers to amino acids and phosphate. Biochim. Biophys. Acta 1111:171-177. [DOI] [PubMed] [Google Scholar]
- 10.Chambers, H. F., D. Moreau, D. Yajko, C. Miick, C. Wagner, C. Hackbarth, S. Kocagoz, E. Rosenberg, W. K. Hadley, and H. Nikaido. 1995. Can penicillins and other beta-lactam antibiotics be used to treat tuberculosis? Antimicrob. Agents Chemother. 39:2620-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers, H. F., J. Turner, G. F. Schecter, M. Kawamura, and P. C. Hopewell. 2005. Imipenem for treatment of tuberculosis in mice and humans. Antimicrob. Agents Chemother. 49:2816-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chouchani, C., R. Berlemont, A. Masmoudi, M. Galleni, J. M. Frere, O. Belhadj, and K. Ben-Mahrez. 2006. A novel extended-spectrum TEM-type beta-lactamase, TEM-138, from Salmonella enterica serovar Infantis. Antimicrob. Agents Chemother. 50:3183-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Converse, S. E., J. D. Mougous, M. D. Leavell, J. A. Leary, C. R. Bertozzi, and J. S. Cox. 2003. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. USA 100:6121-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delogu, G., C. Pusceddu, A. Bua, G. Fadda, M. J. Brennan, and S. Zanetti. 2004. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 52:725-733. [DOI] [PubMed] [Google Scholar]
- 15.De Rossi, E., J. A. Ainsa, and G. Riccardi. 2006. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30:36-52. [DOI] [PubMed] [Google Scholar]
- 16.De Rossi, E., M. C. Blokpoel, R. Cantoni, M. Branzoni, G. Riccardi, D. B. Young, K. A. De Smet, and O. Ciferri. 1998. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob. Agents Chemother. 42:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domenech, P., M. B. Reed, and C. E. Barry III. 2005. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73:3492-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrow, M. F., and E. J. Rubin. 2008. Function of a mycobacterial major facilitator superfamily pump requires a membrane-associated lipoprotein. J. Bacteriol. 190:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores, A. R., L. M. Parsons, and M. S. Pavelka, Jr. 2005. Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology 151:521-532. [DOI] [PubMed] [Google Scholar]
- 20.Franzblau, S. G., R. S. Witzig, J. C. McLaughlin, P. Torres, G. Madico, A. Hernandez, M. T. Degnan, M. B. Cook, V. K. Quenzer, R. M. Ferguson, and R. H. Gilman. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 36:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gokhale, R. S., P. Saxena, T. Chopra, and D. Mohanty. 2007. Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat. Prod. Rep. 24:267-277. [DOI] [PubMed] [Google Scholar]
- 22.Harries, A. D., and C. Dye. 2006. Tuberculosis. Ann. Trop. Med. Parasitol. 100:415-431. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez-Alles, S., M. Conejo, A. Pascual, J. M. Tomas, V. J. Benedi, and L. Martinez-Martinez. 2000. Relationship between outer membrane alterations and susceptibility to antimicrobial agents in isogenic strains of Klebsiella pneumoniae. J. Antimicrob. Chemother. 46:273-277. [DOI] [PubMed] [Google Scholar]
- 24.Hillmann, D., I. Eschenbacher, A. Thiel, and M. Niederweis. 2007. Expression of the major porin gene mspA is regulated in Mycobacterium smegmatis. J. Bacteriol. 189:958-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huda, N., E. W. Lee, J. Chen, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Molecular cloning and characterization of an ABC multidrug efflux pump, VcaM, in Non-O1 Vibrio cholerae. Antimicrob. Agents Chemother. 47:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaps, I., S. Ehrt, S. Seeber, D. Schnappinger, C. Martin, L. W. Riley, and M. Niederweis. 2001. Energy transfer between fluorescent proteins using a co-expression system in Mycobacterium smegmatis. Gene 278:115-124. [DOI] [PubMed] [Google Scholar]
- 27.Koch, A. L. 2000. Penicillin binding proteins, beta-lactams, and lactamases: offensives, attacks, and defensive countermeasures. Crit. Rev. Microbiol. 26:205-220. [DOI] [PubMed] [Google Scholar]
- 28.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 29.Li, X. Z., L. Zhang, and H. Nikaido. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubelski, J., W. N. Konings, and A. J. Driessen. 2007. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol Rev. 71:463-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynett, J., and R. W. Stokes. 2007. Selection of transposon mutants of Mycobacterium tuberculosis with increased macrophage infectivity identifies fadD23 to be involved in sulfolipid production and association with macrophages. Microbiology 153:3133-3140. [DOI] [PubMed] [Google Scholar]
- 32.Mailaender, C., N. Reiling, H. Engelhardt, S. Bossmann, S. Ehlers, and M. Niederweis. 2004. The MspA porin promotes growth and increases antibiotic susceptibility of both Mycobacterium bovis BCG and Mycobacterium tuberculosis. Microbiology 150:853-864. [DOI] [PubMed] [Google Scholar]
- 33.Margolles, A., M. Putman, H. W. van Veen, and W. N. Konings. 1999. The purified and functionally reconstituted multidrug transporter LmrA of Lactococcus lactis mediates the transbilayer movement of specific fluorescent phospholipids. Biochemistry 38:16298-16306. [DOI] [PubMed] [Google Scholar]
- 34.Markham, P. N., and A. A. Neyfakh. 2001. Efflux-mediated drug resistance in gram-positive bacteria. Curr. Opin. Microbiol. 4:509-514. [DOI] [PubMed] [Google Scholar]
- 35.Martinez, M. B., M. Flickinger, L. Higgins, T. Krick, and G. L. Nelsestuen. 2001. Reduced outer membrane permeability of Escherichia coli O157:H7: suggested role of modified outer membrane porins and theoretical function in resistance to antimicrobial agents. Biochemistry 40:11965-11974. [DOI] [PubMed] [Google Scholar]
- 36.McAdam, R. A., T. R. Weisbrod, J. Martin, J. D. Scuderi, A. M. Brown, J. D. Cirillo, B. R. Bloom, and W. R. Jacobs, Jr. 1995. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect. Immun. 63:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta, P. K., A. K. Pandey, S. Subbian, S. H. El-Etr, S. L. Cirillo, M. M. Samrakandi, and J. D. Cirillo. 2006. Identification of Mycobacterium marinum macrophage infection mutants. Microb. Pathog. 40:139-151. [DOI] [PubMed] [Google Scholar]
- 38.Migliori, G. B., R. Loddenkemper, F. Blasi, and M. C. Raviglione. 2007. 125 years after Robert Koch's discovery of the tubercle bacillus: the new XDR-TB threat. Is “science” enough to tackle the epidemic? Eur. Respir. J. 29:423-427. [DOI] [PubMed] [Google Scholar]
- 39.Nikaido, H. 2001. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin. Cell. Dev. Biol. 12:215-223. [DOI] [PubMed] [Google Scholar]
- 40.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat Rev. Microbiol. 4:629-636. [DOI] [PubMed] [Google Scholar]
- 42.Pohl, A., P. F. Devaux, and A. Herrmann. 2005. Function of prokaryotic and eukaryotic ABC proteins in lipid transport. Biochim. Biophys. Acta 1733:29-52. [DOI] [PubMed] [Google Scholar]
- 43.Poole, K. 2004. Resistance to beta-lactam antibiotics. Cell Mol. Life Sci. 61:2200-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prod'hom, G., B. Lagier, V. Pelicic, A. J. Hance, B. Gicquel, and C. Guilhot. 1998. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol. Lett. 158:75-81. [DOI] [PubMed] [Google Scholar]
- 45.Raimondi, A., A. Traverso, and H. Nikaido. 1991. Imipenem- and meropenem-resistant mutants of Enterobacter cloacae and Proteus rettgeri lack porins. Antimicrob. Agents Chemother. 35:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ried, G., I. Hindennach, and U. Henning. 1990. Role of lipopolysaccharide in assembly of Escherichia coli outer membrane proteins OmpA, OmpC, and OmpF. J. Bacteriol. 172:6048-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosas-Magallanes, V., G. Stadthagen-Gomez, J. Rauzier, L. B. Barreiro, L. Tailleux, F. Boudou, R. Griffin, J. Nigou, M. Jackson, B. Gicquel, and O. Neyrolles. 2007. Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect. Immun. 75:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruley, K. M., J. H. Ansede, C. L. Pritchett, A. M. Talaat, R. Reimschuessel, and M. Trucksis. 2004. Identification of Mycobacterium marinum virulence genes using signature-tagged mutagenesis and the goldfish model of mycobacterial pathogenesis. FEMS Microbiol. Lett. 232:75-81. [DOI] [PubMed] [Google Scholar]
- 50.Song, H., Y. Wang, R. Sandie, M. Andrade-Navarro, and M. Niederweis. 24 April 2008, posting date. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 51.Stadthagen, G., M. Jackson, P. Charles, F. Boudou, N. Barilone, M. Huerre, P. Constant, A. Liav, I. Bottova, J. Nigou, T. Brando, G. Puzo, M. Daffe, P. Benjamin, S. Coade, R. S. Buxton, R. E. Tascon, A. Rae, B. D. Robertson, D. B. Lowrie, D. B. Young, B. Gicquel, and R. Griffin. 2006. Comparative investigation of the pathogenicity of three Mycobacterium tuberculosis mutants defective in the synthesis of p-hydroxybenzoic acid derivatives. Microbes Infect. 8:2245-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stahl, C., S. Kubetzko, I. Kaps, S. Seeber, H. Engelhardt, and M. Niederweis. 2001. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 40:451-464 (Authors' correction, 57:1509, 2005). [DOI] [PubMed] [Google Scholar]
- 53.Stephan, J., J. Bender, F. Wolschendorf, C. Hoffmann, E. Roth, C. Mailänder, H. Engelhardt, and M. Niederweis. 2005. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58:714-730. [DOI] [PubMed] [Google Scholar]
- 54.Stephan, J., C. Mailaender, G. Etienne, M. Daffe, and M. Niederweis. 2004. Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:4163-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takiff, H. E., M. Cimino, M. C. Musso, T. Weisbrod, R. Martinez, M. B. Delgado, L. Salazar, B. R. Bloom, and W. R. Jacobs, Jr. 1996. Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 93:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White, V. L., and J. Moore-Gillon. 2000. Resource implications of patients with multidrug resistant tuberculosis. Thorax 55:962-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright, G. D. 2003. Mechanisms of resistance to antibiotics. Curr. Opin. Chem. Biol. 7:563-569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.